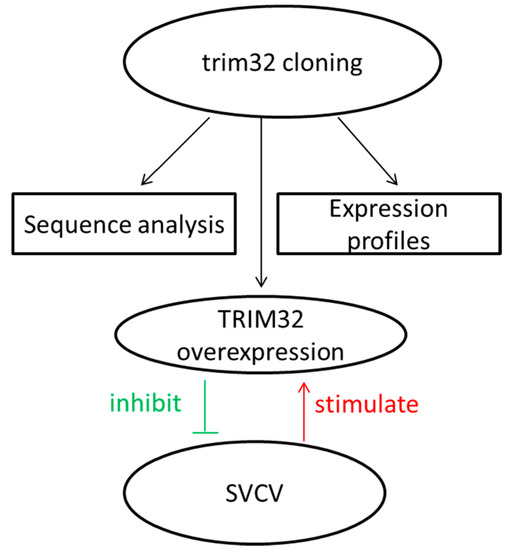
Molecular Characterization, Tissue Distribution and Expression, and Potential Antiviral Effects of TRIM32 in the Common Carp (Cyprinus carpio)
Abstract
:1. Introduction
2. Result
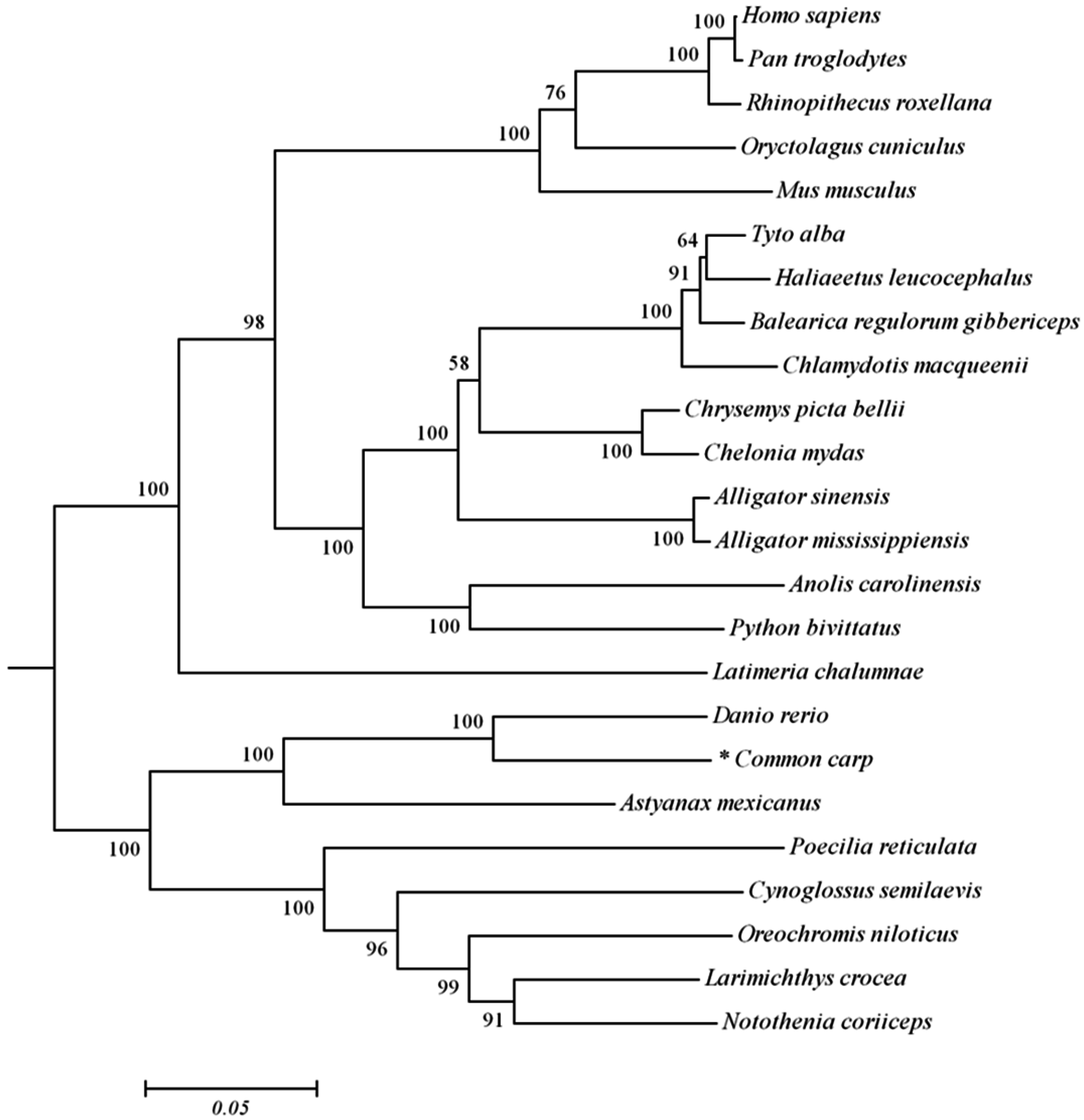
2.1. Analysis of Common Carp trim32 Coding DNA Sequence (CDS) Region
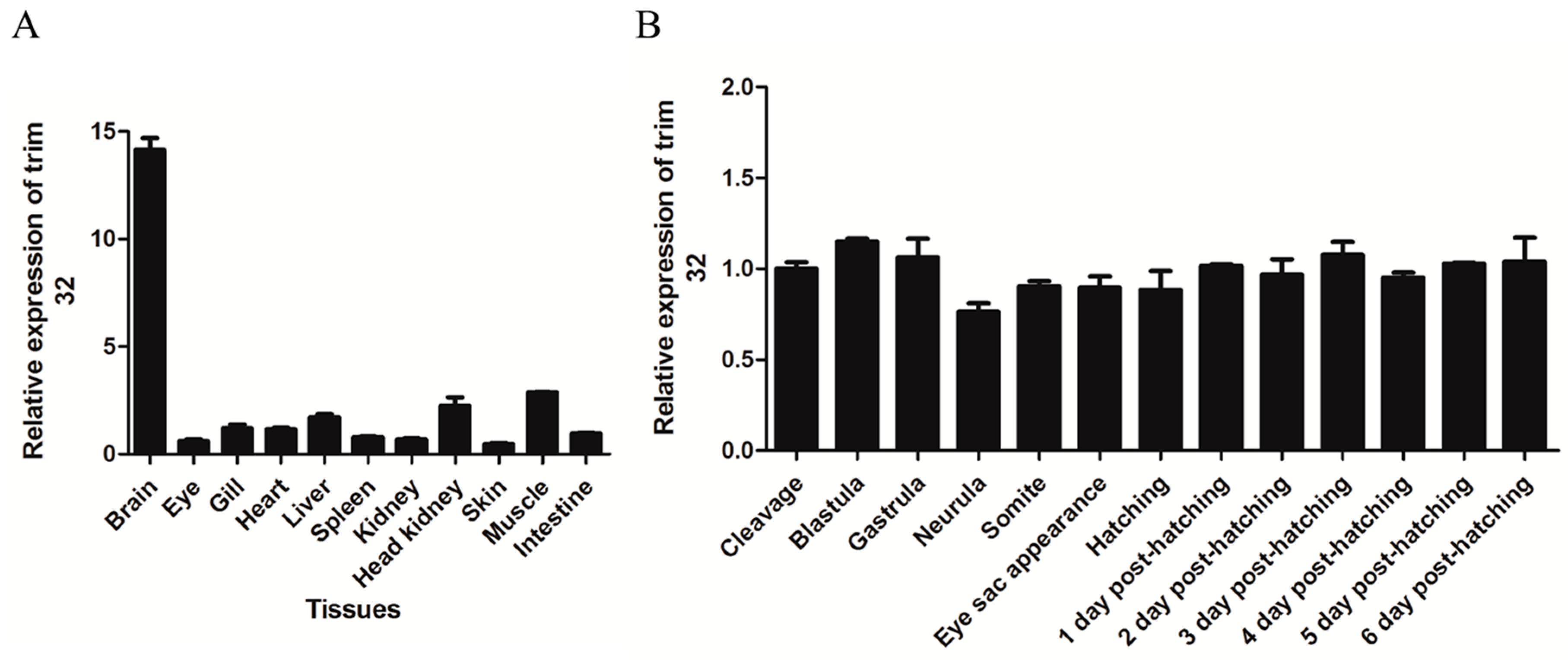
2.2. TRIM32 Tissue-Specific Distribution and Expression during Embryonic Development
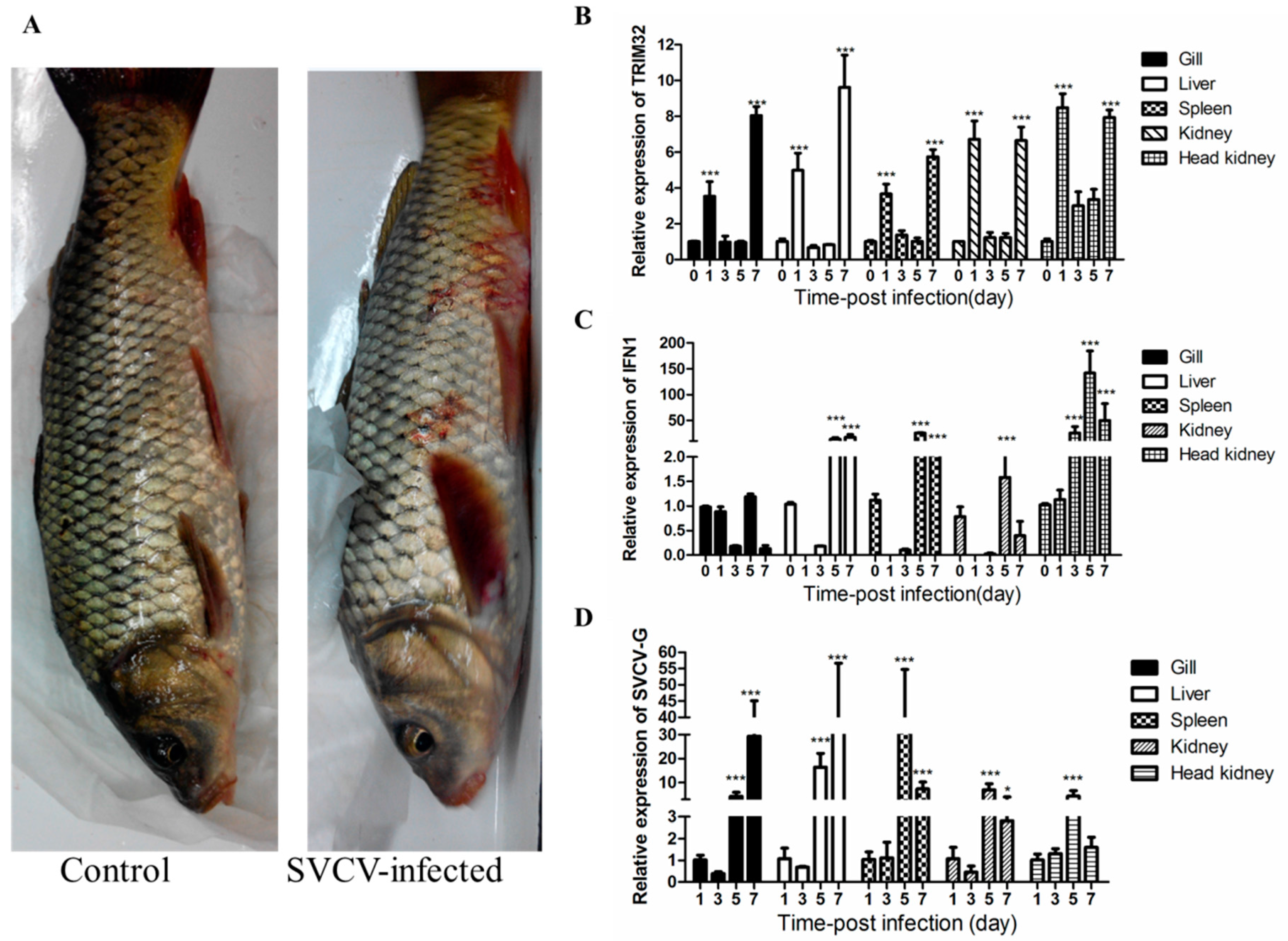
2.3. Changes of TRIM32 Expression after SVCV Infection
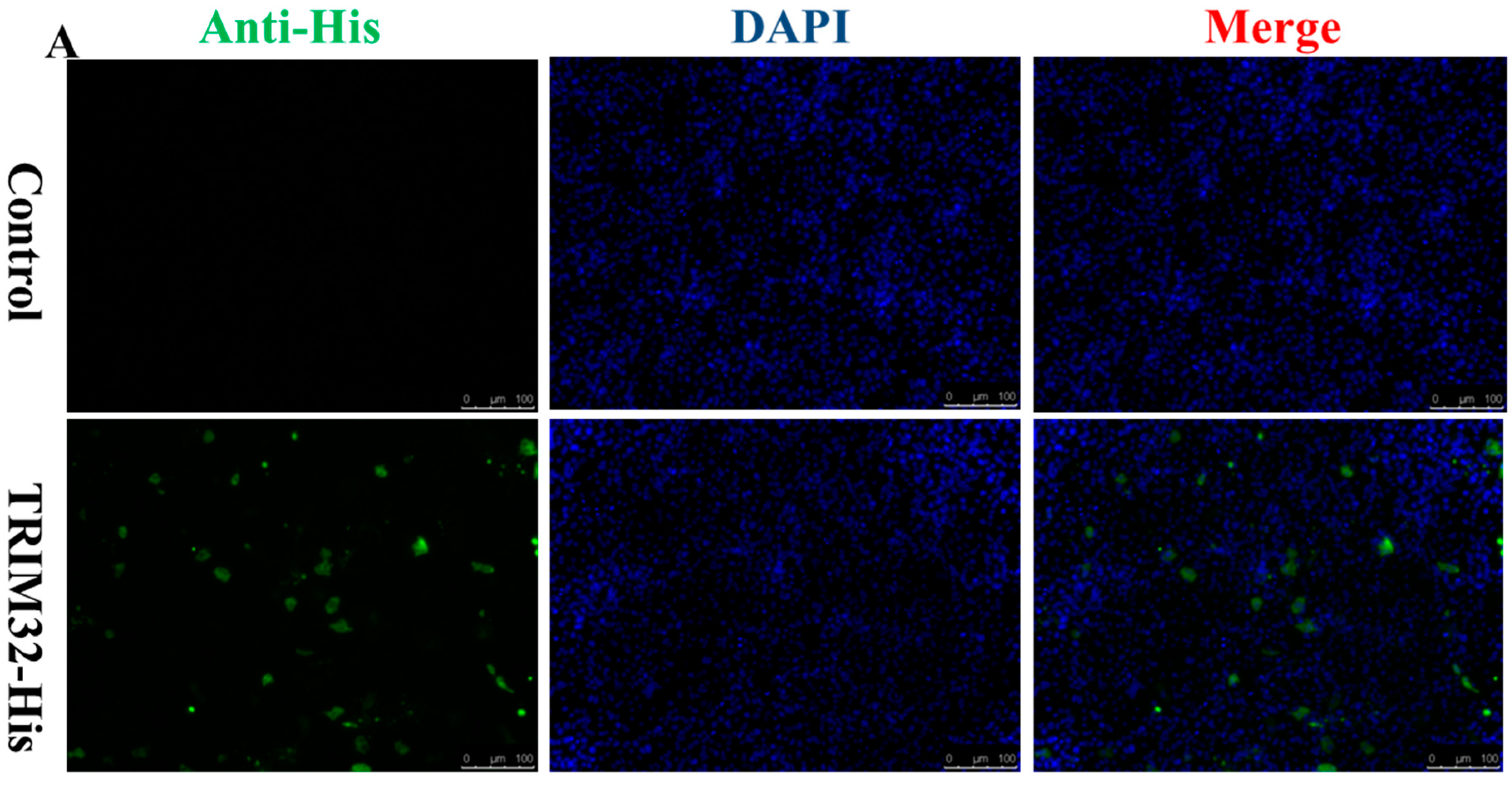
2.4. Expression of TRIM32 in Transfected EPC Cells
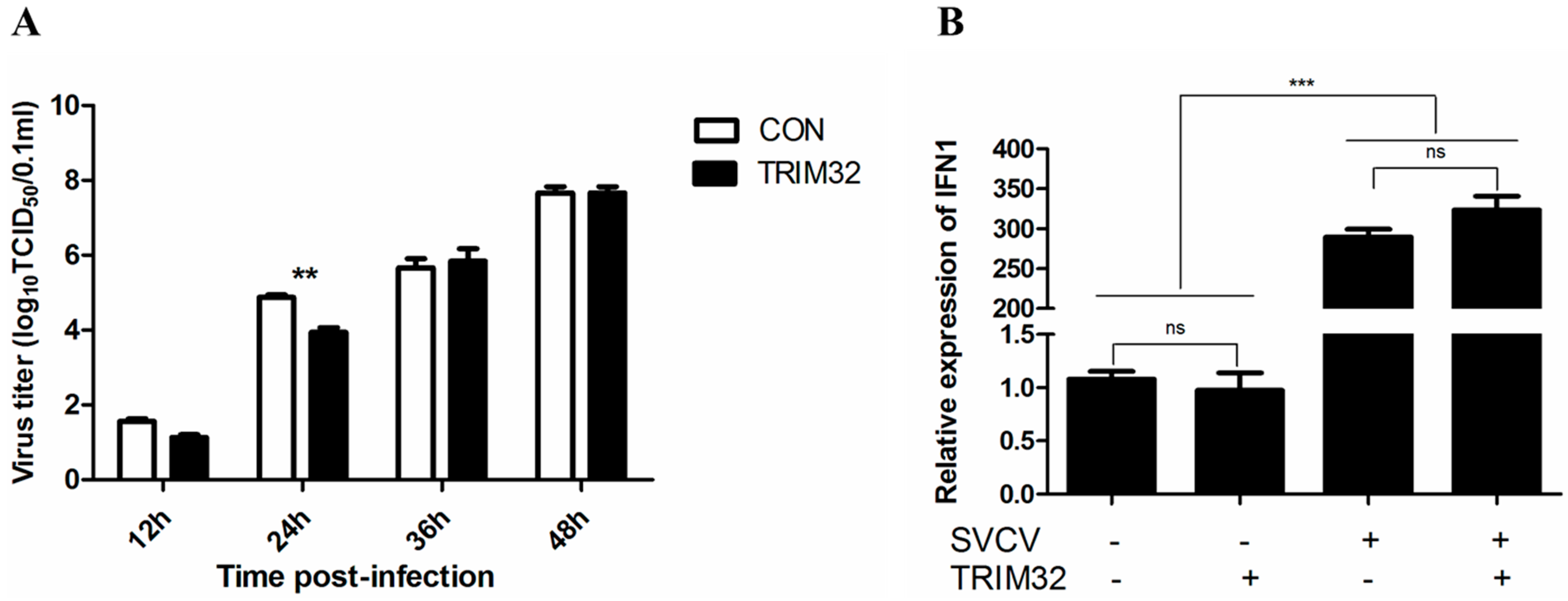
2.5. Overexpression of TRIM32 Decreased the Virus Titer
3. Discussion
4. Materials and Methods
4.1. Cell and Virus
4.2. RNA Extraction and Synthesis of cDNA
4.3. Cloning the Full-Length Coding DNA Sequence (CDS) of Common Carp trim32 Gene
4.4. Sequence Analysis
4.5. Experiment Common Carp and Common Carp Eggs
4.6. Quantitative PCR Analysis of Tissue-Specific Distribution and Expression during Embryonic Development
4.7. Viral Infection
4.8. Plasmid Construction and Transfection
