Selenoprotein R Protects Human Lens Epithelial Cells against D-Galactose-Induced Apoptosis by Regulating Oxidative Stress and Endoplasmic Reticulum Stress
Abstract
:1. Introduction
2. Result
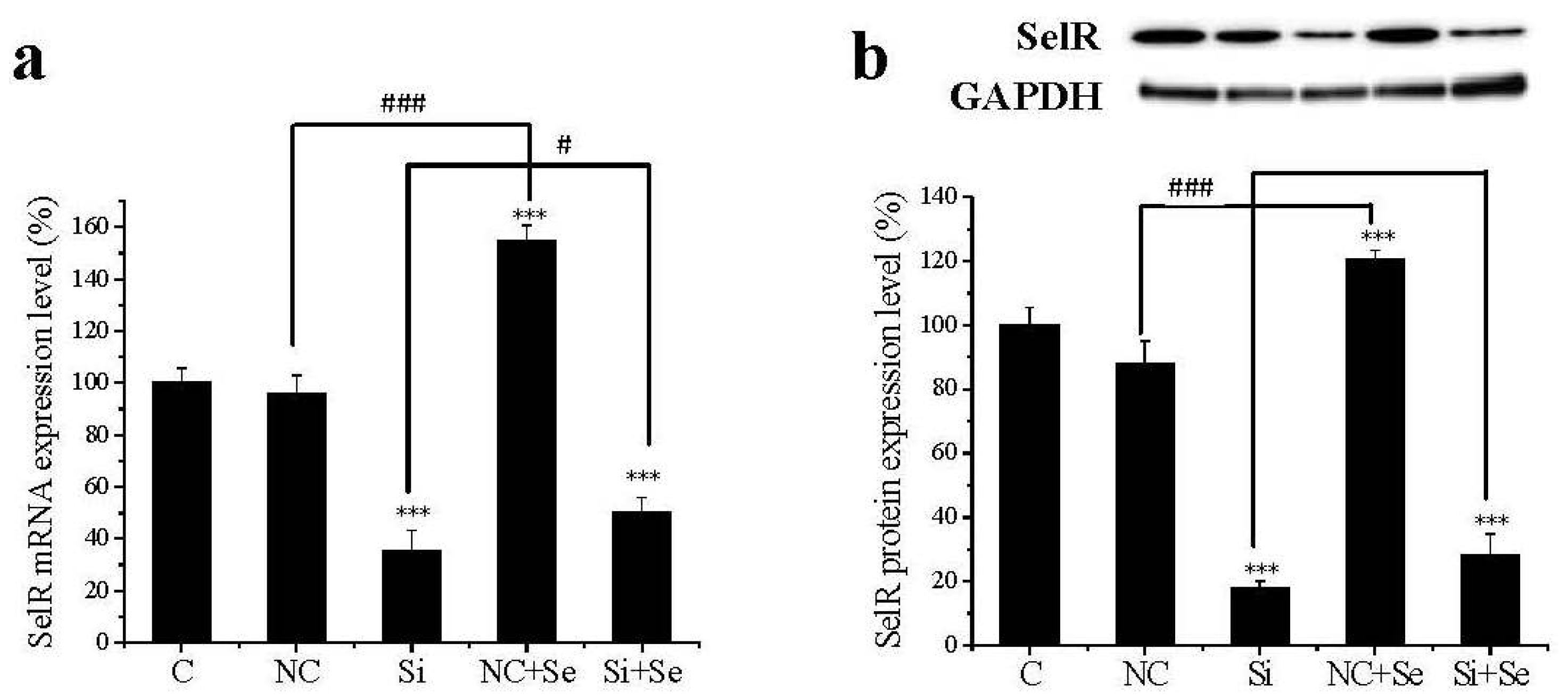
2.1. SelR Gene Silence Effectiveness
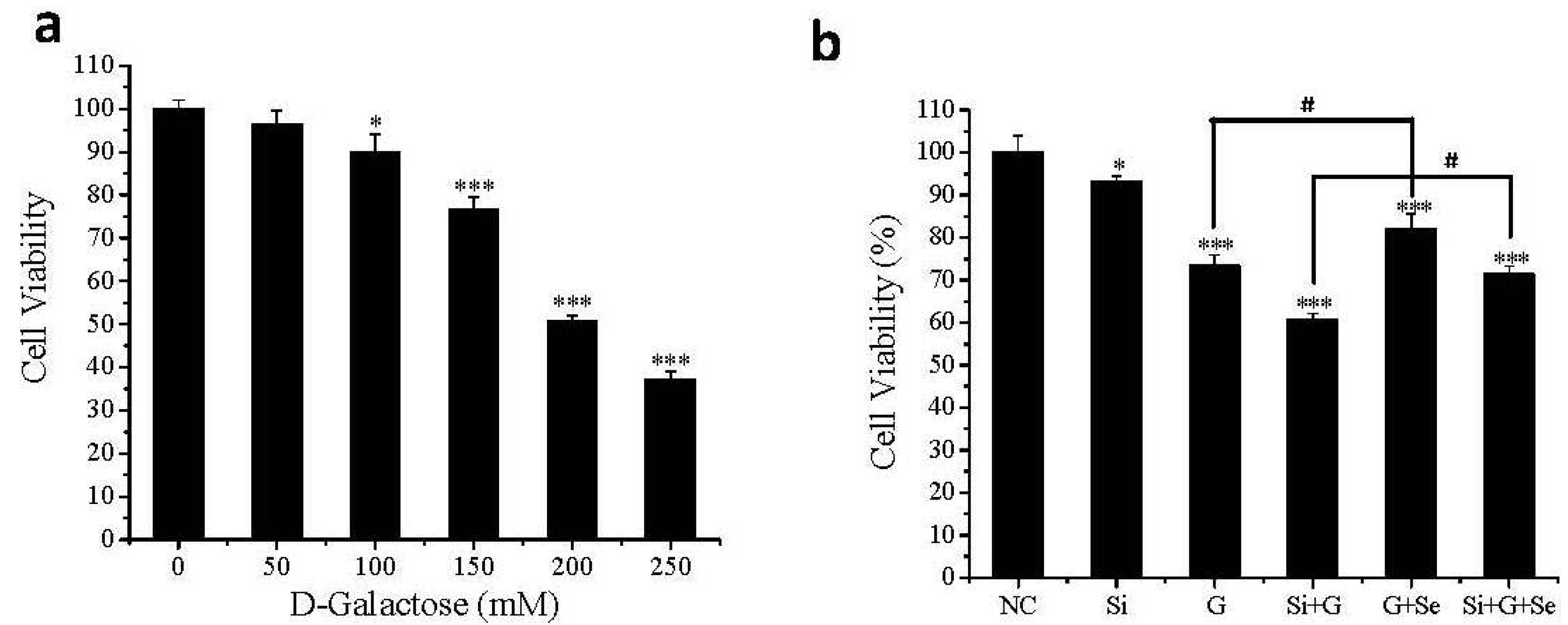
2.2. Effect of SelR Gene Knockdown and Na2SeO3 on Cell Viability in d-Galactose-Treated hLE Cells
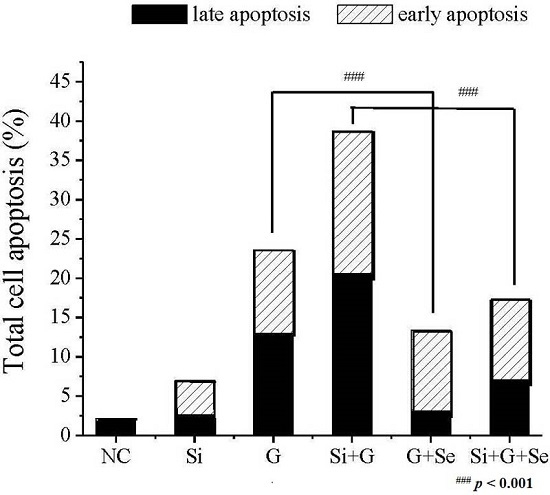
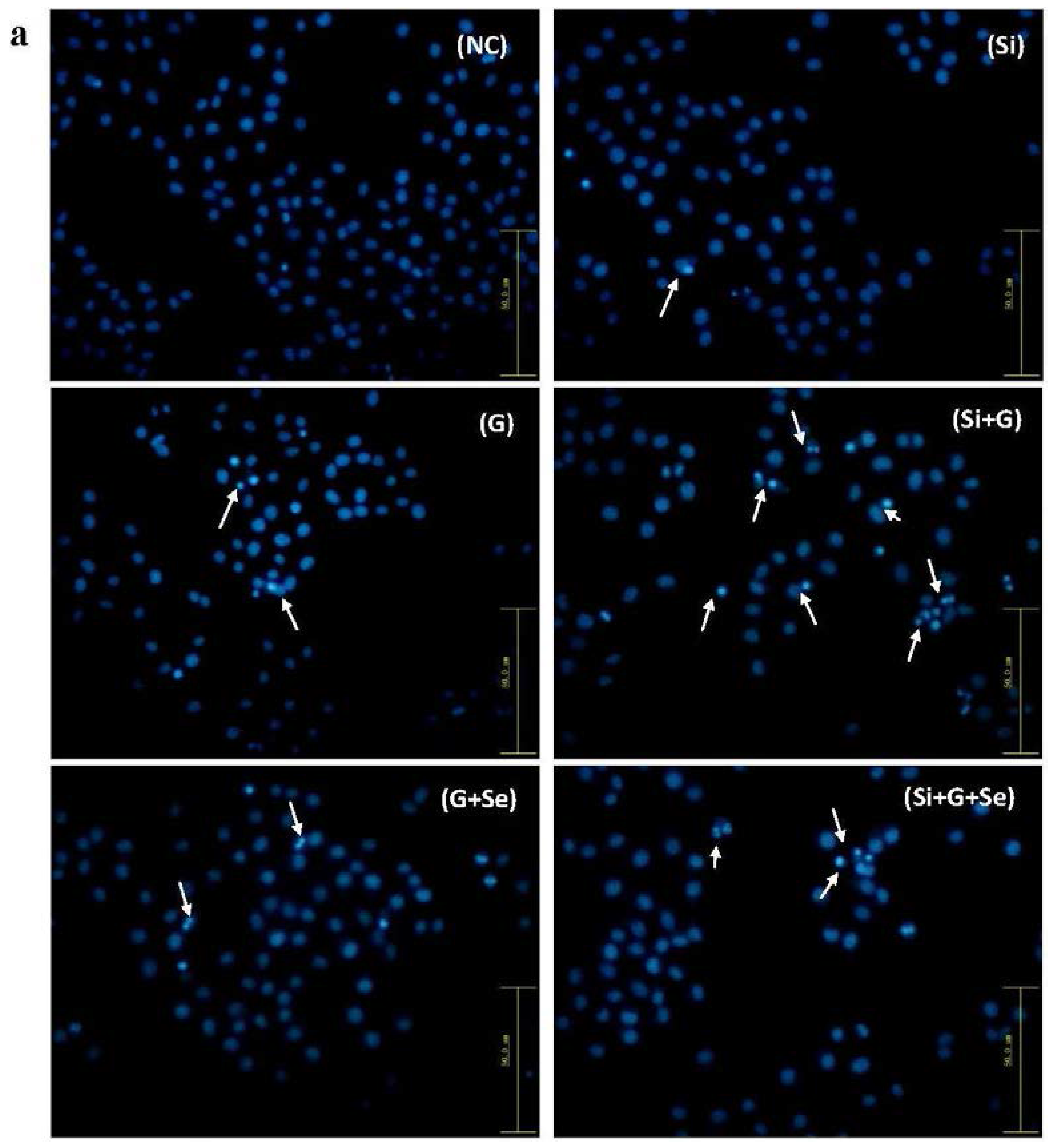
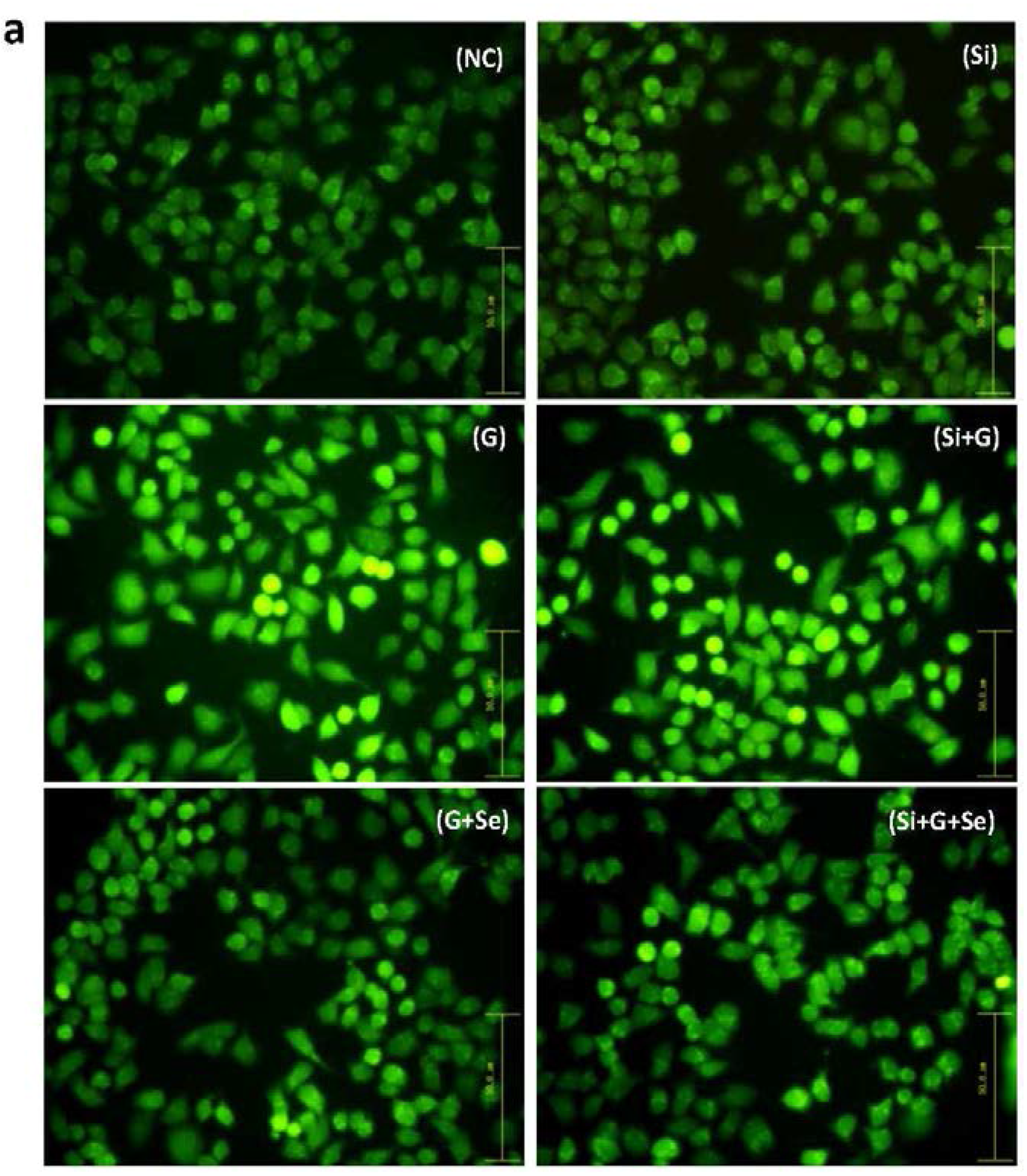
2.3. Effect of SelR Gene Knockdown and Na2SeO3 on d-Galactose-Induced Cell Apoptosis
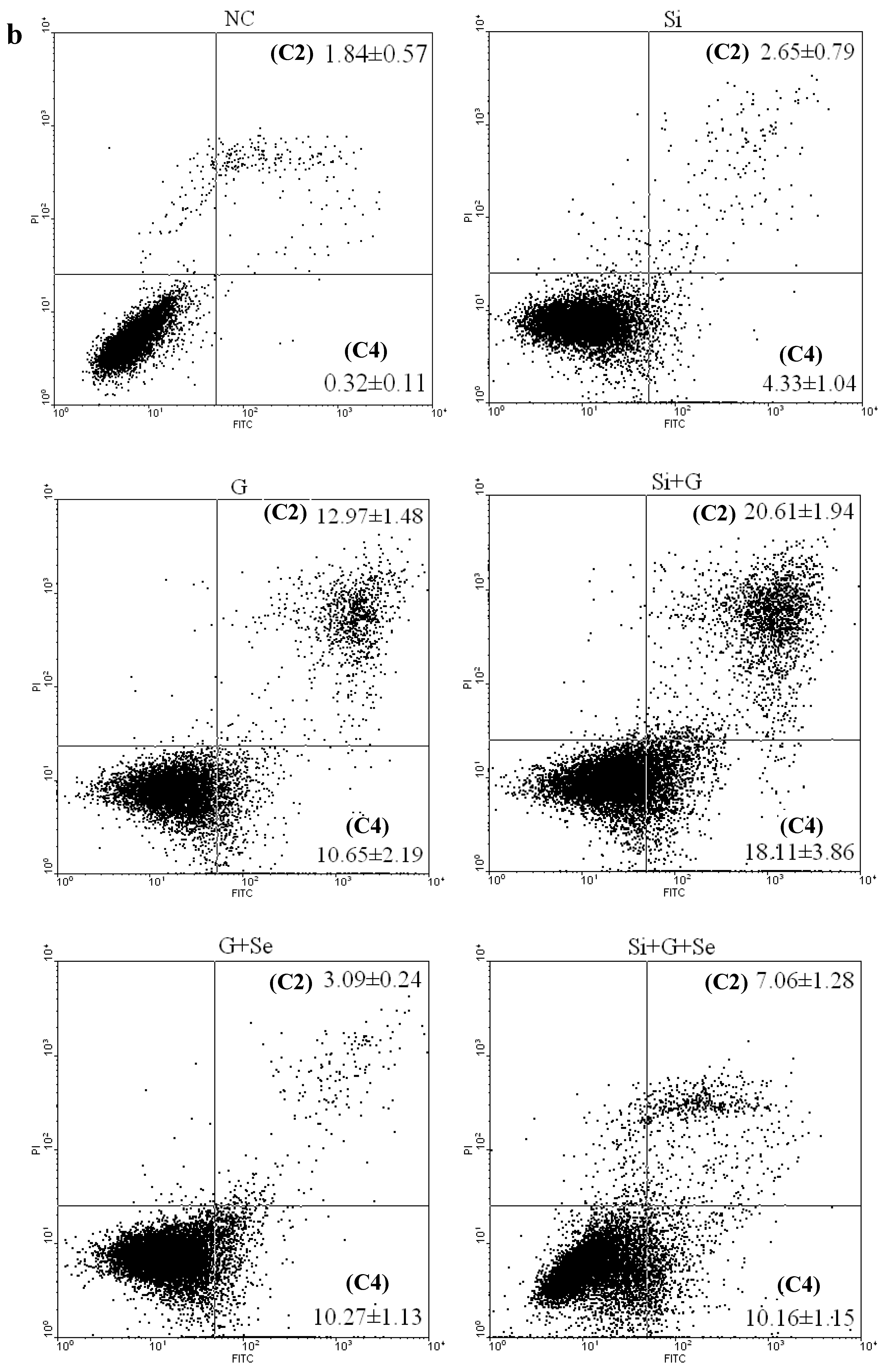
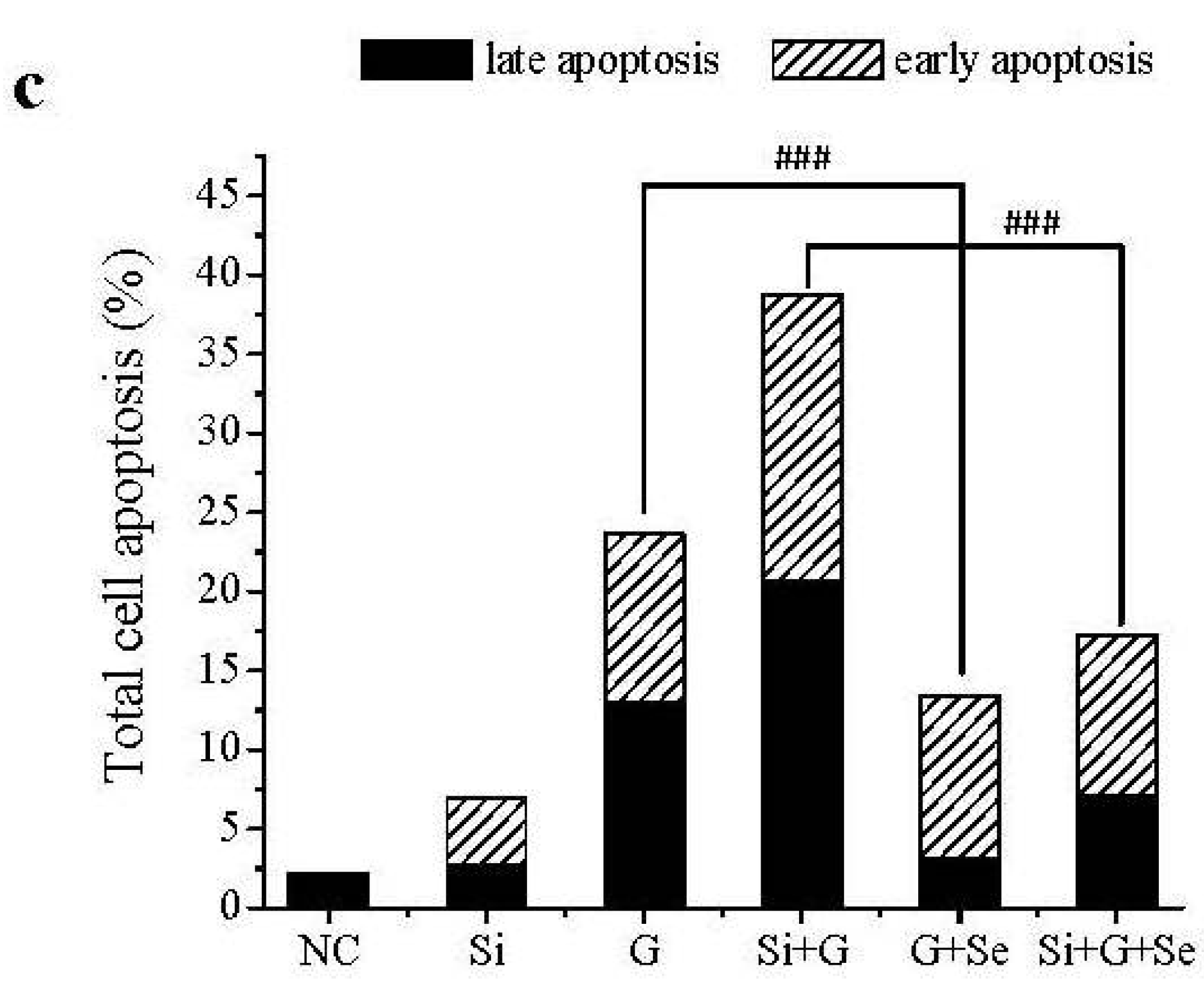
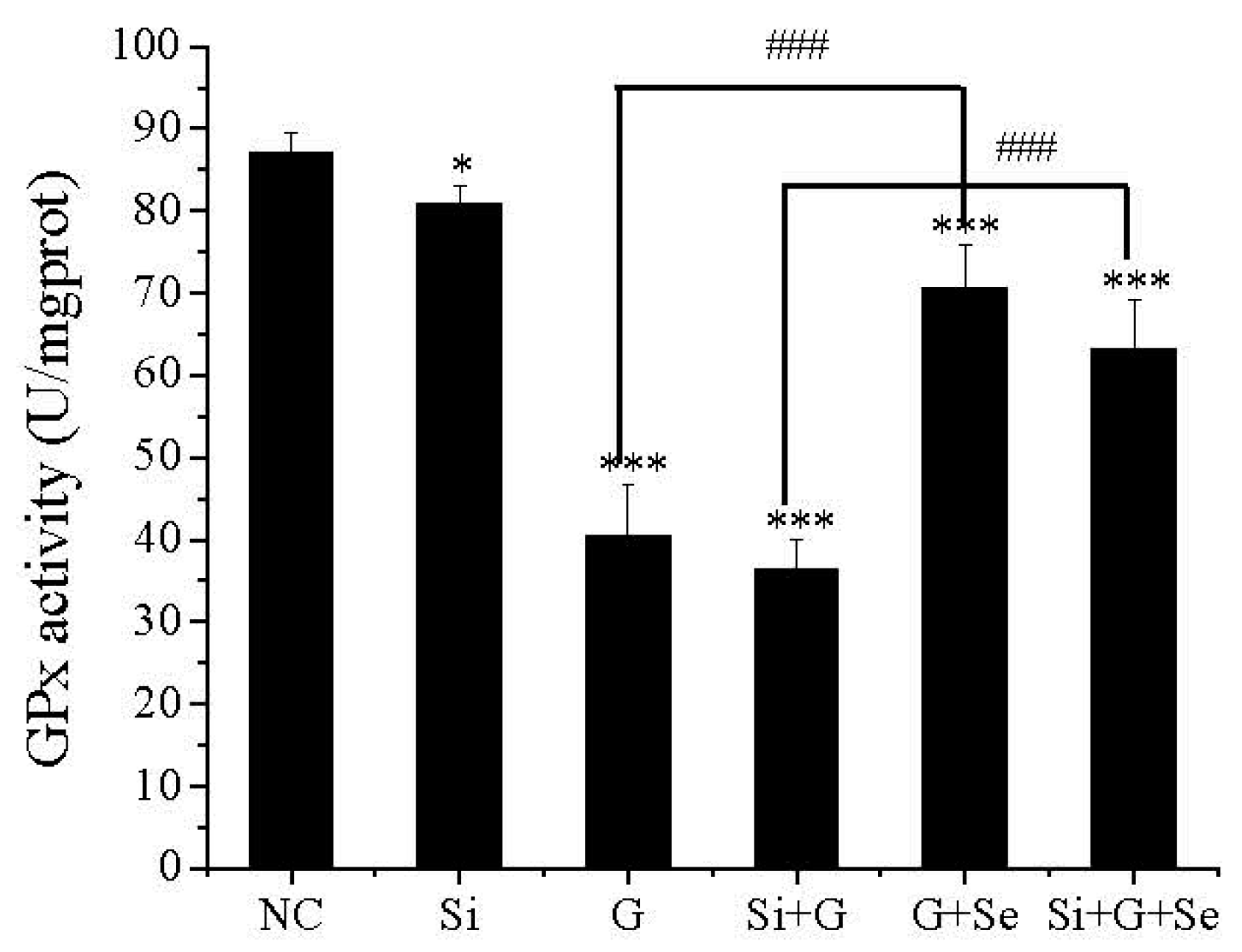
2.4. Effect of SelR Gene Knockdown and Na2SeO3 on GPx Activity in d-Galactose-Treated hLE Cells
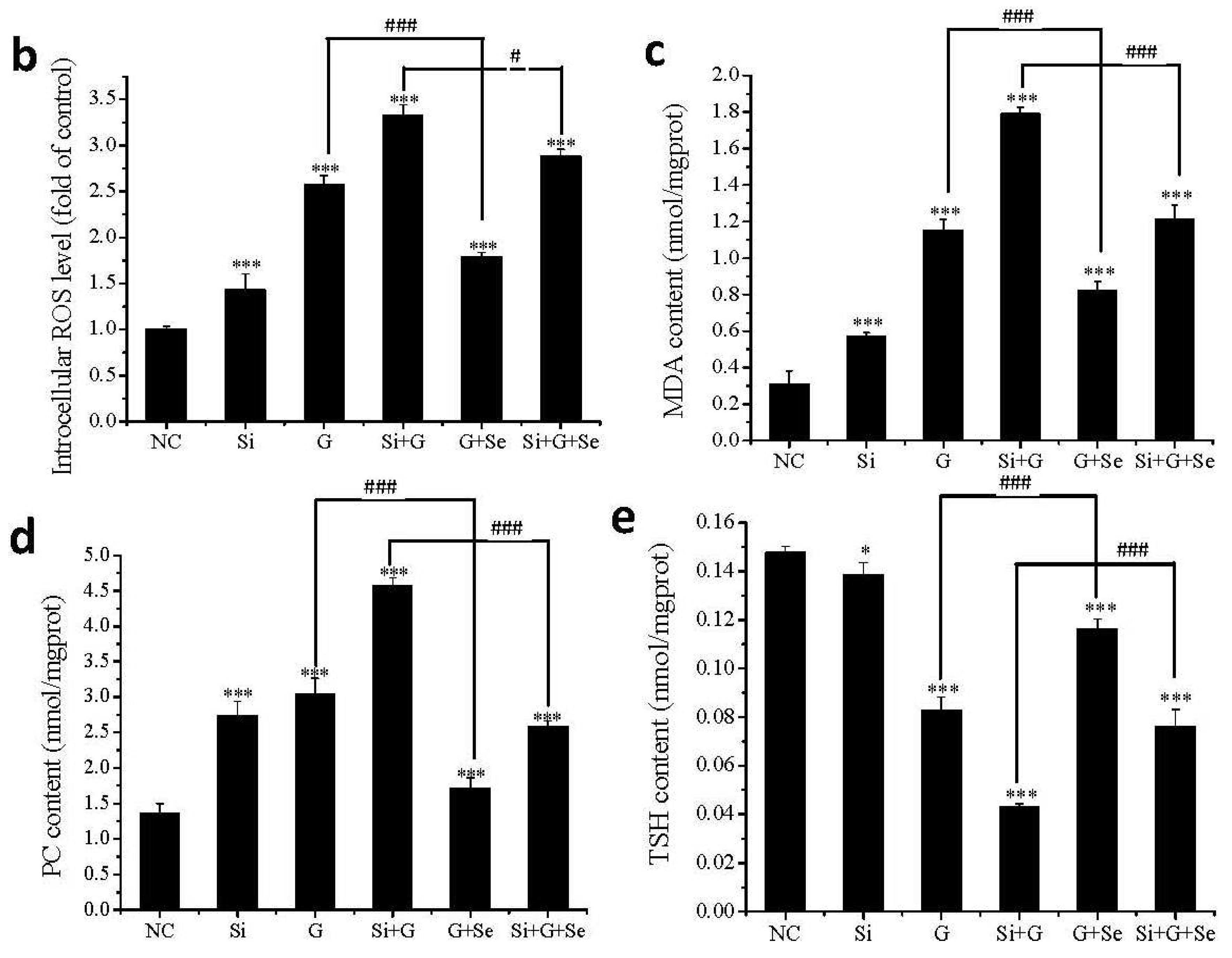
2.5. Effect of SelR Gene Knockdown and Na2SeO3 on d-Galactose-Induced ROS, MDA, PC and TSH Levels
2.6. Effect of SelR Gene Knockdown on d-Galactose-Induced Mitochondrial Dysfunctions in hLE Cells

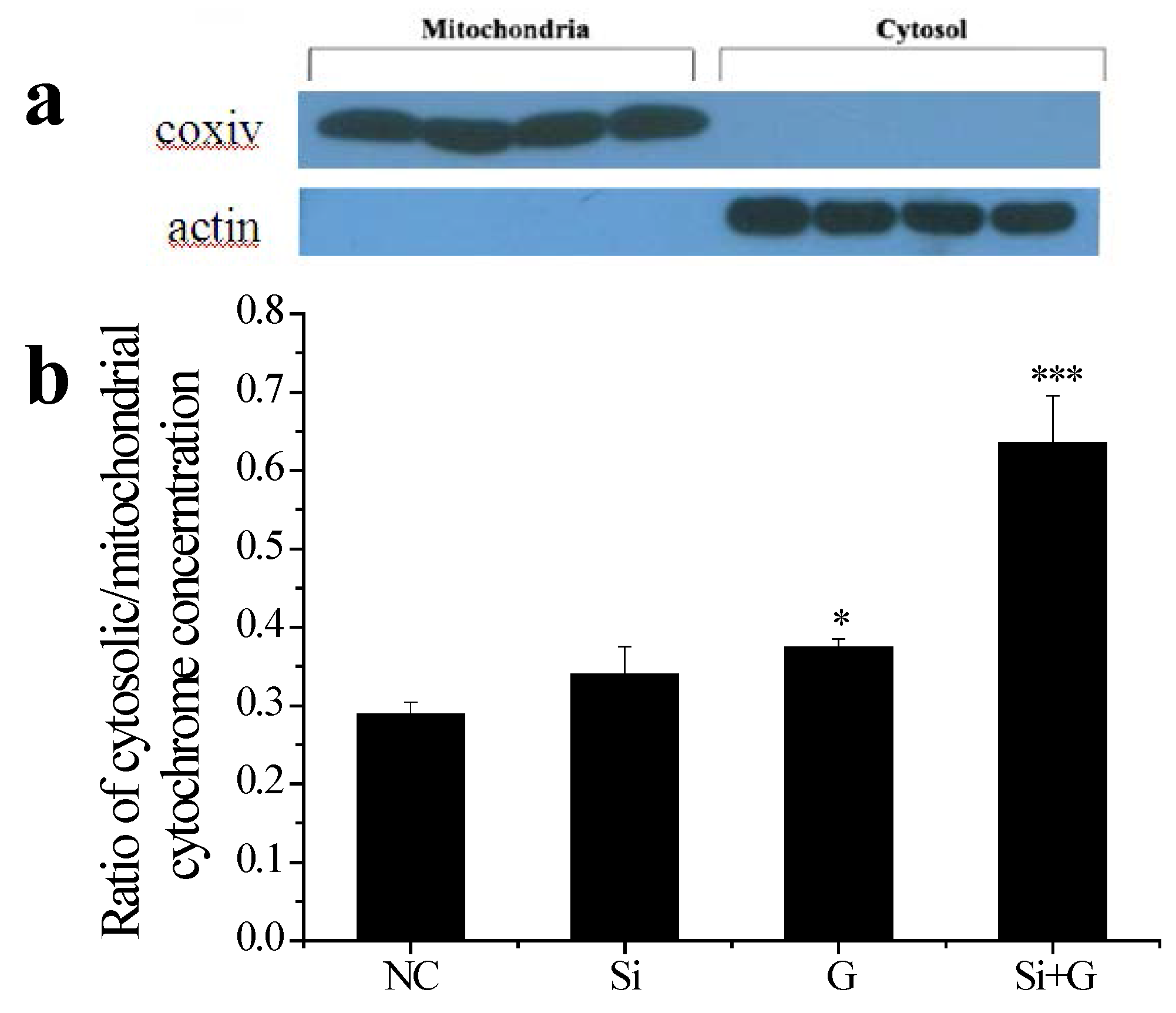
2.7. Effect of SelR Gene Knockdown on Cytochrome c Release in d-Galactose-Treated hLE Cells
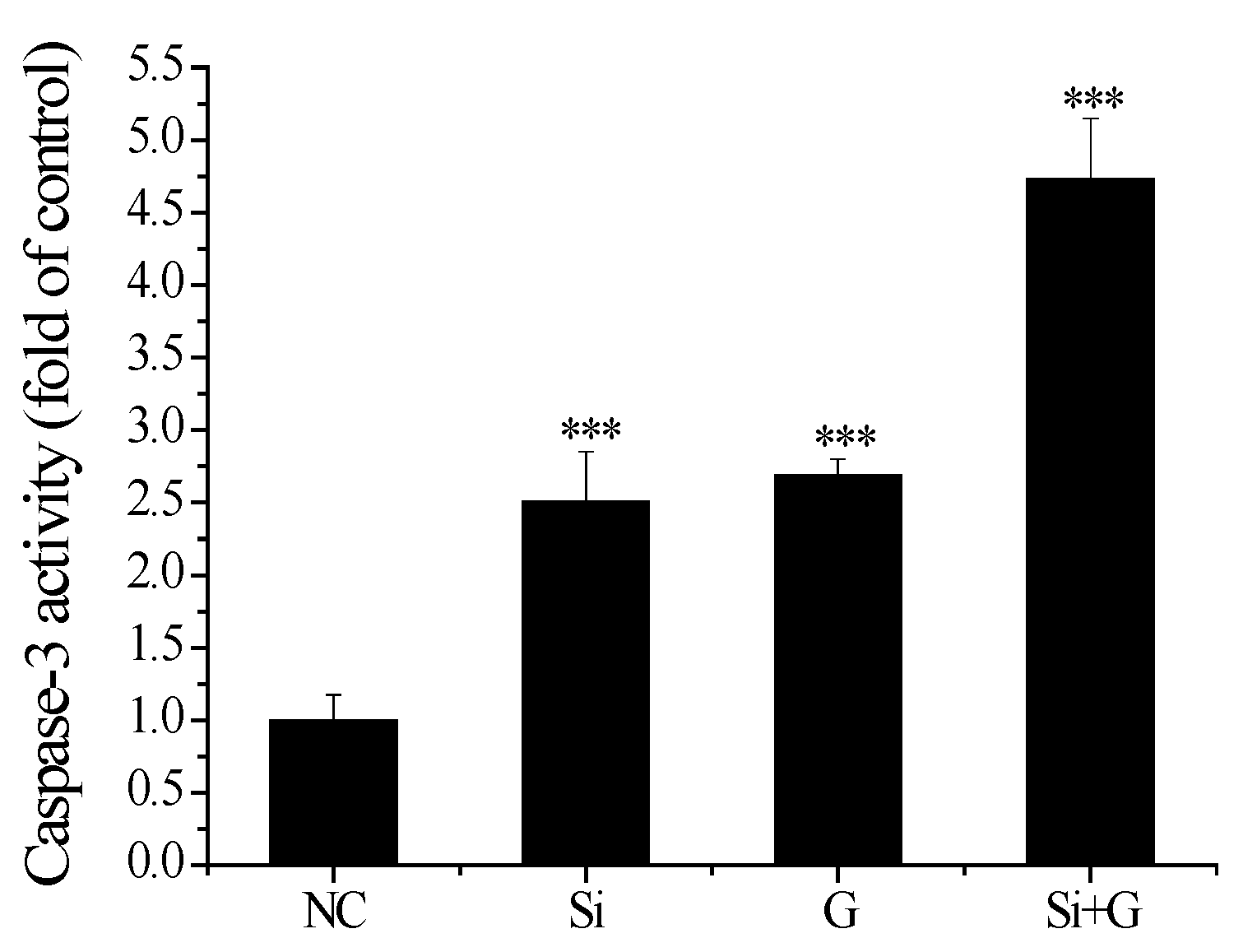
2.8. Effect of SelR Gene Knockdown on Caspase-3 Activity in d-Galactose-Treated hLE Cells
2.9. Effect of SelR Gene Knockdown on GRP78 Protein Expression in Glactose-Treated hLE Cells

3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture
4.3. SelR RNA Interference and Cell Treatment
4.4. Cell Viability Assay
4.5. Isolation of Total RNA and Real-Time PCR
4.6. Western Blot Analysis
4.7. Measurement of GPx Activity
4.8. Measurement of ROS, MDA, PC and TSH
4.9. Detection of Apoptosis
4.10. Assay of Cell Caspase-3 Activity
4.11. Detection of Mitochondrial Membrane Potential (Δψm)
4.12. Release of Cytochrome c from Mitochondria
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| cDNA | Complementary DNA |
| DCFH-DA | 2′,7′-Dichlorodihydro-fluorescein diacetate |
| DMEM | Dulbecco’s modified Eagle’s medium |
| ECL | Enhanced chemiluminescence |
| ER | Endoplasmic reticulum |
| FITC | Fluorescein isothiocyanate |
| GPx | Glutathione peroxidase |
| GRP78 | Glucose-regulated protein |
