Abstract
Shiraia bambusicola is a species of the monotypic genus Shiraia in the phylum Ascomycota. In China, it is known for its pharmacological properties that are used to treat rheumatic arthritis, sciatica, pertussis, tracheitis and so forth. Its major medicinal active metabolite is hypocrellin A, which exhibits excellent antiviral and antitumor properties. However, the genes involved in the hypocrellin A anabolic pathways were still unknown due to the lack of genomic information for this species. To investigate putative genes that are involved in the biosynthesis of hypocrellin A and determine the pathway, we performed transcriptome sequencing for Shiraia bambusicola S4201-W and the mutant S4201-D1 for the first time. S4201-W has excellent hypocrellin A production, while the mutant S4201-D1 does not. Then, we obtained 38,056,034 and 39,086,896 clean reads from S4201-W and S4201-D1, respectively. In all, 17,923 unigenes were de novo assembled, and the N50 length was 1970 bp. Based on the negative binomial distribution test, 716 unigenes were found to be upregulated, and 188 genes were downregulated in S4201-D1, compared with S4201-W. We have found seven unigenes involved in the biosynthesis of hypocrellin A and proposed a putative hypocrellin A biosynthetic pathway. These data will provide a valuable resource and theoretical basis for future molecular studies of hypocrellin A, help identify the genes involved in the biosynthesis of hypocrellin A and help facilitate functional studies for enhancing hypocrellin A production.
1. Introduction
Shiraia bambusicola is a species of the monotypic genus Shiraia in the phylum Ascomycota. It is known to grow in only some provinces south of the Yangtze River of China and in Japan [1,2]. In China, it has long been used as a traditional medicine for the treatment of rheumatic arthritis, sciatica, pertussis, tracheitis and so on. Its major active metabolic product is hypocrellin (hypocrellin A, hypocrellin B, hypocrellin C, hypocrellin D), which is a perylenequinoid pigment. Hypocrellin A (HA) is the main component in hypocrellin, and it has been found that HA has antiviral and antitumor activities [3,4,5,6]. As a photosensitizer, it can be activated and can generate molecular singlet oxygen (1O2) and superoxide radicals (O2−) in the presence of oxygen, which can damage cells [7,8]. HA showed great potential in clinical photodynamic therapy [9]. Although HA has exhibited a variety of pharmacological functions and has been used widely, its anabolic pathway remains unknown.
Perylenequinones, including cercosporin, elsinochrome, phleichrome, cladochrome and hypocrellin, are similar in structure and red in color [10]. The hypocrellin A has been well studied, due to its pharmaceutical potential as anticancer agents through photodynamic tumor therapy. However, cercosporin biosynthesis has been studied long-term and has been used as a model for resistance to 1O2 and other oxidative stress [11]. Therefore, we can use the working model of cercosporin biosynthetic pathway to study the biosynthesis of HA. In the 1970s, using a substrate-feeding study with acetate and malonate, Okubo et al. concluded that cercosporin biosynthesis occurs via a fungal polyketide pathway [12]. Eight co-regulated genes (CTB1–8) were identified in the cluster using a chromosome walking strategy. The CTB1 (Cercosporin Toxin Biosynthesis 1) gene, a polyketide synthase gene, encodes a protein of 2196 amino acids that synthesizes the polyketomethylene backbone of cercosporin [13]. The CTB3 gene includes two putative domains, a flavin adenine dinucleotide (FAD)-dependent monooxygenase domain in the C-terminus and an O-methyltransferase domain in the N-terminus [14]. The CTB4 gene is a major facilitator superfamily (MFS) transporter [15]. The CTB5 and CTB7 genes encode a FMN/FAD-dependent oxidoreductases, and CTB6 encodes a NADPH-dependent oxidoreductase [16]. CTB2 encodes an O-methyltransferase gene involved in cercosporin biosynthesis. The CTB8 protein has similarity to many fungal transcription factors, which contain zinc finger DNA-binding domains, and CTB8 controls the expression of all of the core cercosporin cluster genes [17]. Chen et al. proposed a possible pathway for cercosporin biosynthesis in Cercospora nicotianae and paved the way for future research of other perylenequinones. In addition, the first gene putatively identified in the pathway was cercosporin facilitator protein (CFP), which has high homology with MFS drug resistance transporter proteins and was found to be required for exportation of cercosporin from the mycelium of Cercospora kikuchii [18]. At present, there is little research in the iterature focused on the molecular genetics of Shiraia bambusicola. Chang et al. obtained one fungal polyketide synthase gene named g4 PKS from Shiraia sp. SUPER-H168, and amplified one keto-synthase (PKS) domain fragment of g4 PKS. The expression vector pCold II-KS was constructed and expressed in Escherichia coli BL21 (DE3) [19]. One type III PKS gene was obtained from Shiraia sp. Slf14, which is the key synthase in the product process of the HA [20]. Notwithstanding, the putative genes and the mechanism of the HA biosynthetic pathway remains unclear.
Next-generation sequencing (NGS) technology has recently been widely used in diverse researches to provide a large amount of genetic data of certain species. Compared with time- and energy-consuming traditional methods, RNA-Seq has already been shown to be highly accurate and, with a large dynamic range of expression levels, it requires less RNA to test gene expression of each transcript under different conditions. RNA-Seq technology provides a fast and accurate way to generate large amounts of sequenced transcriptome for these non-model species without reference genome sequence information [21].
To investigate putative genes involved in the biosynthesis of HA and propose a putative biosynthetic pathway, firstly we screened a hypocrellin-producing strain S4201-W, which was isolated from the fruiting bodies of the Shiraia bambusicola P. Henn. Then we irradiated S4201-W by means of ultraviolet ray, and obtained mutant S4201-D1 that did not produce HA. We performed transcriptome sequencing for them for further analysis. In this study, the Illumina HiSeqTM 2500 platform was used to sequence the transcriptome of Shiraia bambusicola to study its molecular genetics and HA biosynthesis. In the future, this study will provide an important resource for gene discovery and increase HA production in Shiraia bambusicola through further genetic manipulation.
