Direct LAMP Assay without Prior DNA Purification for Sex Determination of Papaya
Abstract
:1. Introduction
2. Results and Discussion
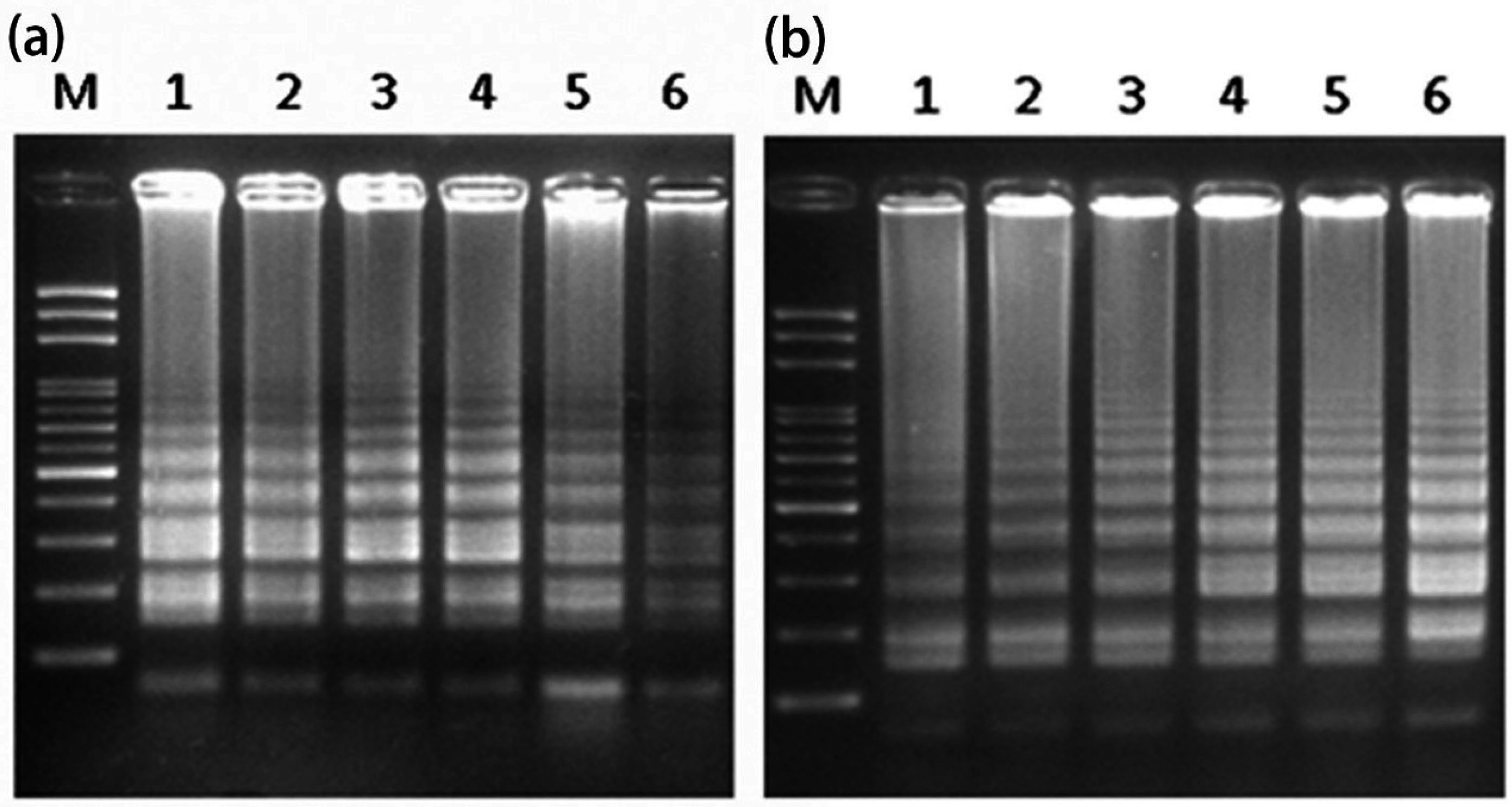
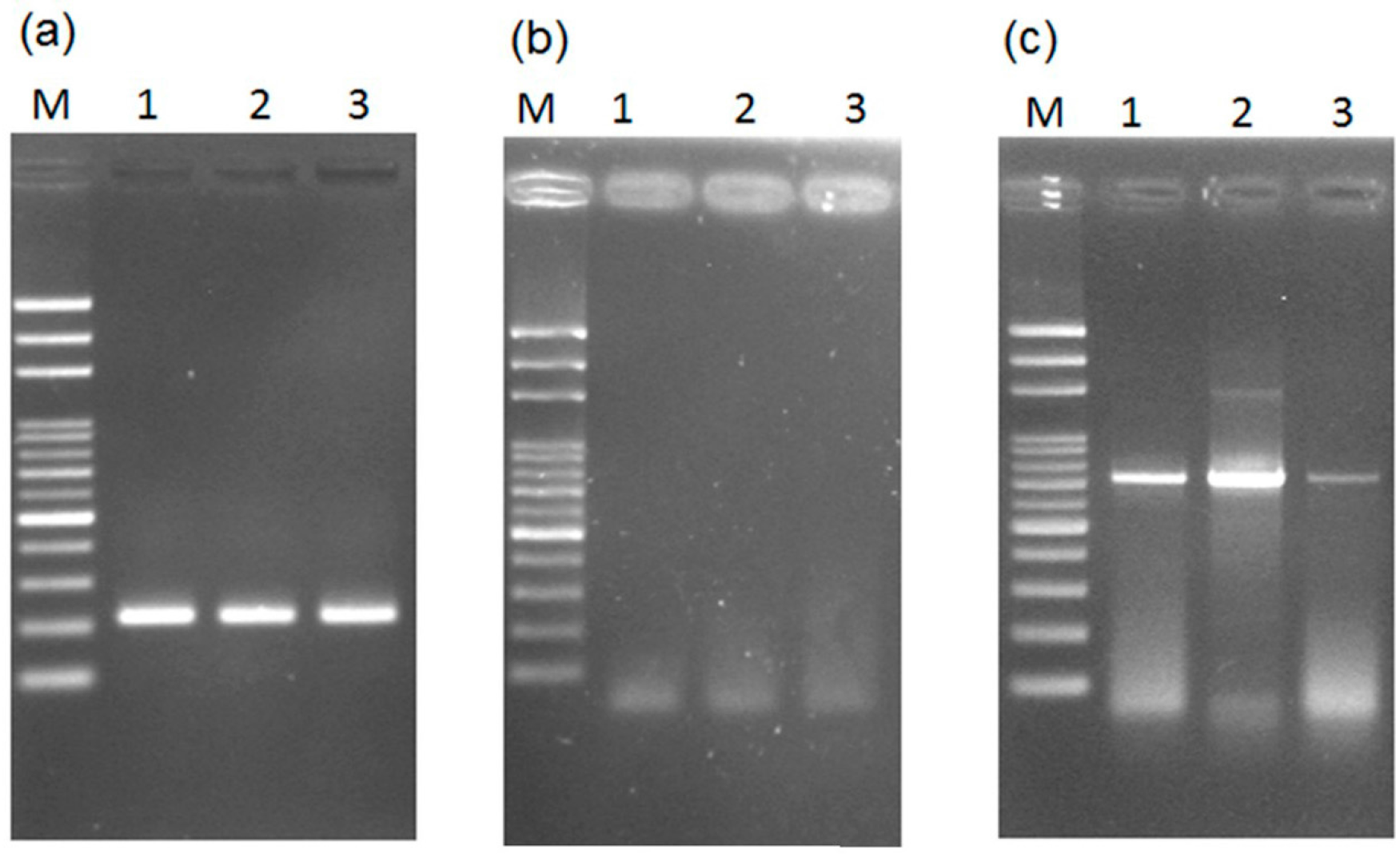
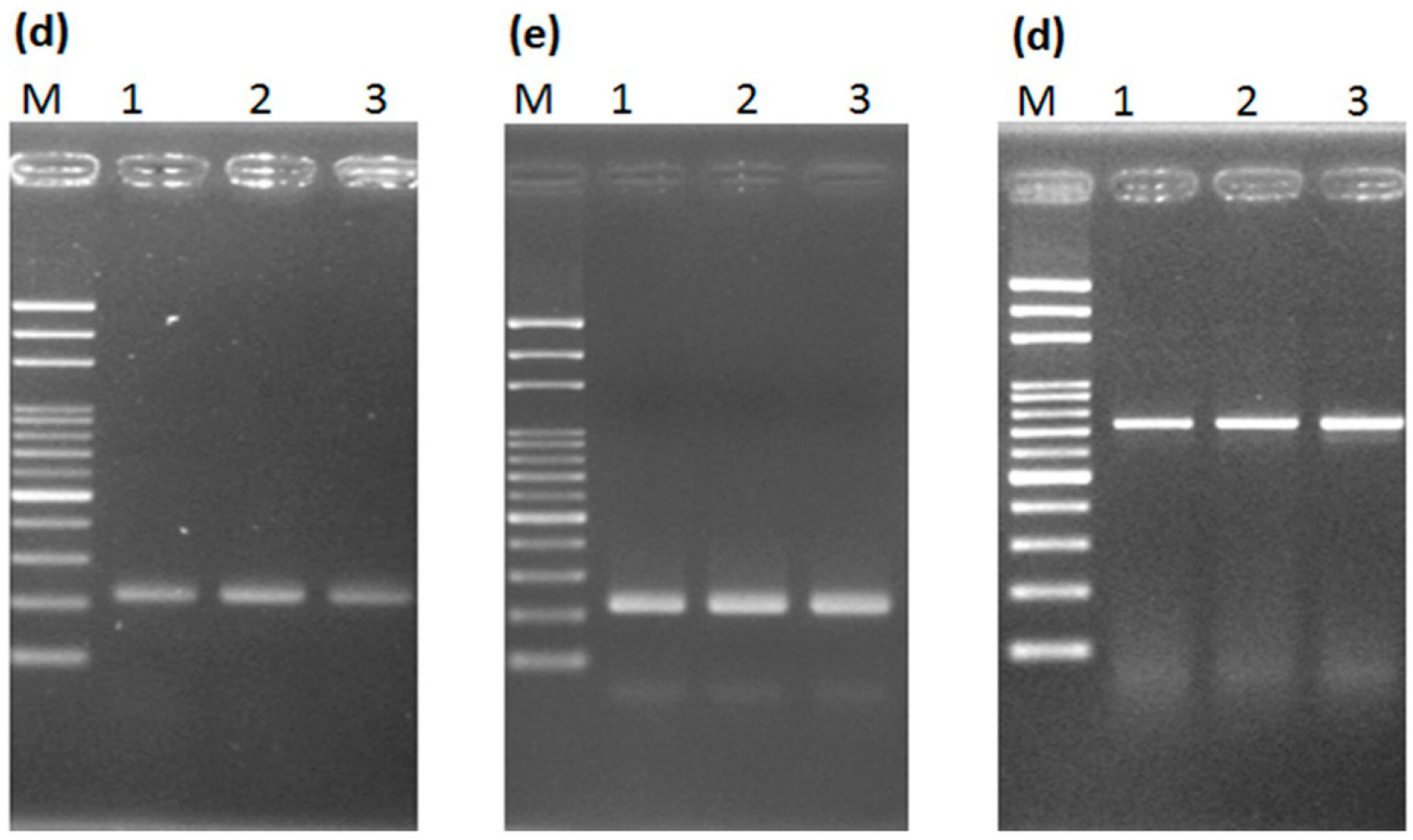
2.1. Loop-Mediated Isothermal Amplification (LAMP) Analysis for the Male-Specific Region of the Y Chromosome Using Intact Plant Material as the DNA Source
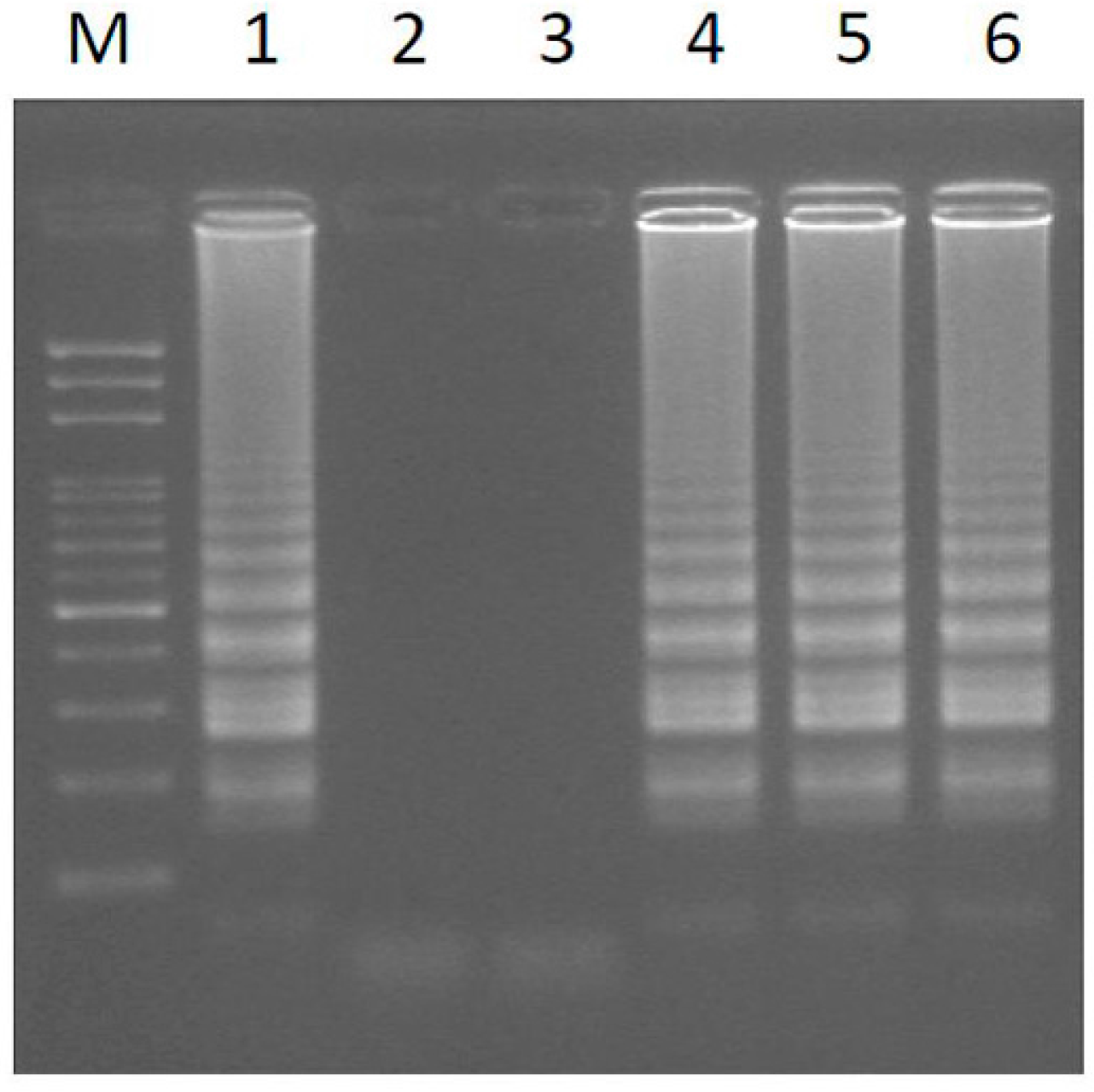
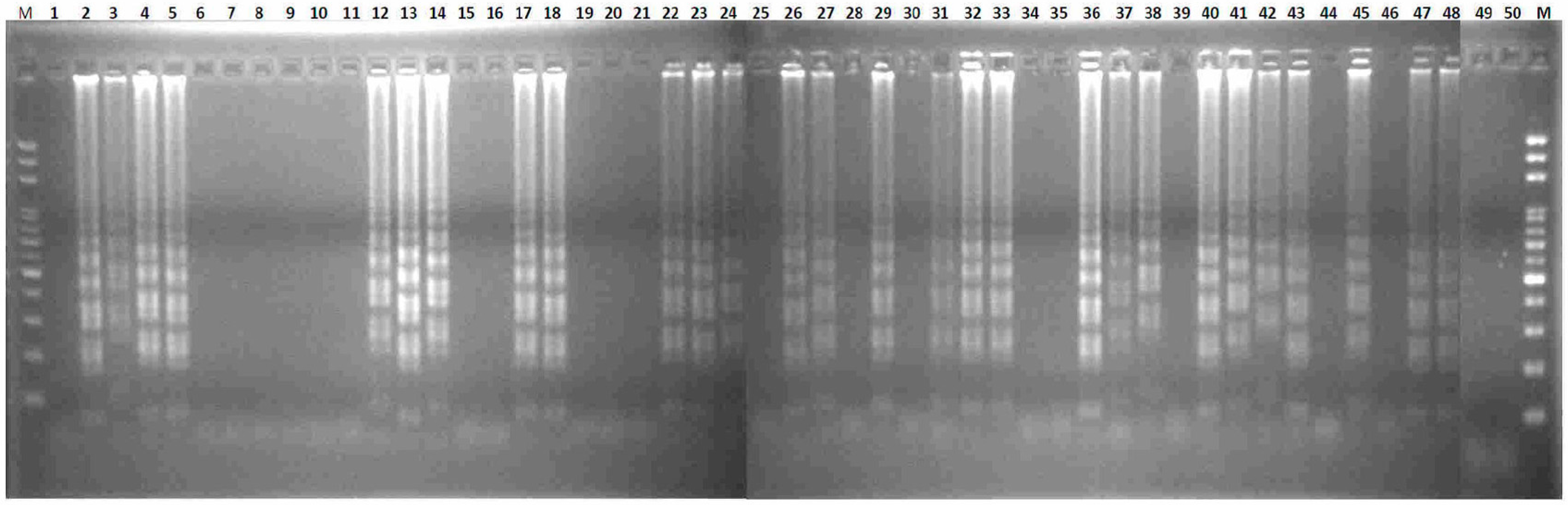
2.2. Transferability of Papaya Varieties and Error Rate of Sex Determination for LAMP without Prior DNA Purification
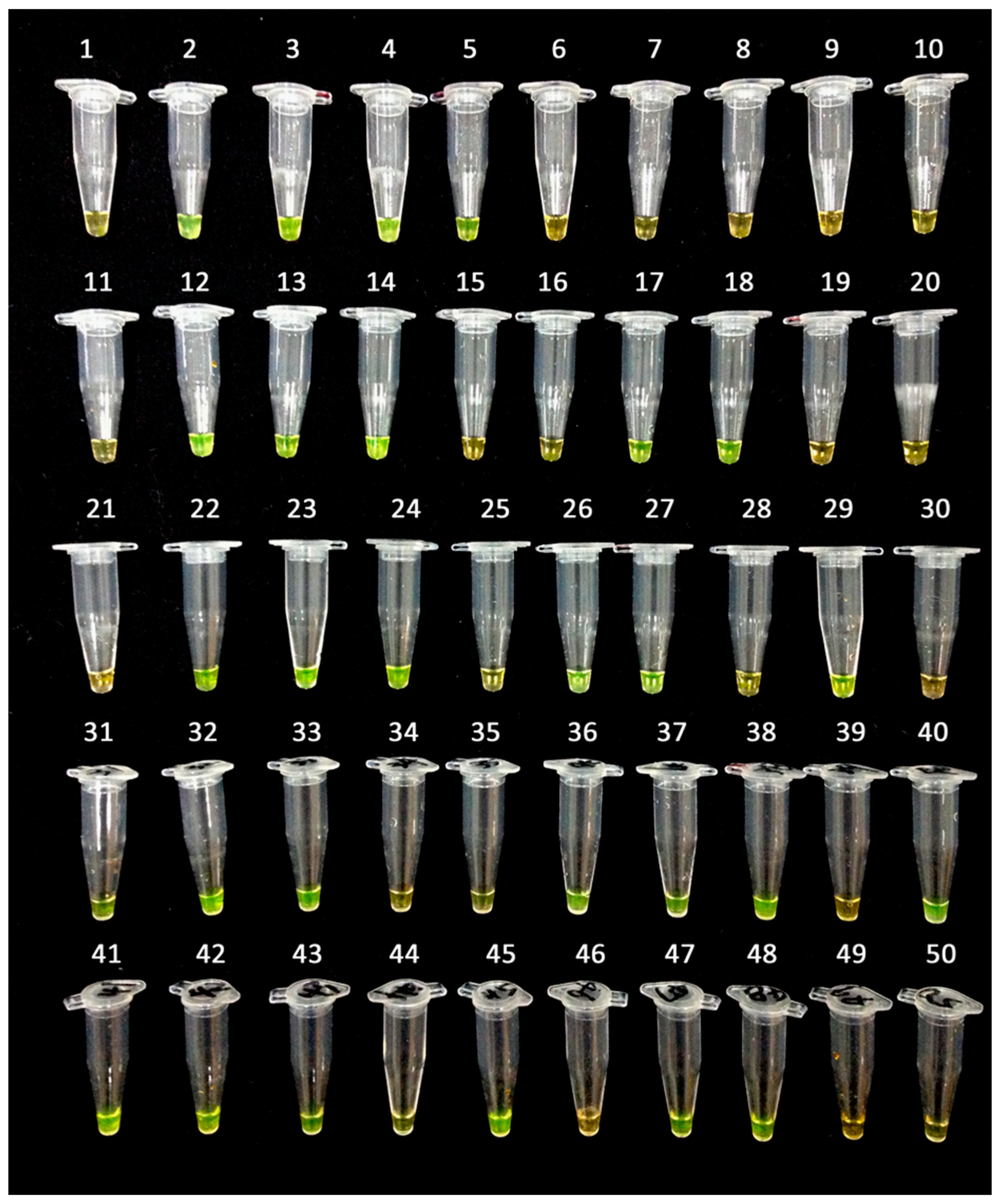
2.3. Mechanism of LAMP Assay without Prior DNA Purification
3. Materials and Methods
3.1. Plant Materials
3.2. Design of the Primers for the Male-Specific Region of Y Chromosome for LAMP Assay