| hLE cells | Human lens epithelial cells |
| JC-1 | 5,5′,6,6′-Tetrachloro-1,1′,3,3′-tetraethyl-imidacarbocyanine iodide |
| MDA | Malondialdehyde |
| MetO | Methionine sulfoxide |
| Msr | Methionine sulfoxide reductase |
| MTT | 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenylte-trazolium bromide |
| NCS | Newborn calf serum |
| PBS | Phosphate-buffered saline |
| PC | Protein carbonyl |
| PI | Propidium iodide |
| ROS | Reactive oxygen species |
| SelR | Selenoprotein R |
| siRNA | Short interfering RNA |
| TBS-T | Tris-buffered saline with Tween |
| TSH | Total mercapto group |
| Δψm | Mitochondrial membrane potential |
References
- Kryukov, G.V.; Castellano, S.; Novoselov, S.V.; Lobanov, A.V.; Zehtab, O.; Guigó, R.; Gladyshev, V.N. Characterization of mammalian selenoproteomes. Science 2003, 300, 1439–1443. [Google Scholar] [CrossRef] [PubMed]
- Kryukov, G.V.; Gladyshev, V.N. The prokaryotic selenoproteome. EMBO Rep. 2004, 5, 538–543. [Google Scholar] [CrossRef] [PubMed]
- Castellano, O.; Bermúdez, Y.; Giffard, M.; Mabon, G.; Cubillan, N.; Sylla, M.; Nguyen-Phu, X.; Hinchliffe, A.; Soscún, H. Ab initio and DFT study of the geometric structures and static dipole (hyper) polarizabilities of aromatic anions. J. Phys. Chem. A 2005, 109, 10380–10387. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, H.; Yan, X.; Zhang, L. Comparison of short-term toxicity between Nano-Se and selenite in mice. Life Sci. 2005, 76, 1099–1109. [Google Scholar] [CrossRef] [PubMed]
- Flohé, L. Selenium, selenoproteins and vision. Dev. Ophthalmol. 2005, 38, 89–102. [Google Scholar] [PubMed]
- Zhu, X.; Lu, Y. Selenium supplementation can slow the development of naphthalene cataract. Curr. Eye Res. 2012, 37, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Garner, M.H.; Spector, A. Selective oxidation of cysteine and methionine in normal and senile cataractous lenses. Proc. Natl. Acad. Sci. USA 1980, 77, 1274–1277. [Google Scholar] [CrossRef] [PubMed]
- Kantorow, M.; Hawse, J.R.; Cowell, T.L.; Benhamed, S.; Pizarro, G.O.; Reddy, V.N.; Hejtmancik, J.F. Methionine sulfoxide reductase A is important for lens cell viability and resistance to oxidative stress. Proc. Natl. Acad. Sci. USA 2004, 101, 9654–9659. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.J. The oxidative environment and protein damage. Biochim. Biophys. Acta 2005, 1703, 93–109. [Google Scholar] [CrossRef] [PubMed]