2. Results and Discussion
2.1. Transcriptome Sequencing and Assembly
The S. bambusicola strains were cultured on potato dextrose agar (PDA, 200 g/L potato extract, 20 g/L glucose, 20 g/L agar, pH 7.0) and potato dextrose broth (PDB, 200 g/L potato extract, 20 g/L glucose, pH 7.0) (shown in Supplementary Figure S1). The concentration of HA was measured by high-performance liquid chromatography (HPLC) with HA standard reagent (Figure S2). As a result, no HA could be detected by HPLC in mutant S4201-D1. According to the standard curve, we know that the yield of HA was about 156.67 mg/L broth in wild type S4201-W. This was prepared for later experiment and analysis. To obtain an overview of the de novo transcriptome of S. bambusicola, cDNA libraries were constructed and sequenced using the Illumina HiSeqTM 2500 platform, generating 38,230,680 and 39,301,794 raw reads. After removing the adapter sequence and low-quality reads, we obtained 38,056,034 and 39,086,896 clean reads from the S4201-W and S4201-D1 transcriptomes, respectively. The Q30 percentages (sequencing error rate < 0.1%) of S4201-W and S4201-D1 were 90.20% and 90.43%, respectively. The GC content of the unigenes of S4201-W and S4201-D1 was 51.00% and 49.00%, respectively (Table 1). Afterwards, all of the high-quality reads were assembled into 17,923 unigenes with an average length of 1287.45 bp and an N50 of 1970 bp using the Trinity software. The length distribution of all unigenes is shown in Figure S3. And the GC content frequency distribution is shown in Figure S4. The sequences from both the libraries were deposited at NCBI and can be accessed in the Short Read Archive (SRA) under the accession numbers SRR2352154 and SRR2153022.

Table 1.
Statistics of transcriptome sequencing.
2.2. Functional Annotation and Classification of Unigenes
Annotation of the unigenes was performed against GenBank non-redundant (NR), SwissProt, Clusters of orthologous groups for eukaryotic complete genomes (KOG), Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases using BLASTX search with a cut-off E value of 10−5. Here, 14,255 unigenes (79.53%) were annotated in these databases (Table 2).

Table 2.
Summary of annotations of unigenes in public databases.
Clusters of orthologous groups for eukaryotic complete genomes (KOG) describesorthologous groups for eukaryotic genomes, and each protein is assumed to be from an ancestor protein. In all, 7225 unigenes were clustered into 25 functional categories (Figure 1). Among the 25 KOG categories, the cluster for ”General function prediction only” (2280, 31.56%) was the largest group, followed by “Posttranslational modification, protein turnover, chaperones” (1472, 20.37%), “Signal transduction mechanisms” (1136, 15.72%), “Lipid transport and metabolism” (883, 12.22%) and “Secondary metabolites biosynthesis, transport and catabolism” (847, 11.72%); only a few unigenes were assigned to the “Extracellular structures” and “Cell motility” groups.
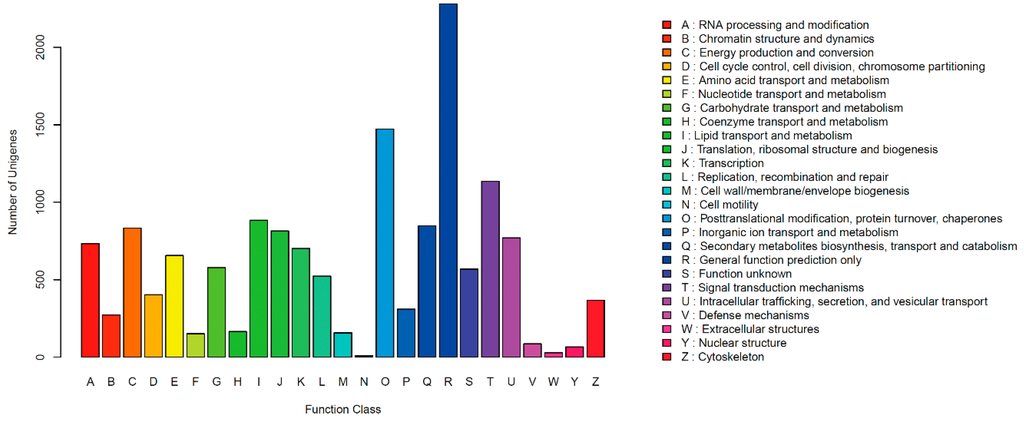
Figure 1.
KOG of unigenes of Shiraia bambusicola. KOG Functional Classification of transcriptome. In all, 7225 unigenes were clustered into 25 functional categories.
Gene ontology (GO) is an international standardized gene functional classification system that provides a controlled vocabulary for describing the properties of genes and their products. The three main independent categories, including “biological process”, “cellular component” and “molecular function”, are shown in Figure 2. A total of 9647 (53.82%) unigenes were assigned to their main GO categories, including 52 sub-categories. Under the biological process group, the largest proportions were metabolic process (GO: 0008152) and cellular process (GO: 0009987). Within the cellular component, cell (GO: 0005623) and cell part (GO: 0044464) were the largest groups by far. Catalytic activity (GO: 0003824) and binding (GO: 0005488) represented the majority of terms in the molecular function category.