4.9. Immune Fluorescence and Western Blotting Assay
4.10. Virus Titration
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Uchil, P.D.; Quinlan, B.D.; Chan, W.T.; Luna, J.M.; Mothes, W. TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog. 2008, 4, e16. [Google Scholar] [CrossRef] [PubMed]
- Boudinot, P.; van der Aa, L.M.; Jouneau, L.; Du Pasquier, L.; Pontarotti, P.; Briolat, V.; Benmansour, A.; Levraud, J.P. Origin and evolution of TRIM proteins: New insights from the complete TRIM repertoire of zebrafish and pufferfish. PLoS ONE 2011, 6, e22022. [Google Scholar] [CrossRef] [PubMed]
- Nisole, S.; Stoye, J.P.; Saïb, A. TRIM family proteins: Retroviral restriction and antiviral defence. Nat. Rev. Microbiol. 2005, 3, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Matthew, S.; Christopher, M.O.; Michel, J.P.; Michael, K.; Patrick, A.; Joseph, S. The cytoplasmic body component TRIM5a restricts HIV-1 infection in Old World monkeys. Lett. Nat. 2004, 427, 848–853. [Google Scholar]
- Narayan, K.; Waggoner, L.; Pham, S.T.; Hendricks, G.L.; Waggoner, S.N.; Conlon, J.; Wang, J.P.; Fitzgerald, K.A.; Kang, J. TRIM13 is a negative regulator of MDA5-mediated type I interferon production. J. Virol. 