3.3. DNA Extraction and Optimization of the LAMP Assay
3.4. LAMP Assay without Prior DNA Purification and Its Compatibility
3.5. PCR Amplification with and without Prior DNA Purification by Two Different DNA Polymerases for Single/Multiple Genes
3.6. DNA Sequencing
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dallwitz, M.J. A general system for coding taxonomic descriptions. Taxon 1980, 29, 41–46. [Google Scholar] [CrossRef]
- Chan, T.C.; Yen, C.R.; Chang, L.S.; Hsiao, C.H.; Ko, T.S. All hermaphrodite progeny are derived by self-pollinating the sunrise papaya mutant. Plant Breed. 2003, 122, 431–434. [Google Scholar] [CrossRef]
- Parasnis, A.S.; Gupta, V.S.; Tamhankar, S.A.; Ranjekar, P.K. A highly reliable sex diagnostic PCR assay for mass screening of papaya seedlings. Mol. Breed. 2000, 6, 337–344. [Google Scholar] [CrossRef]
- Tsai, C.C.; Shih, H.C.; Wang, H.V.; Lin, Y.S.; Chang, C.H.; Chiang, Y.C.; Chou, C.H. RNA-seq SSRs of moth orchid and screening for molecular markers across genus Phalaenopsis (Orchidaceae). PLoS ONE 2015, 10, e0141761. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.C.; Chen, Y.K.H.; Chen, C.H.; Weng, I.S.; Tsai, C.M.; Lee, S.R.; Lin, Y.S.; Chiang, Y.C. Cultivar identification and genetic relationship of mango (Mangifera indica) in Taiwan using 37 SSR markers. Sci. Hortic. 2013, 164, 196–201. [Google Scholar] [CrossRef]
- Lai, J.M.; Tsai, C.C.; Yen, C.R.; Ko, Y.Z.; Chen, S.R.; Weng, I.S.; Lin, Y.S.; Chiang, Y.C. Molecular characterization of twenty polymorphic microsatellite markers in the polyploid fruit tree species Syzygium samarangense (Myrtaceae). Genet. Mol. Res. 2015, 14, 13013–13021. [Google Scholar] [CrossRef] [PubMed]