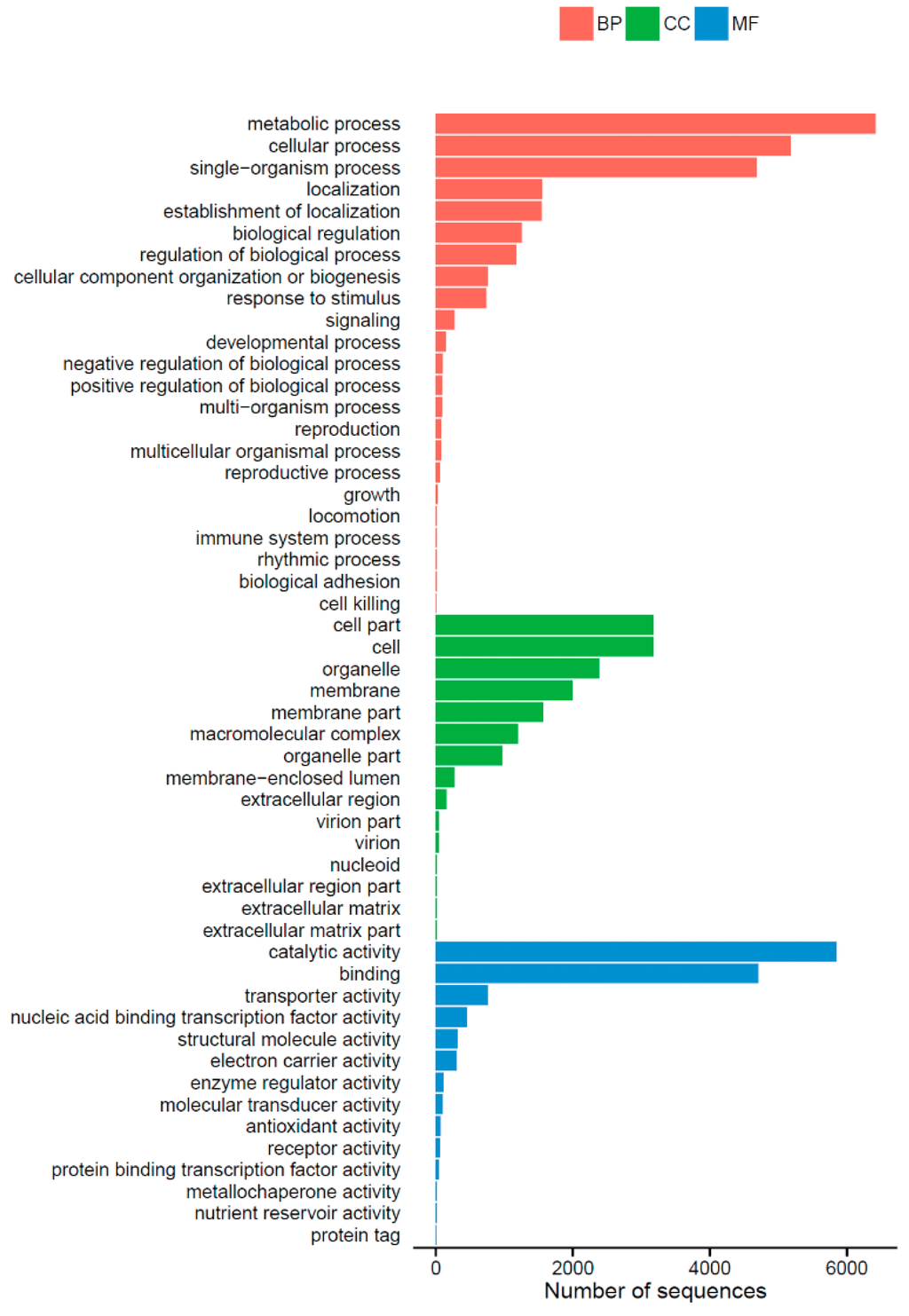
Figure 2.
Gene Ontology classification of Shiraia bambusicola transcriptome. A total of 9647 (53.82%) unigenes were assigned to their main GO categories (Biological processes (BP), Cellular components (CC), molecular functions (MF)), including 52 sub-categories.
To further identify the active biological pathways, unigenes were searched against pathway collections in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database, which can analyze gene functions systematically and integrate chemical, genomic and systemic functional information [22,23]. A total of 3581 unigenes were annotated and assigned to 323 different pathways (Table S1). “Ribosome” (ko03010, 253 unigenes) was the most enriched pathway, followed by “Biosynthesis of amino acids” (ko01230, 238 unigenes), “Carbon metabolism” (ko01200, 231 unigenes) and “Oxidative phosphorylation” (ko00190, 168 unigenes).
2.3. Transcripts Differentially Expressed between the Wild Type and Mutant
Based on the criteria of FDR ≤ 0.05 and |log2ratio| ≥ 1, the results show that 904 unigenes were found to be differentially expressed; 716 unigenes were up regulated in S4201-D1, and 188 unigenes were down regulated. We selected 97 significant differentially expressed genes, the detailed lists are given in Table 3. GO-enrichment was performed against all of the DEGs to research the relationship between genetics and phenotype. We then mapped all DEGs to each term of the GO database and calculated the gene numbers from each GO term.

Table 3.
Significant differentially expressed genes between S4201-W and S4201-D1.
Oxidation-reduction process and transmembrane transport were prominently enriched in biological process. Integral components of the membrane and ribosome were significantly enriched in cellular component. Oxidoreductase activity and heme binding were the majority of terms in molecular function (Table S2). Taken together, these results reveal that during HA production, the main processes are oxidation-reduction and biosynthesis; however, transmembrane transport processes are active when HA needs to be exported.
In the meantime, we identified significantly enriched metabolic pathways by enrichment analysis of the DEGs and mapped 904 unigenes to 201 pathways (Table S3). Among them, the pathways with the most representation by unigenes were “Ribosome” and “Oxidative phosphorylation”.
In the KEGG pathway enrichment analysis, genes of certain pathway in S4201-W showed upregulation compared with S4201-D1, and these pathways included “Glycolysis/Gluconeogenesis” (ko00010), “Fructose and mannose metabolism” (ko00051), “Tyrosine metabolism” (ko00350), “Phenylalanine metabolism” (ko00360), “Inositol phosphate metabolism” (ko00562), “Pyruvate metabolism” (ko00620), “Methane metabolism” (ko00680) and “Carbon metabolism” (ko01200). The upregulated unigenes will likely increase production of the two substrates acetyl-CoA and malonyl CoA, either directly or indirectly, which can provide enough raw material for HA biosynthesis.