2014, 88, 10748–10757. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.M.; Yang, Q.; Zhang, J.; Liu, S.M.; Zhang, Y.; Lin, H.; Huang, Z.F.; Wang, Y.Y.; Zhang, X.D.; Zhong, B.; et al. TRIM38 inhibits TNFα- and IL-1β-triggered NF-κB activation by mediating lysosome-dependent degradation of TAB2/3. Proc. Natl. Acad. Sci. USA 2014, 111, 1509–1514. [Google Scholar] [CrossRef] [PubMed]
- Tabah, A.A.; Tardif, K.; Mansky, L.M. Anti-HIV-1 activity of Trim 37. J. Gen. Virol. 2014, 95, 960–967. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hu, M.M.; Wang, Y.Y.; Shu, H.B. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J. Biol. Chem. 2012, 287, 28646–28655. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, Y.; Hu, W.; Feng, Z. TRIM32 is a novel negative regulator of p53. Mol. Cell. Oncol. 2014, 2, e970951. [Google Scholar] [CrossRef] [PubMed]
- Ransom, D.G.; Bahary, N.; Niss, K.; Traver, D.; Burns, C.; Trede, N.S.; Paffett-Lugassy, N.; Saganic, W.J.; Lim, C.A.; Hersey, C.; et al. The zebrafish moonshine gene encodes transcriptional intermediary factor 1γ, an essential regulator of hematopoiesis. PLoS Biol. 2004, 2, E237. [Google Scholar] [CrossRef] [PubMed]