- Reeves, M.A.; Hoffmann, P.R. The human selenoproteome: Recent insights into functions and regulation. Cell Mol. Life Sci. 2009, 66, 2457–2478. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.L.; Mosoni, L.; Berlett, B.S.; Stadtman, E.R. Methionine residues as endogenous antioxidants in proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 15036–15040. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Gladyshev, V.N. Methionine sulfoxide reductases: Selenoprotein forms and roles in antioxidant protein repair in mammals. Biochem. J. 2007, 407, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, M.A.; Pizarro, G.O.; Sagher, D.; DeAmicis, C.; Brot, N.; Hejtmancik, J.F.; Weissbach, H.; Kantorow, M. Methionine sulfoxide reductases B1, B2, and B3 are present in the human lens and confer oxidative stress resistance to lens cells. Investig. Ophthalmol. Vis. Sci. 2005, 46, 2107–2112. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jia, Y.; Zhou, J.; Huang, K.X. Effect of methionine sulfoxide reductase B1 silencing on high-glucose-induced apoptosis of human lens epithelial cells. Life Sci. 2013, 92, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Li, Y.; Du, S.Q.; Huang, K.X. Involvement of MsrB1 in the regulation of redox balance and inhibition of peroxynitrite-induced apoptosis in human lens epithelial cells. Exp. Eye Res. 2012, 100, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Zhou, J.; Du, S.Q.; Liu, H.M.; Huang, K.X. Effect of methionine sulfoxide reductase B1 (SelR) gene silencing on peroxynitrite-induced F-actin disruption in human lens epithelial cells. Biochem. Biophys. Res. Commun. 2014, 443, 876–881. [Google Scholar] [CrossRef] [PubMed]
- Acosta, P.B.; Gross, K.C. Hidden sources of galactose in the environment. Eur. J. Pediatr. 1995, 154, 87–92. [Google Scholar] [CrossRef]
