Molecular Detection of Two Potential Probiotic Lactobacilli Strains and Evaluation of Their Performance as Starter Adjuncts in Yogurt Production
Abstract
:1. Introduction
2. Results and Discussion
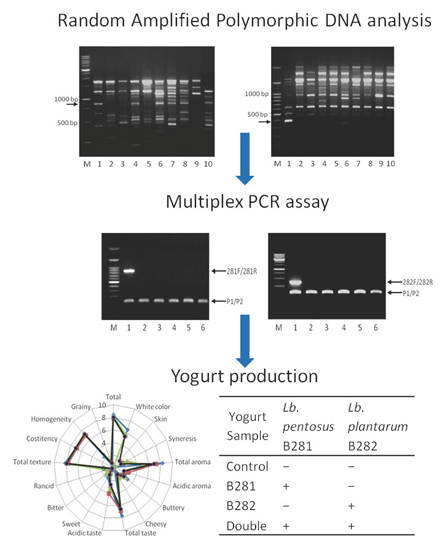
2.1. Screening of Random Amplified Polymorphic DNA (RAPD) Primers, Isolation of Sequence-Characterized Amplified Regions (SCAR) Markers, and Design of Novel Primers for Multiplex Polymerase Chain Reaction (PCR)
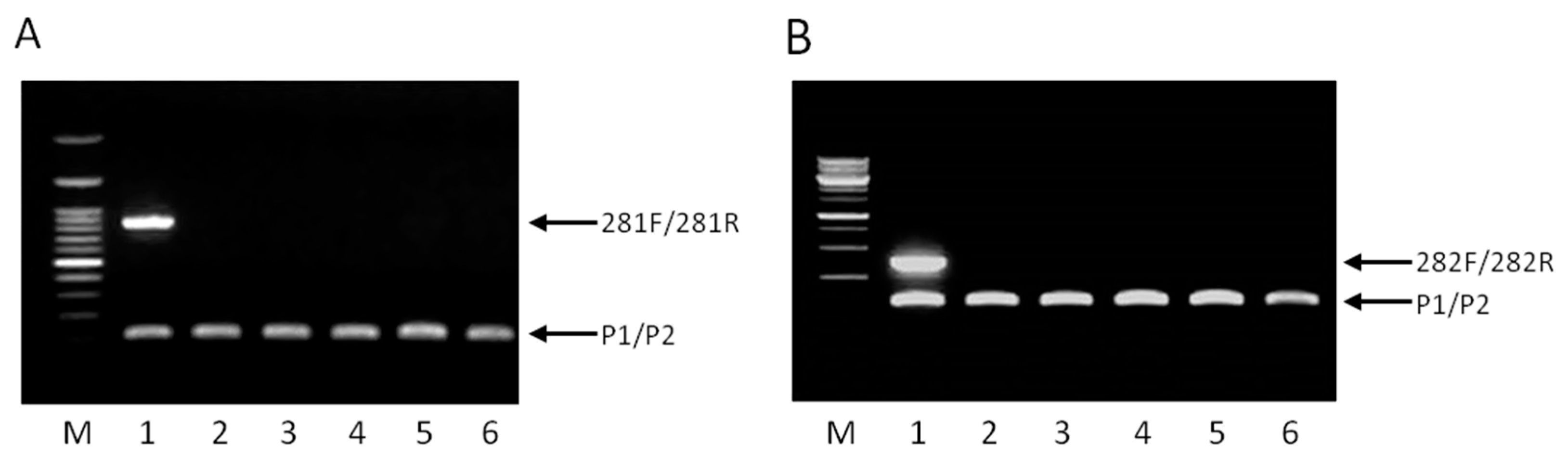
2.2. Specificity of the Multiplex PCR
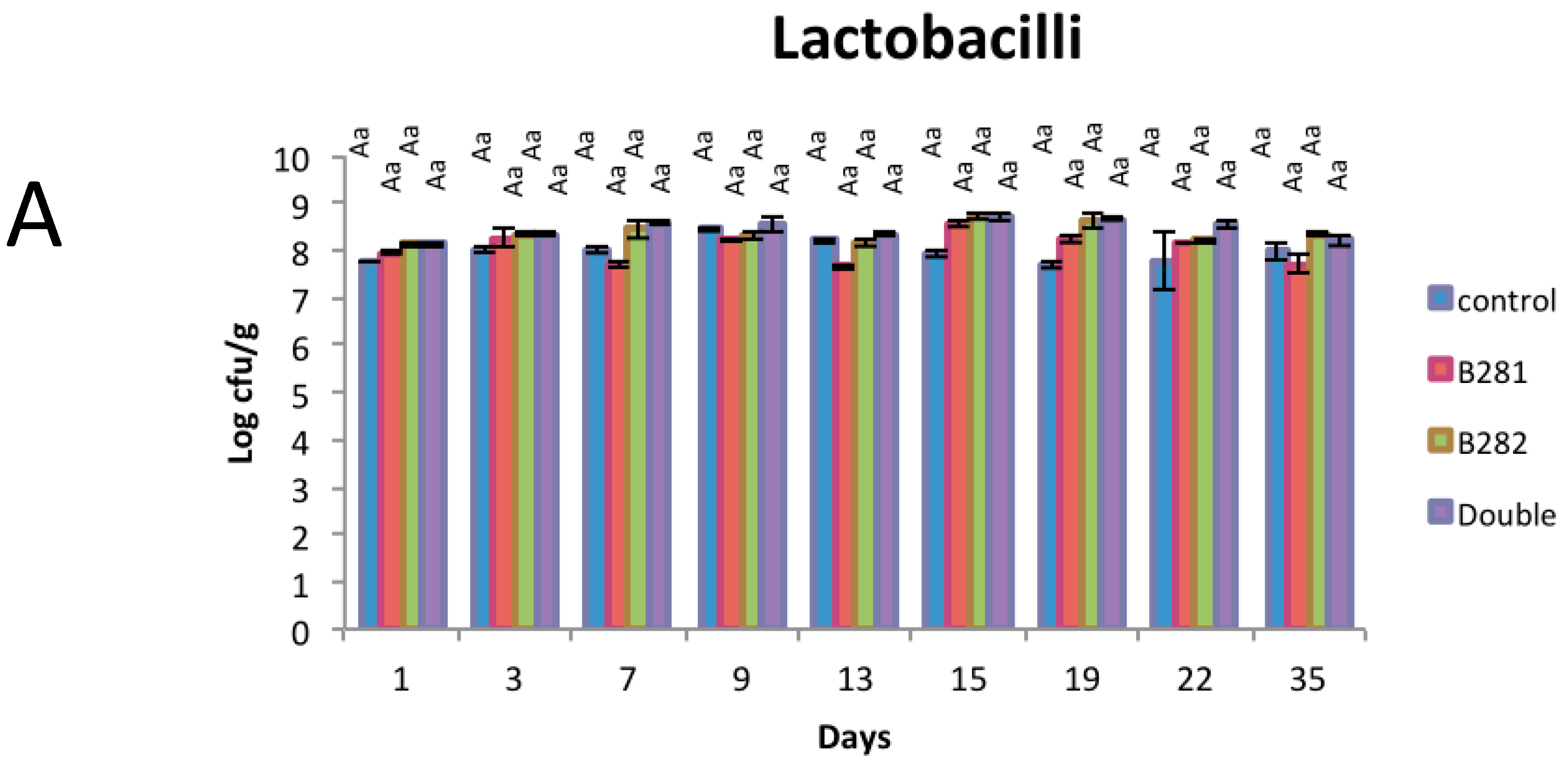
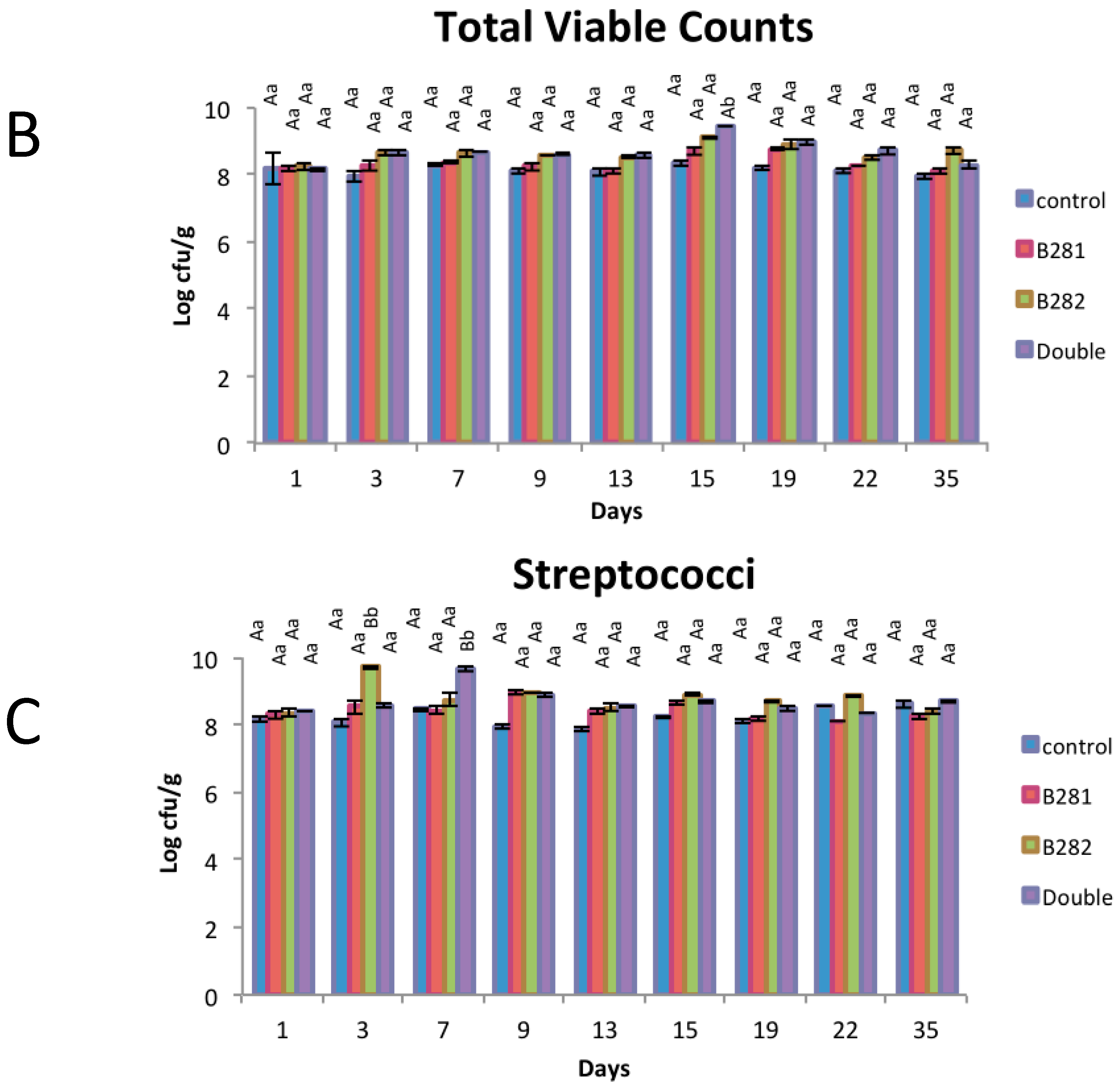
2.3. Μicrobiological Analysis
2.4. Monitoring the Presence of Lb. pentosus B281 and Lb. plantarum B282 in Yogurt
2.5. pH Determination
2.6. Sensory Evaluation
3. Experimental Section
3.1. Bacterial Strains and Culture Conditions
3.2. RAPD PCR
3.3. Cloning and Sequencing
3.4. Multiplex PCR
3.5. Yogurt Production
3.6. Microbiological Analysis
3.7. Molecular Detection of Lb. pentosus B281 and Lb. plantarum B282 in Yogurt
3.8. pH Determination
3.9. Sensory Evaluation
3.10. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Varankovich, N.V.; Nickerson, M.T.; Korber, D.R. Probiotic-based strategies for therapeutic and prophylactic use against multiple gastrointestinal diseases. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, W.; Wang, C.; Yu, Q.; Dai, R.; Pei, X. Lactococcus lactis expressing food-grade β-galactosidase alleviates lactose intolerance symptoms in post-weaning Balb/c mice. Appl. Microbiol. Biotechnol. 2012, 96, 1499–1506. [Google Scholar] [CrossRef] [PubMed]