The metabolism of S4201-D1, which lacks HA production ability, is more vigorous than S4201-W in fatty acid, amino acid and most saccharide synthesis. This suggests that in S4201-D1, it is more helpful to store nutrition and defend against environmental stresses. There are various possible reasons for this phenomenon: (1) Because the secondary metabolism in S4201-D1, which produced HA, was diminished or eliminated, the primary metabolism that produced carbohydrates, amino acids and lipids was reinforced; (2) HA is cytotoxic and can damage cells and decrease cellular activity. Without HA, the cellular activity of S4201-D1 increased, and the primary metabolism also increased; (3) Without HA, the defense mechanism of S4201-D1 was also diminished or eliminated. This saved materials and energy for other metabolic processes; (4) To repair UV damage, S4201-D1 increased its metabolism of carbohydrates, amino acids and lipids. The possible reasons are based on the speculation that they might be cause and effect relationship, but they may also be correlations. We need further studies to identify whether the relationship between these up regulated genes and HA production is correlation or a cause and effect relationship.
2.4. Candidate Genes Involved in HA Biosynthesis and a Putative Biosynthetic Pathway
The genes involved in the biosynthesis and modification of many secondary metabolites in fungi are organized in clusters [24]. We found seven of the 904 differentially expressed genes may be candidate genes involved in HA biosynthesis. These transcripts showed significantly higher expression in S4201-W (with HA production) compared with S4201-D1 (without HA production). They were not expressed or were at very low level in S4201-D1. These seven unigenes encode multicopper oxidase, fasciclin, polyketide synthase, o-methyltransferase/FAD-dependent monooxygenase, o-methyltransferase, hydroxylase and FAD/FMN-dependent oxidoreductase respectively. They were found to have high identity with their corresponding homologous genes through annotation using NR databases. Finally, their sequence information was completely matched with the “Shiraia sp. slf14 hypocrellin A biosynthesis gene cluster, complete sequence” (GenBank: KM434884.1, unpublished results) in the NCBI database. Based on the above results, we identified these seven genes as the putative genes of HA biosynthesis.
We proposed a putative HA biosynthetic pathway, shown in Figure 3. It is based on the model of cercosporin biosynthesis, which shares a similar structure, coding genes and is also a perylenequinoid pigment. Similar to cercosporin biosynthesis [16], acetyl-CoA and malonyl-CoA are the precursor and substrate of HA. The first step of the biosynthesis pathway has been predicted to be condensation and decarboxylation of acetyl-CoA (starter) and malonyl-CoA (extender) by the functional domains of the polyketide synthase. The chain of carbon elongates to form a pentaketide molecule that subsequently undergoes cyclization, then o-methyltransferase/FAD-dependent monooxygenase, FAD/FMN-dependent oxidoreductase and o-methyltransferase catalyze a series of oxidation, hydration and methylation reactions to yield cyclized polyhydroxynaphthalene units. Thus, HA is likely formed by dimerizing two polyhydroxynaphthalene units, and then undergoes other unknown modification reactions. Cercosporin has a bilateral symmetric structure, but HA does not. Hence, it is likely that before dimerization, HA has formed two different polyhydroxynaphthalene units, or the final form of HA is accomplished by dimerization of the two identical polyhydroxynaphthalene units which were similar to cercosporin. Once synthesized, HA must be exported out of cells, presumably by a major facilitator superfamily (MFS) transporter, but our RNA-Seq data showed complete opposite expression patterns that were not concordant with the MFS transporter in Cercospora species. In addition, other DEGs are likely to be involved in and regulate the biosynthesis of HA, but this still needs further research. In all, we identified a large number of candidate genes involved in HA biosynthesis by analyzing the RNA-Seq data. To identify whether a gene takes part in the biosynthesis of HA or not and to understand its function, we need to do future studies, such as gene cloning, analysis of its structure, and function verification.
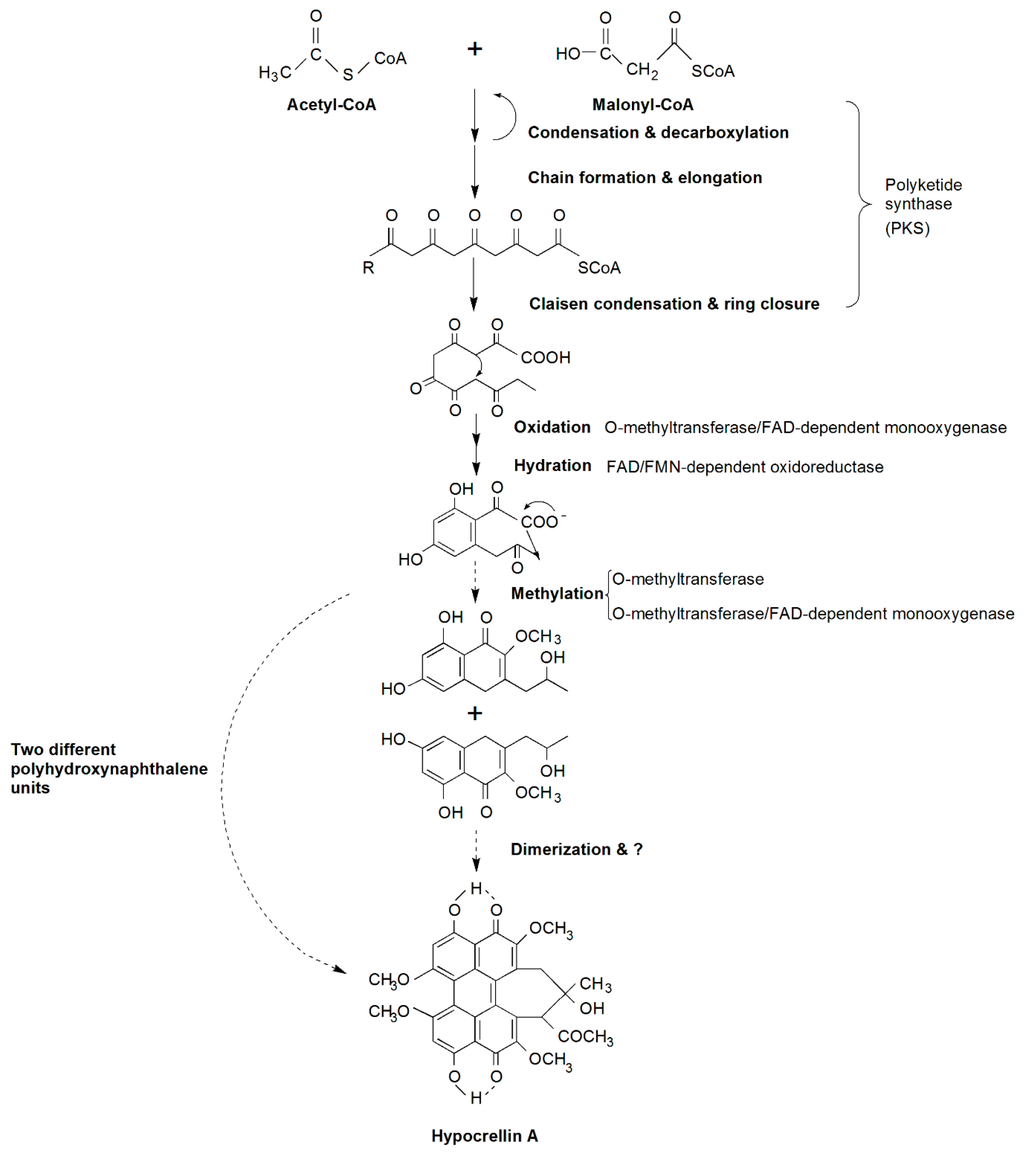
Figure 3.