- Van der Aa, L.M.; Levraud, J.P.; Yahmi, M.; Lauret, E.; Briolat, V.; Herbomel, P.; Benmansour, A.; Boudinot, P. A large new subset of TRIM genes highly diversified by duplication and positive selection in teleost fish. BMC Biol. 2009, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Van der Aa, L.M.; Jouneau, L.; Laplantine, E.; Bouchez, O.; van Kemenade, L.; Boudinot, P. FinTRIMs, fish virus-inducible proteins with E3 ubiquitin ligase activity. Dev. Comp. Immunol. 2012, 36, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Huang, Y.; Yu, Y.; Yang, Y.; Xu, M.; Chen, X.; Ni, S.; Qin, Q.; Huang, X. Fish TRIM39 regulates cell cycle progression and exerts its antiviral function against iridovirus and nodavirus. Fish Shellfish Immunol. 2016, 50, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Teng, Y.; Xie, X.; Li, H.; Lv, J.; Gao, L.; Tian, F.; Jiang, Y.; Chu, Z.; Xie, C.; et al. Development and evaluation of a one-step loop-mediated isothermal amplification for detection of spring viraemia of carp virus. J. Appl. Microbiol. 2008, 105, 1220–1226. [Google Scholar] [CrossRef] [PubMed]