- Cuello-Garcia, C.A.; Brożek, J.L.; Fiocchi, A.; Pawankar, R.; Yepes-Nuñez, J.J.; Terracciano, L.; Gandhi, S.; Agarwal, A.; Zhang, Y.; Schünemann, H.J. Probiotics for the prevention of allergy: A systematic review and meta-analysis of randomized controlled trials. J. Allergy Clin. Immunol. 2015, 136, 952–961. [Google Scholar] [CrossRef] [PubMed]
- Park, D.Y.; Ahn, Y.T.; Park, S.H.; Huh, C.S.; Yoo, S.R.; Yu, R.; Sung, M.K.; McGregor, R.; Choi, M.S. Supplementation of Lactobacillus curvatus HY7601 and Lactobacillus plantarum KY1032 in diet-induced obese mice is associated with gut microbial changes and reduction in obesity. PLoS ONE 2013, 8, e59470. [Google Scholar]
- Poutahidis, T.; Kleinewietfeld, M.; Smillie, C.; Levkovich, Τ.; Perrotta, Α.; Bhela, S.; Varian, B.J.; Ibrahim, Y.M.; Lakritz, J.R.; Kearney, S.M.; et al. Microbial reprogramming inhibits western diet-associated obesity. PLoS ONE 2013, 8, e685961. [Google Scholar] [CrossRef] [PubMed]
- Britton, R.A.; Irwin, R.; Quach, D.; Schaefer, L.; Zhang, J.; Lee, T.; Parameswaran, N.; McCabe, L.R. Probiotic L. reuteri treatment prevents bone loss in a menopausal ovariectomized mouse model. J. Cell. Physiol. 2014, 229, 1822–1830. [Google Scholar] [CrossRef] [PubMed]
- Uccello, M.; Malaguarnera, G.; Basile, F.; D’agata, V.; Malaguarnera, M.; Bertino, G.; Vacante, M.; Drago, F.; Biondi, A. Potential role of probiotics on colorectal cancer prevention. BMC Surg. 2012, 12, S35. [Google Scholar] [CrossRef] [PubMed]
- Kaga, C.; Takagi, A.; Kano, M.; Kado, S.; Kato, I.; Sakai, M.; Miyazaki, K.; Nanno, M.; Ishikawa, F.; Ohashi, Y.; et al. Lactobacillus casei Shirota enhances the preventive efficacy of soymilk in chemically induced breast cancer. Cancer Sci. 2013, 104, 1508–1514. [Google Scholar] [CrossRef] [PubMed]