- Wu, D.M.; Lu, J.; Zheng, Y.L.; Zhou, Z.; Shan, Q.; Ma, D.F. Purple sweet potato color repairs d-galactose-induced spatial learning and memory impairment by regulating the expression of synaptic proteins. Neurobiol. Learn. Mem. 2008, 90, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, R.; Lezhitsa, I. Effects of magnesium taurate on the onset and progression of d-galactose-induced experimental cataract: In vivo and in vitro evaluation. Exp. Eye Res. 2013, 110, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Datiles, M.B.; Kador, P.F.; Fukui, H.N.; Hu, T.S.; Kinoshita, J.H. Corneal re-epithelialization in d-galactosemic rats. Investig. Ophthalmol. Vis. Sci. 1983, 24, 563–569. [Google Scholar]
- Hu, T.S.; Datiles, M.; Kinoshita, J.H. Reversal of d-galactose cataract with Sorbinil in rats. Investig. Ophthalmol. Vis. Sci. 1983, 24, 640–644. [Google Scholar]
- Sakthivel, M.; Geraldine, P.; Thomas, P.A. Alterations in the lenticular protein profile in experimental selenite-induced cataractogenesis and prevention by ellagic acid. Graef Arch. Clin. Exp. 2011, 249, 1201–1210. [Google Scholar] [CrossRef] [PubMed]
- Li, W.C.; Kuszak, J.R.; Dunn, K.; Wang, R.R.; Ma, W.; Wang, G.M.; Spector, A.; Leib, M.; Cotliar, A.M.; Weiss, M. Lens epithelial cell apoptosis appears to be a common cellular basis for noncongenital cataract development in humans and animals. J. Cell Biol. 1995, 130, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Devi, V.G.; Rooban, B.N.; Sasikala, V.; Sahasranamam, V.; Abraham, A. Isorhamnetin-3-glucoside alleviates oxidative stress and opacification in selenite cataract in vitro. Toxicol. In Vitro 2010, 24, 1662–1669. [Google Scholar] [CrossRef] [PubMed]
- Ohta, Y.; Torii, H.; Yamasaki, T.; Niwa, T.; Majima, Y.; Ishiguro, I. Preventive action of vitamin E-containing liposomes on cataractogenesis in young adult rats fed a 25% d-galactose diet. J. Ocul. Pharmacol. Ther. 1997, 13, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Ohta, Y.; Yamasaki, T.; Niwa, T.; Goto, H.; Majima, Y.; Ishiguro, I. Cataract development in 12-Month-Old Rats Fed a 25% d-Galactose Diet and Its Relation to Osmotic Stress and Oxidative Damage. Ophthalmic Res. 1999, 31, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.P.; Pandey, D.J.; Sharma, A.L.; Strivastava, R.K.; Mishra, S.S. Prevention of experimental cataract by α-tocopherol. Indian J. Exp. Biol. 1984, 22, 620–622. [Google Scholar] [PubMed]
- Hirai, T.; Majima, Y.; Ohta, Y.; Ishiguro, I. Relationship between lenticular vitamin E level and cataract formation in d-galactosemic rats. Folia Ophthalmol. Jpn. 1986, 37, 1266–1270. [Google Scholar]