Putative HA biosynthetic pathway-related genes and products in Shiraia bambusicola. Reactions catalyzed at every step are shown. The broken arrow represents putative steps, in which the enzymes involved are not yet clear.
By comparing these gene sequences between the wild type and the mutant, we found that there are a total of three single nucleotides polymorphisms, of which one resulted in amino acid polymorphisms. This gene encodes o-methyltransferase/FAD-dependent monooxygenase and its N-terminal amino acid sequence is similar to that of the transcription enhancer AFLS (formerly AFLJ) involved in aflatoxin biosynthesis. Unfortunately, the mutation site may not be in the functional domains. Therefore, we suggested that the cause of the lack of HA biosynthesis might be in transcriptional regulation, rather than genetic mutations in candidate genes involved in HA biosynthesis. Gene expression in eukaryotes is regulated primarily at the level of transcription and we proposed the following reasons why S4201-D1 cannot produce HA at transcription level: (1) The mutations in the specific transcription factor of the HA biosynthesis gene cluster may lead to that the transcription complexes cannot transcribe normally; (2) Trans-acting factors cannot bind to the mutant cis-acting elements and this blocks the initiation of transcription; (3) UV might cause the formation of new silencers which can affect these genes, or cause the formation of binding sites for repressor proteins in the upstream region of the transcription start sites; (4) Moreover, we found that there are no significant differences in expression of transcription factors between the wild type and the mutant, and the cause of the lack of HA biosynthesis may not be the change in numbers of transcription factors. Therefore we still need to study the gene expression mechanism of HA biosynthesis at the transcriptional level.
2.5. Autodefense by Shiraia bambusicola against HA
Apart from studying the biosynthesis of HA, autodefense against HA also needs to be understood to improve HA production and S. bambusicola self-resistance. The candidate genes that are involved in HA biosynthesis were not expressed, this means S4201-D1 cannot produce HA. Because of the absence of HA, S4201-D1 would not generate reactive oxygen species (ROS), and further related oxidative damages would not happen yet. Meanwhile, the redox reaction in S4201-D1 was not as remarkable as it was in S4201-W, and we will discuss the self-defense mechanism of S. bambusicola as follows. In the presence of light and oxygen, photosensitizers can generate ROS, such as singlet oxygen (1O2) and superoxide radical (O2−), which can damage proteins, lipids and nucleic acids [25,26]. Due to its mode of action, HA has almost universal toxicity to cells, and it can damage not only plants but also bacteria, fungi, viruses and tumors. S. bambusicola, however, not only synthesizes, but also is resistant to, high concentrations of HA in culture. Cellular resistance mechanisms against perylenequinone photosensitizers such as cercosporin are a basic biological phenomenon. Cercosporin serves as a model system in which to study cellular resistance to 1O2 in HA. Several 1O2 quenchers have been characterized in previous research [27]. Carotenoids are one of the most effective quenchers of cellular 1O2 that exist in biological systems, and they are one of the most effective defenses against the compounds producing 1O2 [28,29,30]. In our RNA-Seq data, two genes involved in carotenoid biosynthesis were upregulated in S4201-W compared with S4201-D1. And we found that the two genes encoded lycopene beta-cyclase and torulene dioxygenase respectively. Therefore, carotenoid is likely to be involved in autodefense mechanisms of HA. PDX1 and PDX2, two genes that encode enzymes involved in the vitamin B6 biosynthesis pathway, were identified as essential for cercosporin resistance [31,32]. After this discovery, vitamin B6 was found to quench 1O2 superoxide effectively and to have antioxidant activity [33,34]. Cercospora nicotianae mutants, which do not have a functional vitamin B6 biosynthetic gene, lose their cellular resistance to cercosporin [31]. Our RNA-Seq data showed that the gene, in S. bambusicola S4201-W, encoding pyridoxine 4-dehydrogenase involved in vitamin B6 biosynthesis was expressed at higher levels than in mutant S4201-D1. Therefore, VB6 is likely to be involved in the autodefense mechanism of HA. Many photosensitizers are converted to a non-phototoxic form when reduced, and colorless forms lack normal visible light absorption. Cercosporin in these reduced derivatives absorb about half the amount of light compared with cercosporin [35]. The cercosporin in the fungal cell is reduced to a nontoxic mode. When the cercosporin has been released, it reoxidizes to its photoactive form spontaneously [36]. Therefore, reduced cercosporin is used as a cellular resistance mechanism in Cercospora species. Meanwhile, our DEG data mainly found enrichment in oxidation-reduction processes within the biological process group. In addition, dioxin degradation (ko00621), polycyclic aromatic hydrocarbon degradation (ko00624), naphthalene degradation (ko00626) and degradation of aromatic compounds (ko01220) were significantly enriched pathways, in which certain unigenes were upregulated in S4201-W compared with S4201-D. These pathways refer to strong reduction processes that can decrease cell damage from an oxidizing agent. These results are in accordance with biological characteristics of HA.