- Naito, S.; Koga, H.; Yamaguchi, A.; Fujimoto, N.; Hasui, Y.; Kuramoto, H.; Iguchi, A.; Kinukawa, N. Prevention of recurrence with epirubicin and lactobacillus casei after transurethral resection of bladder cancer. J. Urol. 2008, 179, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Granato, D.; Branco, G.; Gomes Cruz, A.; de Assis Fonseca Faria, J.; Shah, N. Probiotic dairy products as functional foods. Compr. Rev. Food Sci. Food Saf. 2010, 9, 455–470. [Google Scholar] [CrossRef]
- Rivera-Espinoza, Y.; Gallardo-Navarro, Y. Non-dairy probiotic products. Food Microbiol. 2010, 27, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Céspedes, M.; Cárdenas, P.; Staffolani, M.; Ciappini, M.C.; Vinderola, G. Performance in nondairy drinks of probiotic L. casei strains usually employed in dairy products. J. Food Sci. 2013, 78, M756–M762. [Google Scholar] [CrossRef] [PubMed]
- Blaiotta, G.; di Capua, M.; Coppola, R.; Aponte, M. Production of fermented chestnut purees by lactic acid bacteria. Int. J. Food Microbiol. 2012, 158, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Sidira, M.; Karapetsas, A.; Galanis, A.; Kanellaki, M.; Kourkoutas, Y. Effective survival of immobilized Lactobacillus casei during ripening and heat treatment of probiotic dry-fermented sausages and investigation of the microbial dynamics. Meat Sci. 2014, 96, 948–955. [Google Scholar] [CrossRef] [PubMed]