- Srivastava, S.K.; Ansari, N.H. Prevention of sugar-induced cataractogenesis in rats by butylated hydroxytoluene. Diabetes 1988, 37, 1505–1508. [Google Scholar] [CrossRef] [PubMed]
- Takamura, Y.; Sugimoto, Y.; Kubo, E.; Takahashi, Y.; Akagi, Y. Immunohistochemical study of apoptosis of lens epithelial cells inhumanand diabetic rat cataracts. Jpn. J. Ophthalmol. 2001, 45, 559–563. [Google Scholar] [CrossRef]
- Mulhern, M.L.; Madson, C.J.; Danford, A.; Ikesugi, K.; Kador, P.F.; Shinohara, T. The unfolded protein response in lens epithelial cells from d-galactosemic rat lenses. Investig. Ophthalmol. Vis. Sci. 2006, 47, 3951–3959. [Google Scholar] [CrossRef] [PubMed]
- Mia, A.; Zhang, X.; Jiang, Y.; Chen, Y.; Fang, Y.; Ye, H.; Chu, R.; Lu, Y. Proteomic analysis of SRA01/04 transfected with wild-type and mutant HSF4b identified from a Chinese congenital cataract family. Mol. Vis. 2012, 18, 694–704. [Google Scholar]
- Ceriello, A.; Bortolotti, N.; Crescentini, A.; Motz, E.; Lizzio, S.; Russo, A.; Ezsol, Z.; Tonutti, L.; Taboga, C. Antioxidant defences are reduced during the oral glucose tolerance test in normal and non-insulin-dependent diabetic subjects. Eur. J. Clin. Investig. 1998, 28, 329–333. [Google Scholar] [CrossRef]
- Bloemendal, H.; Benedetti, E.L.; Ramaekers, F.; Dunia, I. The lens cytoskeleton. Intermediate-sized filaments, their biosynthesis and association with plasma membranes. Mol. Biol. Rep. 1981, 7, 167–168. [Google Scholar] [CrossRef] [PubMed]
- Spector, A. On presentation of the proctor medal of the association for research in vision and ophthalmology to Dr. Hans Bloemendal. Investig. Ophthalmol. Vis. Sci. 1991, 32, 443–444. [Google Scholar]
- Wang, X.; Wang, Z.; Liu, J.Z.; Hu, J.X.; Chen, H.L.; Li, W.L.; Hai, C.X. Double antioxidant activities of rosiglitazone against high glucose-induced oxidative stress in hepatocyte. Toxicol. In Vitro 2011, 25, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Ou, Y.; Liao, G.Y.; Wu, W.T. Potential use of hirudin in diabetic cataract: A study of d-galactose mediated human lens epithelial cells injury. Chem. Biol. Interact. 