- Deputy, J.C.; Ming, R.; Ma, H.; Liu, Z.; Fitch, M.; Wang, M.; Manshardt, R.; Stiles, J.L. Molecular markers for sex determination in papaya (Carica papaya L.). Theor. Appl. Genet. 2002, 106, 107–111. [Google Scholar] [PubMed]
- Urasaki, N.; Tokumoto, M.; Tarora, K.; Ban, Y.; Kayano, T.; Tanaka, H.; Oku, H.; Chinen, I.; Terauchi, R. A male and hermaphrodite specific RAPD marker for papaya (Carica papaya L.). Theor. Appl. Genet. 2002, 104, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhang, Y.; Liu, Z.Y. Development of sex linked AFLP-derived SCAR markers in Cairca papaya. Sci. Agric. Sin. 2009, 42, 967–973. [Google Scholar]
- Sobir, S.S.; Pandia, E.C. Development of SCAR marker for detection of sex expression in papaya (Carica papaya L.) from several genetic backgrounds. Bul. Agron. 2008, 36, 236–240. [Google Scholar]
- Rigano, L.A.; Marano, M.R.; Castagnaro, A.P.; Do Amaral, A.M.; Vojnov, A.A. Rapid and sensitive detection of Citrus Bacterial Canker by loop-mediated isothermal amplification combined with simple visual evaluation methods. BMC Microbiol. 2010, 10. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28. [Google Scholar] [CrossRef]
- Fukuta, S.; Ohishi, K.; Yoshida, K.; Mizukami, Y.; Ishida, A.; Kanbe, M. Development of immunocapture reverse transcription loop-mediated isothermal amplification for the detection of tomato spotted wilt virus from chrysanthemum. J. Virol. Methods 2004, 121, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Buates, S.; Bantuchai, S.; Sattabongkot, J.; Han, E.T.; Tsuboi, T.; Udomsangpetch, R.; Sirichaisinthop, J.; Tan-ariya, P. Development