- Argyri, A.; Zoumpopoulou, G.; Karatzas, K.A.; Tsakalidou, E.; Nychas, G.J.; Panagou, E.; Tassou, C. Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food Microbiol. 2013, 33, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Argyri, A.; Nisiotou, A.; Malouchos, A.; Panagou, E.; Tassou, C. Performance of two potential probiotic Lactobacillus strains from the olive microbiota as starters in the fermentation of heat shocked green olives. Int. J. Food Microbiol. 2014, 171, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Blana, V.; Grounta, A.; Tassou, C.; Nychas, G.J.; Panagou, E. Inoculated fermentation of green olives with potential probiotic Lactobacillus pentosus and Lactobacillus plantarum starter cultures isolated from industrially fermented olives. Food Microbiol. 2014, 38, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Mohania, D.; Nagpal, R.; Kumar, M.; Bhardwaj, A.; Yadav, M.; Jain, S.; Marotta, F.; Singh, V.; Parkash, O.; Yadav, H. Molecular approaches for identification and characterization of lactic acid bacteria. J. Dig. Dis. 2008, 9, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Karapetsas, A.; Vavoulidis, E.; Galanis, A.; Sandaltzopoulos, R.; Kourkoutas, Y. Rapid Detection and Identification of Probiotic Lactobacillus casei ATCC 393 by Multiplex PCR. J. Mol. Microbiol. Biotechnol. 2010, 18, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Nikolaou, A.; Saxami, G.; Kourkoutas, Y.; Galanis, A. A new methodology for rapid detection of Lactobacillus delbrueckii subsp. bulgaricus based on multiplex PCR. J. Microbiol. Meth. 2011, 84, 362–364. [Google Scholar] [CrossRef] [PubMed]
- Cremonesi, P.; Vanoni, L.; Morandi, S.; Silvetti, T.; Castiglioni, B.; Brasca, M. Development of a pentaplex PCR assay for the simultaneous detection of Streptococcus thermophilus, Lactobacillus delbrueckii subsp. bulgaricus, L. delbrueckii subsp. lactis, L. helveticus, L. fermentum in whey starter for Grana Padano cheese. Int. J. Food Microbiol. 2011, 146, 207–211. [Google Scholar]