- Ahne, W.; Bjorklund, H.V.; Essbauer, S.; Fijan, N.; Kurath, G.; Winton, J.R. Spring viremia of carp (SVC). Anat. Notes 2002, 52, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhu, B.; Wu, S.; Lin, L.; Liu, G.; Zhou, Y.; Wang, W.; Asim, M.; Yuan, J.; Li, L.; et al. Spring viraemia of carp virus induces autophagy for necessary viral replication. Cell. Microbiol. 2015, 17, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, H.; Chen, Y.; Liu, C.; Meng, K.; Yang, P.; Wang, Y.; Wang, G.; Yao, B. Characterization and biological function analysis of the trim3a gene from zebrafish (Danio rerio). Fish Shellfish Immunol. 2012, 32, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Boulay, J.L.; Stiefel, U.; Taylor, E.; Dolder, B.; Merlo, A.; Hirth, F. Loss of heterozygosity of TRIM3 in malignant gliomas. BMC Cancer 2009, 9, 71. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Wang, L.; Ding, H.; Schwamborn, J.C.; Li, S.; Dorf, M.E. TRIM32 Senses and Restricts Influenza A Virus by Ubiquitination of PB1 Polymerase. PLoS Pathog. 2015, 11, e1004960. [Google Scholar] [CrossRef] [PubMed]