2.6. Real-Time PCR Analysis of Several Transcripts
We selected six unigenes involved in HA biosynthesis and four DEGs randomly chosen through quantitative real time PCR (qRT-PCR) to validate the changes in gene expression identified by RNA-Seq. The results showed that the unigenes that encode multicopper oxidase, fasciclin, polyketide synthase, o-methyltransferase/FAD-dependent monooxygenase, hydroxylase FAD/FMN-dependent oxidoreductase and pyridoxal reductase, were significantly higher in S4201-W, whereas aldehyde dehydrogenase (NAD+), cytochrome P450, alcohol dehydrogenase encoding genes were mainly expressed in S4201-D (Figure 4). The expression levels of these 10 samples were highly consistent with the RNA-Seq data, suggesting that the RNA-Seq data are reliable.
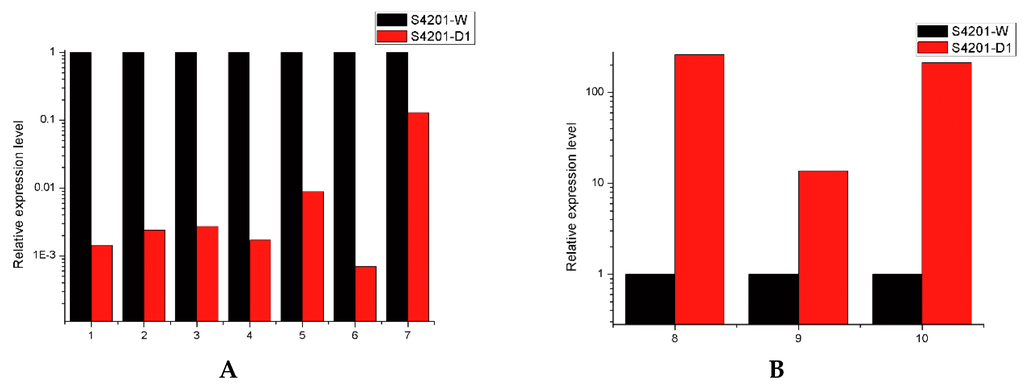
Figure 4.
Quantitative RT-PCR validation. The relative expression levels of mRNA were normalized with internal reference gene (GAPDH) and relative expression to the corresponding values of S4201-W (control) were given an arbitrary value of 1. (A) 1: Multicopper oxidase, 2: Fasciclin, 3: Polyketide synthase, 4: O-methyltransferase/FAD-dependent monooxygenase, 5: Hydroxylase, 6: FAD/FMN-dependent oxidoreductase, 7: Pyridoxal reductase; (B) 8: Aldehyde dehydrogenase (NAD+), 9: Cytochrome P450, 10: Alcohol dehydrogenase.
3. Experimental Section
3.1. Materials
The wild strain S4201-W was isolated from the fruiting bodies of the Shiraia bambusicola P. Henn collected in Anji in the Zhejiang Province of China, and screened as a hypocrellin-producing strain. No specific permissions were required for the described field studies. The location is not privately-owned or protected, and the field studies did not involve endangered or protected species. After molecular identification based on nuclear ribosomal internal transcribed spacer (rDNA ITS, the ITS region is the most frequently sequenced genetic marker of fungi), the strain was preserved in the Microbial Culture Center of College of Life Sciences, Nanjing Normal University, China. The strain S4201-W was cultured on potato dextrose agar (PDA) and washed with sterile water to prepare the spore suspension, and the concentration was adjusted to 1 × 106 spores/mL. The spore suspension was irradiated by a 800 mj/cm2 dose of ultraviolet (UV) radiation treatment with a UV-light cross-linker (UV light source). We selected the sample that did not produce HA on PDA. HA was measured by high-performance liquid chromatography (HPLC) with HA standard reagent. S. bambusicola S4201-W has excellent HA production, while the mutant S4201-D1 does not. Then, S4201-W and S4201-D1 were inoculated into potato dextrose broth (PDB) medium with Triton X-100 and cultured in the dark at 28 °C on a shaker at 150 rpm/min for 82 h [37]. Fermentation liquid was centrifuged at 3000 rpm for 3 min at room temperature in a microcentrifuge, and the supernatant was removed. Mycelia of each sample were immediately frozen and stored at −80 °C.
3.2. RNA Isolation and Illumina Sequencing
The samples were collected from 82-h-old mycelia, respectively. Total RNA from each sample was extracted using the mirVana™ miRNA Isolation Kit without phenol (Ambion, Inc., Austin, TX, USA) and treated with RNase free DNaseI according to manufacturer’s instructions. The quality of RNA was confirmed by spectrophotometer (NanoDrop ND-1000, NanoDrop Technologies, Wilmington, DE, USA) and agarose gel electrophoresis before further processing. Poly (A) mRNA from total RNA was enriched by oligo (dT) magnetic beads and then broken into short fragments to synthesize the first strand of cDNA by random hexamer-primer. DNA polymerase I and RNaseH were used to synthesize the second strand of cDNA and repair the ends, which were connected using sequencing adapters. Afterwards, cDNA libraries were constructed by PCR, and the sequences were analyzed using the Illumina HiSeqTM 2500 sequencing platform (commercial service) at Shanghai OE Biotech Co., Ltd. (Shanghai, China).
3.3. De Novo Transcriptome Assembly and Annotation
The raw reads were cleaned by removing adaptor sequences, empty reads and low-quality reads. In our research, the Trinity (vesion: trinityrnaseq_r20131110) program was used to assemble high quality reads for each sample [38]. Because S. bambusicola has no reference genome in public databases, reads needed to be assembled de novo. Based on a BLASTX search (E-value < 10−5), the functional annotation of these unigenes were performed using different protein databases, including the NCBI NR, Kyoto Encyclopedia of Genes and Genomes (KEGG), SwissProt and KOG databases. GO annotations based on the BLAST2GO program were used to further analyze the functional classifications of these unique sequences [39].