- Tilsala-Timisjärvi, A.; Tapani Alatossava, T. Strain-specific identification of probiotic Lactobacillus rhamnosus with randomly amplified polymorphic DNA-derived PCR primers. Appl. Environ. Microbiol. 1998, 64, 4816–4819. [Google Scholar] [PubMed]
- Maruo, T.; Sakamoto, M.; Toda, T.; Benno, Y. Monitoring the cell number of Lactococcus lactis subsp. cremoris FC in human feces by real-time PCR with strain-specific primers designed using the RAPD technique. Int. J. Food Microbiol. 2006, 110, 69–76. [Google Scholar] [PubMed]
- Galanis, A.; Kourkoutas, Y.; Tassou, C.C.; Chorianopoulos, N. Detection and identification of probiotic Lactobacillus plantarum strains by multiplex PCR using RAPD-derived primers. Int. J. Mol. Sci. 2015, 16, 25141–25153. [Google Scholar] [CrossRef] [PubMed]
- Klijn, N.; Weerkamp, A.H.; de Vos, W.M. Identification of mesophilic lactic acid bacteria by using polymerase chain reaction-amplified variable regions of 16S rRNA and specific DNA probes. Appl. Environ. Microbiol. 1991, 57, 3390–3393. [Google Scholar] [PubMed]
- Henegariu, O.; Heerema, N.A.; Dlouhy, S.R.; Vance, G.H.; Vogt, P.H. Multiplex PCR: Critical parameters and step-by-step protocol. Biotechniques 1997, 23, 504–511. [Google Scholar] [PubMed]
- Doulgeraki, A.; Pramateftaki, P.; Argyri, A.; Nychas, G.J.; Tassou, C.; Panagou, E. Molecular characterization of lactic acid bacteria isolated from industrially fermented Greek table olives. LWT Food Sci. Technol. 2013, 50, 353–356. [Google Scholar] [CrossRef]