- Perumal, T.M.; Gonzalez-Cano, L.; Hillje, A.-L.; Taher, L.; Makalowski, W.; Suzuki, Y.; Fuellen, G.; del Sol, A.; Schwamborn, J.C. TRIM32 modulates pluripotency entry and exit by directly regulating Oct4 stability. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef]
- Reed, L.; Muench, H. Measurement of viruses by end-point dilution assay. Am. J. Hyg. 1938, 27, 493–497. [Google Scholar]


| Primer | Sequence (5’ to 3’) | Application |
|---|---|---|
| Core segment Forward | TCCCAGCTGGCTGACaayytnacngt | PCR |
| Core segment Reverse | AGGCTTGATCAGCTGGTTCTtrtgrcangcna | PCR |
| TRIM32ORF Forward | AAAGGATCCATGGCCACACCAACATCTTTAG | Plasmid construction |
| TRIM32ORF Reverse | AAACTCGAGACATGTAGAGGAACGTCTCCTT | Plasmid construction |
| TRIM32 Forward | GGGTGGCAAGGGAAGT | qPCR |
| TRIM32 Reverse | GAGGAAGGGAGCAACAAT | qPCR |
| TBP Forward | TTACCCACCAGCAGTTTAG | qPCR |
| TBP Reverse | ACCTTGGCACCTGTGAGTA | qPCR |
| SVCV-G Forward | CGACCTGGATTAGACTTG | qPCR |
| SVCV-G Reverse | AATGTTCCGTTTCTCACT | qPCR |
| IFN1-Forward | GGTGAAGTTTCTTGCCCTGACCTTAG | qPCR |
| IFN1-Reverse | CCTTATGTGATGGCTGGTATCGGG | qPCR |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Li, Z.; Lu, Y.; Hu, G.; Lin, L.; Zeng, L.; Zhou, Y.; Liu, X. Molecular Characterization, Tissue