2008, 173, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Hunt, J.V.; Wolff, S.P. Oxidative glycation and free radical production: A causal mechanism of diabetic complications. Free Radic. Res. Commun. 1991, 12, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.Y.W.; Chung, S.S.M. Contributions of polyol pathway to oxidative stress in diabetic cataract. FEBS Lett. 1999, 13, 23–30. [Google Scholar]
- Kubo, E.; Miyoshi, N.; Fukuda, M.; Akagi, Y. Cataract formation through polyol pathway is associated with free radical production. Exp. Eye Res. 1999, 68, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Yokouchi, M.; Hiramatsu, N.; Hayakawa, K.; Okamura, M.; Du, S.; Kasai, A.; Takano, Y.; Shitamura, A.; Shimada, T.; Yao, J.; et al. Involvement of selective reactive oxygen species upstream of proapoptotic branches of unfolded protein response. J. Biol. Chem. 2008, 283, 4252–4260. [Google Scholar] [CrossRef] [PubMed]
- Tagawa, Y.; Hiramatsu, N.; Kasai, A.; Hayakawa, K.; Okamura, M.; Yao, J.; Kitamura, M. Induction of apoptosis by cigarette smoke via ROS-dependent endoplasmic reticulum stress and CCAAT/enhancer-binding protein-homologous protein (CHOP). Free Radic. Biol. Med. 2008, 45, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Giraldo, G.S.; Prentice, H.; Wu, J.Y. Taurine protection of PC12 cells against endoplasmic reticulum stress induced by oxidative stress. J. Biomed. Sci. 2012, 17. [Google Scholar] [CrossRef] [PubMed]
- Dejeans, N.; Tajeddine, N.; Beck, R.; Verrax, J.; Taper, H.; Gailly, P.; Calderon, P.B. Endoplasmic reticulum calcium release potentiates the ER stress and cell death caused by an oxidative stress in MCF-7 cells. Biochem. Pharmacol. 2010, 79, 1221–1230. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, K.; Wierda, W.G.; Keating, M.J.; Gandhi, V. Gossypol, a BH3 mimetic, induces apoptosis in chronic lymphocytic leukemia cells. Blood 2008, 112, 1971–1980. [Google Scholar] [CrossRef] [PubMed]
- Xu, A.; Prophete, C.; Chen, L.C.; Emala, C.W.; Cohen, M.D. Interactive effect of cigarette smoke extract and world trade center dust particles on airway cell cytotoxicity. J. Toxicol. Environ. Health 2011, 74, 887–902. [Google Scholar] [CrossRef] [PubMed]