- Sidira, M.; Kiourtzidis, M.; Argyri, A.; Papadopoulou, O.; Chorianopoulos, N.; Tassou, C.; Kaloutsas, S.; Galanis, A.; Kourkoutas, Y. Evaluation of immobilized Lactobacillus plantarum 2035 on whey protein as adjunct probiotic culture in yoghurt production. LWT Food Sci. Technol. 2016. submitted. [Google Scholar]
- Boylston, T.D.; Vinderola, C.G.; Ghoddusi, H.B.; Reinheimer, J.A. Incorporation of bifidobacteria into cheeses: Challenges and rewards. Int. Dairy J. 2004, 19, 315–387. [Google Scholar] [CrossRef]
- Oliveira, M.; Sodini, I.; Remeuf, R.; Tissier, J.P.; Corrieu, G. Manufacture of fermented lactic beverages containing probiotic cultures. J. Food Sci. 2002, 67, 2336–2341. [Google Scholar] [CrossRef]
- Donkor, O.N.; Henriksson, A.; Vasiljevic, T.; Shah, N.P. Rheological properties and sensory characteristics of set-type soy yogurt. J. Agric. Food Chem. 2007, 55, 9868–9876. [Google Scholar] [CrossRef] [PubMed]
- Sidira, M.; Saxami, G.; Dimitrellou, D.; Santarmaki, V.; Galanis, A.; Kourkoutas, Y. Monitoring survival of Lactobacillus casei ATCC 393 in probiotic yogurts using an efficient molecular tool. J. Dairy Sci. 2013, 96, 3369–3377. [Google Scholar] [CrossRef] [PubMed]
- Maragkoudakis, P.A.; Zoumpopoulou, G.; Miaris, C.; Kalantzopoulos, G.; Pot, B.; Tsakalidou, E. Probiotic potential of Lactobacillus strains isolated from dairy products. Int. Dairy J. 2006, 16, 189–199. [Google Scholar] [CrossRef]
- Vinderola, C.G.; Prosello, W.; Ghiberto, T.D.; Reinheimer, J.A. Viability of probiotic (Bifidobacterium, Lactobacillus acidophilus and Lactobacillus casei) and nonprobiotic microflora in Argentinian Fresco cheese. J. Dairy Sci. 2000, 83, 1905–1911. [Google Scholar] [CrossRef]