Distribution and Expression, and Potential Antiviral Effects of TRIM32 in the Common Carp (Cyprinus carpio). Int. J. Mol. Sci. 2016, 17, 1693. https://doi.org/10.3390/ijms17101693
Wang Y, Li Z, Lu Y, Hu G, Lin L, Zeng L, Zhou Y, Liu X. Molecular Characterization, Tissue Distribution and Expression, and Potential Antiviral Effects of TRIM32 in the Common Carp (Cyprinus carpio). International Journal of Molecular Sciences. 2016; 17(10):1693. https://doi.org/10.3390/ijms17101693
Chicago/Turabian StyleWang, Yeda, Zeming Li, Yuanan Lu, Guangfu Hu, Li Lin, Lingbing Zeng, Yong Zhou, and Xueqin Liu. 2016. "Molecular Characterization, Tissue Distribution and Expression, and Potential Antiviral Effects of TRIM32 in the Common Carp (Cyprinus carpio)" International Journal of Molecular Sciences 17, no. 10: 1693. https://doi.org/10.3390/ijms17101693
APA StyleWang, Y., Li, Z., Lu, Y., Hu, G., Lin, L., Zeng, L., Zhou, Y., & Liu, X. (2016). Molecular Characterization, Tissue Distribution and Expression, and Potential Antiviral Effects of TRIM32 in the Common Carp (Cyprinus carpio). International Journal of Molecular Sciences, 17(10), 1693. https://doi.org/10.3390/ijms17101693