- Brennan, L.A.; Lee, W.; Cowell, T.; Giblin, F.; Kantorow, M. Deletion of mouse MsrA results in HBO-induced cataract: MsrA repairs mitochondrial cytochrome c. Mol. Vis. 2009, 15, 985–999. [Google Scholar] [PubMed]
- Zeng, J.H.; Du, S.Q.; Zhou, J.; Huang, K.X. Role of SelS in lipopolysaccharide-induced inflammatory response in hepatoma HepG2 cells. Arch. Biochem. Biophys. 2008, 478, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ohguro, N.; Fukuda, M.; Sasabe, T.; Tano, Y. Concentration dependent effects of hydrogen peroxide on lens epithelial cells. Br. J. Ophthalmol. 1999, 83, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Luca, A.D.; Sacchetta, P.; Nieddu, M.; Ilio, C.D.; Favaloro, B. Important roles of multiple Sp1 binding sites and epigenetic modifications in the regulation of the methionine sulfoxide reductase B1 (MsrB1) promoter. BMC Mol. Biol. 2007, 8, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Du, S.Q.; Zhou, J.; Jia, Y.; Huang, K.X. SelK is a novel ER stress-regulated protein and protects HepG2 cells from ER stress agent-induced apoptosis. Arch Biochem. Biophys. 2010, 502, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, W.; Li, P.; Huang, K. Effect of streptozocin-induced diabetes mellitus on expression of methionine sulfoxide reductases and accumulation of their substrates in mouse lenses. Exp. Eye Res. 2011, 92, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Cao, G.; Cutler, R.G. Protein oxidation and aging. I. difficulties in measuring reactive protein carbonyls in tissues using 2,4-dinitrophenylhydrazine. Arch Biochem. Biophys. 1995, 320, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Bejma, J.; Ji, L.L. Aging and acute exercise enhance free radical generation in rat skeletal muscle. J. Appl. Physiol. 1999, 87, 465–470. [Google Scholar] [PubMed]
- Agrawal, S.S.; Saraswati, S.; Mathur, R.; Pandey, M. Cytotoxic and antitumor effects of brucine on Ehrlich ascites tumor and human cancer cell line. Life Sci. 2011, 89, 147–158. [Google Scholar] [CrossRef] [PubMed]
- McElnea, E.M.; Quill, B.; Docherty, N.G.; Irnaten, M.; Siah, W.F.; Clark, A.F.; O’Brien, C.J.; Wallace, D.M. Oxidative stress, mitochondrial dysfunction and calcium overload in human lamina cribrosa cells from glaucoma donors. Mol. Vis. 2011, 17, 1182–1191. [Google Scholar] [PubMed]