- Picot, A.; Lacroix, C. Encapsulation of bifidobacteria in whey protein-based microcapsules and survival in simulated gastrointestinal conditions and in yoghurt. Int. Dairy J. 2004, 14, 505–515. [Google Scholar] [CrossRef]
- RAPD-Primer Generator by J. Wöstemeyer, Institute of General Microbiology and Microbial Genetics, Germany. Available online: http://www2.uni-jena.de/biologie/mikrobio/tipps/rapd.html (accessed on 29 April 2016).
- Zhou, M.Y.; Celso, E.; Gomez-Sanchez, C. Universal TA cloning. Curr. Issues Mol. Biol. 2000, 2, 1–7. [Google Scholar] [PubMed]

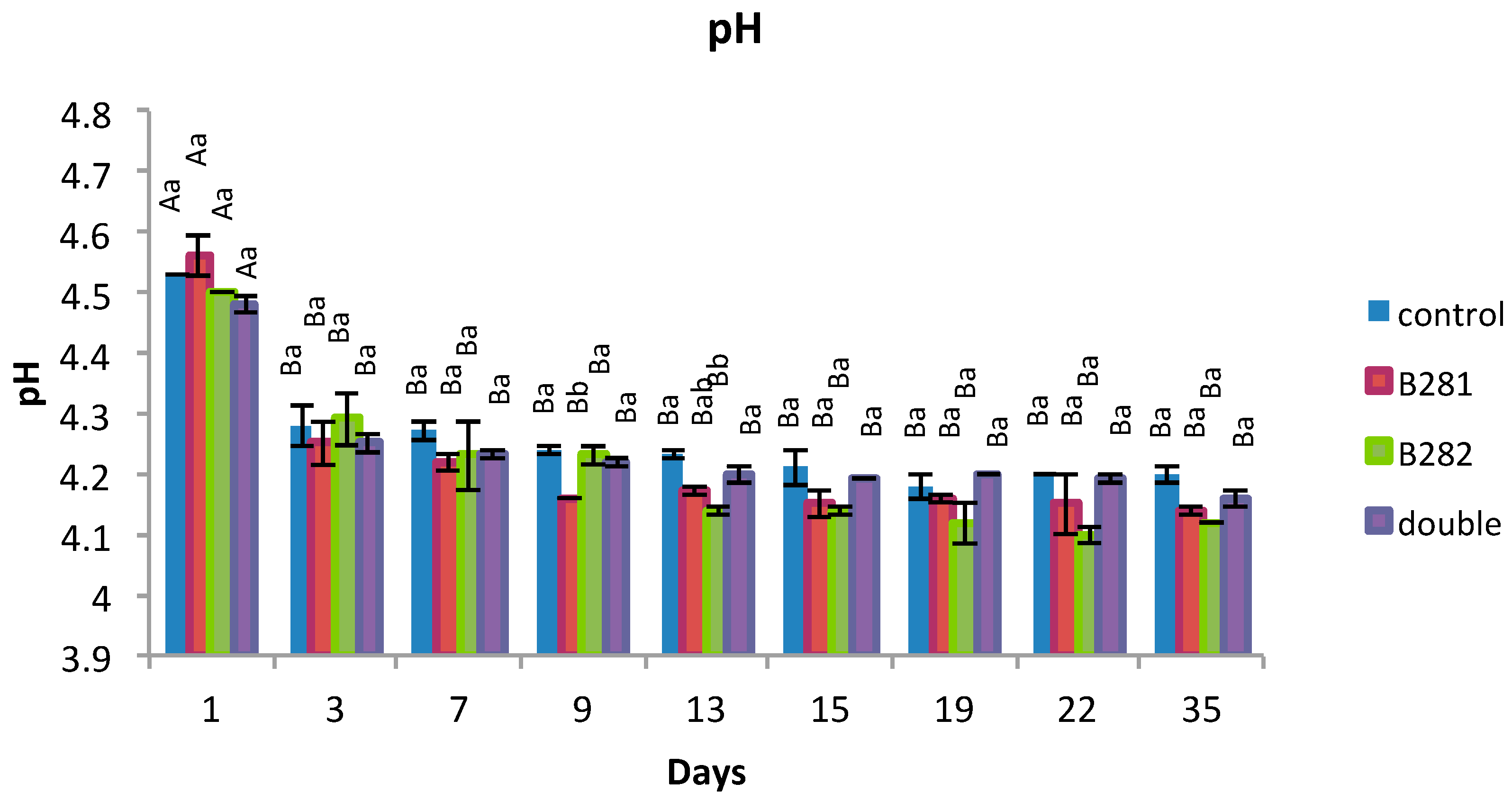
 ): Yogurt produced with no probiotic culture; (
): Yogurt produced with no probiotic culture; (  ): Yogurt containing Lb. pentosus B281; (
): Yogurt containing Lb. pentosus B281; (  ): Yogurt containing Lb. plantarum B282; (
): Yogurt containing Lb. plantarum B282; (  ): Yogurt containing Lb. pentosus B281 and Lb. plantarum B282.
): Yogurt containing Lb. pentosus B281 and Lb. plantarum B282.
 ): Yogurt produced with no probiotic culture; (
): Yogurt produced with no probiotic culture; (  ): Yogurt containing Lb. pentosus B281; (
): Yogurt containing Lb. pentosus B281; (  ): Yogurt containing Lb. plantarum B282; (
): Yogurt containing Lb. plantarum B282; (  ): Yogurt containing Lb. pentosus B281 and Lb. plantarum B282.
): Yogurt containing Lb. pentosus B281 and Lb. plantarum B282.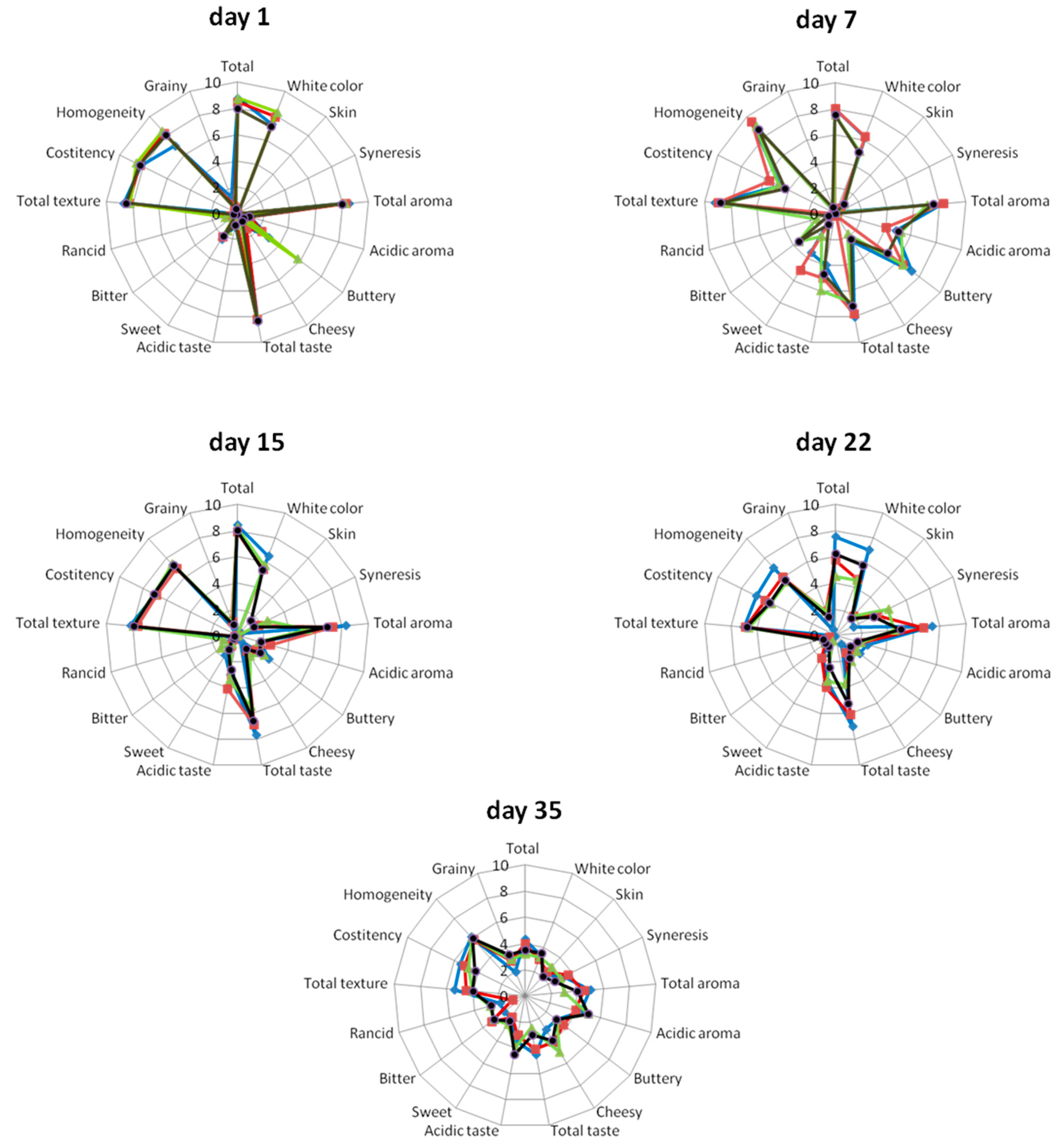
| Yogurt Sample | Day 1 | Day 15 | Day 35 | |||
|---|---|---|---|---|---|---|