3.4. Analysis of Differential Gene Expression
The unigene expression was determined by using the reads per million kilobases per million mapped reads (RPKM) method to normalize the read counts between the samples [40,41]. Differentially expressed genes (DEGs) were identified with a false discovery rate (FDR) ≤ 0.05 and |log2ratio| ≥ 1. To find out what was significantly differentially expressed between S4201-W and S4201-D1, the p-values ≤ 0.05 and fold changes ≥ 2 were used as the estimate threshold.
3.5. Quantitative Real-Time PCR Analysis
Quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) of differentially expressed genes in S4201-W and S4201-D1 was used to evaluate the quality of the sequence assembly. The expression patterns of ten transcripts were monitored, with three independent replicates of each transcript. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal reference gene. RNA from each sample was isolated using the mirVana™ miRNA Isolation Kit without phenol and treated with RNase-free DNaseI according to the manufacturer’s instructions. The first strand of cDNA was synthesized by the use of PrimeScriptTM RT Master Mix (TaKaRa, Dalian, China). qRT-PCR was performed in an StepOnePlusTM Real-Time PCR System (Applied Biosystems, USA) using a SYBR Premix Ex TaqTM II (TaKaRa, Dalian, China) and the conditions as follows: 95 °C for 30 s followed by 40 cycles of 95 °C for 5 s, 60 °C for 30 s. A melting curve was generated to test specificity of the products produced by the qRT-PCR. The results were calculated using the 2−ΔΔCt method [42]. Primer sequences are presented in the supporting information (Table S4).
4. Conclusions
Hypocrellin compounds are perylenequinonoid pigments with excellent antiviral and antitumor properties. However, their complex metabolic pathway remains unknown. Our study is the first attempt to use the Illumina/Solexa deep sequencing platform for de novo sequencing and assembly of S. bambusicola without a reference genome. A large number of transcripts differentially expressed between the wild strain S4201-W and mutant S4201-D1 have been identified, especially the ones involved in the biosynthesis of HA. In addition, we proposed a putative HA biosynthetic pathway. This study facilitates understanding of the gene expression and functional genomics of S. bambusicola and provides a valuable set of public information that can be used to further explore the molecular mechanisms of various metabolic pathways. The transcriptome sequence data will allow for increased HA production using biotechnological applications and metabolic engineering in the near future.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/17/3/311/s1.
Acknowledgments
We are sincerely grateful to the anonymous referees whose comments made our results more accessible to the readers. This study was supported by the funds of the National Key Technology Research and Development Program of the Ministry of Science and Technology of China (No.2012BAD36B05) & the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Author Contributions
Conceived and designed the experiments: Shuang-Lin Chen, Shu-Zhen Yan and Ning Zhao. Performed the experiments: Ning Zhao, Xi Lin and Zhi-Mei Luo. Analyzed the data: Ning Zhao. Contributed reagents/materials/analysis tools: Shan-Shan Qi. Wrote the manuscript: Ning Zhao.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ali, S.M.; Olivo, M. Efficacy of hypocrellin pharmacokinetics in phototherapy. Int. J. Oncol. 2002, 21, 1229–1237. [Google Scholar] [CrossRef] [PubMed]
- Morakotkarn, D.; Kawasaki, H.; Seki, T. Molecular diversity of bamboo-associated fungi isolated from Japan. FEMS Microbiol. Lett. 2007, 266, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Zhenjun, D.; Lown, J.W. Hypocrellins and their use in photosensitization*. Photochem. Photobiol. 1990, 52, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, J.; Ikebuchi, K.; Abe, H.; Kwon, K.W.; Ohnishi, Y.; Horiuchi, M.; Shinagawa, M.; Ikuta, K.; Kamo, N.; Sekiguchi, S. Photoinactivation of virus infectivity by hypocrellin A. Photochem. Photobiol. 1997, 66, 697–700. [Google Scholar] [CrossRef] [PubMed]
- Hudson, J.; Zhou, J.; Chen, J.; Harris, L.; Yip, L.; Towers, G. Hypocrellin, from Hypocrella bambuase, is phototoxic to human immunodeficiency virus. Photochem. Photobiol. 1994, 60, 253–255. [Google Scholar] [CrossRef] [PubMed]
- Longsheng, C.; Jiazhen, W.; Shimi, F. Effect of hypocrellin A sensitization on the lateral mobility of cell membrane proteins. J. Photochem. Photobiol. B 1988, 2, 395–398. [Google Scholar] [CrossRef]
- Oleinick, N.L.; Morris, R.L.; Belichenko, I. The role of apoptosis in response to photodynamic therapy: What, where, why, and how. Photochem. Photobiol. Sci. 2002, 1, 1–21. [Google Scholar] [PubMed]
- Robertson, C.; Evans, D.H.; Abrahamse, H. Photodynamic therapy (PDT): A short review on cellular mechanisms and cancer research applications for PDT. J. Photochem. Photobiol. B 2009, 96, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.S.; Lin, X.; Zhang, M.M.; Yan, S.Z.; Yu, S.Q.; Chen, S.L. Preparation and evaluation of hypocrellin A loaded poly (lactic-co-glycolic acid) nanoparticles for photodynamic therapy. RSC Adv. 2014, 4, 40085–40094. [Google Scholar] [CrossRef]
- Diwu, Z.; Lown, J.W. Phototherapeutic potential of alternative photosensitizers to porphyrins. Pharmacol. Ther. 1994, 63, 1–35. [Google Scholar] [CrossRef]