- Kowluru, R.A.; Abbas, S.N. Diabetes-induced mitochondrial dysfunction in the retina. Investig. Ophthalmol. Vis. Sci. 2003, 44, 5327–5334. [Google Scholar] [CrossRef]
- Yamada, T.; Goto, M.; Punj, V.; Zaborina, O.; Chen, M.L.; Kimbara, K.; Majumdar, D.; Cunningham, E.; Das Gupta, T.K.; Chakrabarty, A.M. Bacterial redox protein azurin, tumor suppressor protein p53, and regression of cancer. Proc. Natl. Acad. Sci. USA 2002, 99, 14098–14103. [Google Scholar] [CrossRef] [PubMed]
- Ventéo, L.; Bourlet, T.; Renois, F.; Douche-Aourik, F.; Mosnier, J.F.; Maison, G.L.; Pluot, M.; Pozzetto, B.; Andreoletti, L. Enterovirus-related activation of the cardiomyocyte mitochondrial apoptotic pathway in patients with acute myocarditis. Eur. Heart J. 2010, 31, 728–36. [Google Scholar] [CrossRef] [PubMed]
- Sommer, S.P.; Sommer, S.; Sinha, B.; Walter, D.; Aleksic, I.; Gohrbandt, B.; Otto, C.; Leyh, R.G. Glutathione preconditioning ameliorates mitochondria dysfunction during warm pulmonary ischemia-reperfusion injury. Eur. J. Cardiothorac. Surg. 2011, 41, 140–148. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, J.; Liu, H.; Zhou, J.; Huang, K. Selenoprotein R Protects Human Lens Epithelial Cells against D-Galactose-Induced Apoptosis by Regulating Oxidative Stress and Endoplasmic Reticulum Stress. Int. J. Mol. Sci. 2016, 17, 231. https://doi.org/10.3390/ijms17020231
Dai J, Liu H, Zhou J, Huang K. Selenoprotein R Protects Human Lens Epithelial Cells against D-Galactose-Induced Apoptosis by Regulating Oxidative Stress and Endoplasmic Reticulum Stress. International Journal of Molecular Sciences. 2016; 17(2):231. https://doi.org/10.3390/ijms17020231
Chicago/Turabian StyleDai, Jie, Hongmei Liu, Jun Zhou, and Kaixun Huang. 2016. "Selenoprotein R Protects Human Lens Epithelial Cells against D-Galactose-Induced Apoptosis by Regulating Oxidative Stress and Endoplasmic Reticulum Stress" International Journal of Molecular Sciences 17, no. 2: 231. https://doi.org/10.3390/ijms17020231
APA StyleDai, J., Liu, H., Zhou, J., & Huang, K. (2016). Selenoprotein R Protects Human Lens Epithelial Cells against D-Galactose-Induced Apoptosis by Regulating Oxidative Stress and Endoplasmic Reticulum Stress. International Journal of Molecular Sciences, 17(2), 231. https://doi.org/10.3390/ijms17020231