| Lb. pentosus B281 | Lb. plantarum B282 | Lb. pentosus B281 | Lb. plantarum B282 | Lb. pentosus B281 | Lb. plantarum B282 | |
| Control | − | − | − | − | − | − |
| B281 | + | − | + | − | + | − |
| B282 | − | + | − | + | − | + |
| Double | + | + | + | + | + | + |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saxami, G.; Papadopoulou, O.S.; Chorianopoulos, N.; Kourkoutas, Y.; Tassou, C.C.; Galanis, A. Molecular Detection of Two Potential Probiotic Lactobacilli Strains and Evaluation of Their Performance as Starter Adjuncts in Yogurt Production. Int. J. Mol. Sci. 2016, 17, 668. https://doi.org/10.3390/ijms17050668
Saxami G, Papadopoulou OS, Chorianopoulos N, Kourkoutas Y, Tassou CC, Galanis A. Molecular Detection of Two Potential Probiotic Lactobacilli Strains and Evaluation of Their Performance as Starter Adjuncts in Yogurt Production. International Journal of Molecular Sciences. 2016; 17(5):668. https://doi.org/10.3390/ijms17050668
Chicago/Turabian StyleSaxami, Georgia, Olga S. Papadopoulou, Nikos Chorianopoulos, Yiannis Kourkoutas, Chrysoula C. Tassou, and Alex Galanis. 2016. "Molecular Detection of Two Potential Probiotic Lactobacilli Strains and Evaluation of Their Performance as Starter Adjuncts in Yogurt Production" International Journal of Molecular Sciences 17, no. 5: 668. https://doi.org/10.3390/ijms17050668
APA StyleSaxami, G., Papadopoulou, O. S., Chorianopoulos, N., Kourkoutas, Y., Tassou, C. C., & Galanis, A. (2016). Molecular Detection of Two Potential Probiotic Lactobacilli Strains and Evaluation of Their Performance as Starter Adjuncts in Yogurt Production. International Journal of Molecular Sciences, 17(5), 668. https://doi.org/10.3390/ijms17050668