- Daub, M.E.; Ehrenshaft, M. The photoactivated Cercospora toxin cercosporin: Contributions to plant disease and fundamental biology. Annu. Rev. Phytopathol. 2000, 38, 461–490. [Google Scholar] [CrossRef] [PubMed]
- Okubo, A.; Yamazaki, S.; Fuwa, K. Biosynthesis of cercosporin. Agric. Biol. Chem. 1975, 39, 1173–1175. [Google Scholar] [CrossRef]
- Choquer, M.; Dekkers, K.L.; Chen, H.Q.; Cao, L.; Ueng, P.P.; Daub, M.E.; Chung, K.R. The ctb1 gene encoding a fungal polyketide synthase is required for cercosporin biosynthesis and fungal virulence of Cercospora nicotianae. Mol. Plant Microbe Interact. 2005, 18, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Dekkers, K.L.; You, B.J.; Gowda, V.S.; Liao, H.L.; Lee, M.H.; Bau, H.J.; Ueng, P.P.; Chung, K.R. The Cercospora nicotianae gene encoding dual O-methyltransferase and FAD-dependent monooxygenase domains mediates cercosporin toxin biosynthesis. Fungal Genet. Biol. 2007, 44, 444–454. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Choquer, M.; Lee, M.H.; Bau, H.J.; Chung, K.R. Deletion of a MFS transporter-like gene in Cercospora nicotianae reduces cercosporin toxin accumulation and fungal virulence. FEBS Lett. 2007, 581, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Lee, M.H.; Daub, M.E.; Chung, K.R. Molecular analysis of the cercosporin biosynthetic gene cluster in Cercospora nicotianae. Mol. Microbiol. 2007, 64, 755–770. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Q.; Lee, M.H.; Chung, K.R. Functional characterization of three genes encoding putative oxidoreductases required for cercosporin toxin biosynthesis in the fungus Cercospora nicotianae. Microbiology 2007, 153, 2781–2790. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Callahan, T.M.; Rose, M.S.; Meade, M.J.; Ehrenshaft, M.; Upchurch, R.G. Cfp, the putative cercosporin transporter of Cercospora kikuchii, is required for wild type cercosporin production, resistance, and virulence on soybean. Mol. Plant Microbe Interact. 1999, 12, 901–910. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.M.; Deng, H.X.; Guan, Z.B.; Liao, X.R.; Cai, Y.J. Preliminary study on one polyketide synthase from Shiraia sp. SUPER-H168. Biotechnology 2014, 1–4. [Google Scholar]
- Peng, S.L.; Yang, H.L.; Li, E.H.; Wang, X.L.; Zhu, D. The prokaryotic expression, purification and bioinformatics of type III polyketide synthase from Shiraia sp. Slf14, which is an endophytic fungus of Huperzia serrata. J. Jiangxi Norm. Univ. 2015, 39, 430–434. [Google Scholar]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Daub, M.E.; Chung, K.R. Photoactivated perylenequinone toxins in plant pathogenesis. Plant Relatsh. 2009, 5, 201–219. [Google Scholar]
- Valenzeno, D.P.; Pooler, J.P. Photodynamic action. BioScience 1987, 37, 270–276. [Google Scholar] [CrossRef]
- Beseli, A.; Amnuaykanjanasin, A.; Herrero, S.; Thomas, E.; Daub, M.E. Membrane transporters in self resistance of Cercospora nicotianae to the photoactivated toxin cercosporin. Curr. Genet. 2015, 61, 601–620. [Google Scholar] [CrossRef] [PubMed]
- Belluš, D. Physical quenchers of singlet molecular oxygen. Adv. Photochem. 1978, 11, 105–205. [Google Scholar]
- Krinsky, N.I. Carotenoid protection against oxidation. Pure Appl. Chem. 1979, 51, 649–660. [Google Scholar] [CrossRef]
- Krinsky, N.I. Actions of carotenoids in biological systems. Annu. Rev. Nutr. 1993, 13, 561–587. [Google Scholar] [CrossRef] [PubMed]
- Young, A.J. The photoprotective role of carotenoids in higher plants. Physiol. Plant. 1991, 83, 702–708. [Google Scholar] [CrossRef]
- Ehrenshaft, M.; Bilski, P.; Li, M.Y.; Chignell, C.F.; Daub, M.E. A highly conserved sequence is a novel gene involved in de novo vitamin b6 biosynthesis. Proc. Natl. Acad. Sci. USA 1999, 96, 9374–9378. [Google Scholar] [CrossRef] [PubMed]
- Ehrenshaft, M.; Daub, M.E. Isolation of pdx2, a second novel gene in the pyridoxine biosynthesis pathway of eukaryotes, archaebacteria, and a subset of eubacteria. J. Bacteriol. 2001, 183, 3383–3390. [Google Scholar] [CrossRef] [PubMed]
- Bilski, P.; Li, M.; Ehrenshaft, M.; Daub, M.; Chignell, C. Vitamin b6 (pyridoxine) and its derivatives are efficient singlet oxygen quenchers and potential fungal antioxidants. Photochem. Photobiol. 2000, 71, 129–134. [Google Scholar] [CrossRef]
- Denslow, S.A.; Walls, A.A.; Daub, M.E. Regulation of biosynthetic genes and antioxidant properties of vitamin B6 vitamers during plant defense responses. Physiol. Mol. Plant Pathol. 2005, 66, 244–255. [Google Scholar] [CrossRef]
- Leisman, G.B.; Daub, M.E. Singlet oxygen yields, optical properties, and phototoxicity of reduced derivatives of the photosensitizer cercosporin. Photochem. Photobiol. 1992, 55, 373–379. [Google Scholar] [CrossRef]
- Sollod, C.C.; Jenns, A.E.; Daub, M.E. Cell surface redox potential as a mechanism of defense against photosensitizers in fungi. Appl. Environ. Microbiol. 1992, 58, 444–449. [Google Scholar] [PubMed]
- Cai, Y.; Liao, X.; Liang, X.; Ding, Y.; Sun, J.; Zhang, D. Induction of hypocrellin production by Triton X-100 under submerged fermentation with Shiraia sp. SUPER-H168. N. Biotechnol. 2011, 28, 588–592. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-seq data. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Nagalakshmi, U.; Wang, Z.; Waern, K.; Shou, C.; Raha, D.; Gerstein, M.; Snyder, M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 2008, 320, 1344–1349. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using Real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).