of a reverse transcription-loop-mediated isothermal amplification (RT-LAMP) for clinical detection of Plasmodium falciparum gametocytes. Parasitol. Int. 2010, 59, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.H.; Lai, G.H.; Lee, M.S.; Lin, W.; Lien, Y.Y.; Hsueh, S.C.; Kao, J.Y.; Chang, W.T.; Lu, T.C.; Lin, W.N.; et al. Development and evaluation of a loop-mediated isothermal amplification assay for rapid detection of chicken anaemia virus. J. Appl. Microbiol. 2010, 108, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Hsu, T.H.; Gwo, J.C.; Lin, K.H. Rapid sex identification of papaya (Carica papaya) using multiplex loop-mediated isothermal amplification (mLAMP). Planta 2012, 236, 1239–1246. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.W.; Liu, P.C.; Yang, W.C.; Kuo, J.; Chang, C.L.; Wang, C.Y. A novel loop-mediated isothermal amplification approach for sex identification of Columbidae birds. Theriogenology 2012, 78, 1329–1338. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Moore, P.H.; Ma, H.; Ackerman, C.M.; Ragiba, M.; Yu, Q.; Pearl, H.M.; Kim, M.S.; Charlton, J.W.; Stiles, J.I.; et al. A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature 2004, 427, 348–352. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Hou, S.; Hobza, R.; Feltus, F.A.; Wang, X.; Jin, W.; Skelton, R.S.; Blas, A.; Lemke, C.; Saw, J.H.; et al. Chromosomal location and gene paucity of the male specific region on papaya Y chromosome. Mol. Genet. Genom. 2007, 278, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Friar, E.A. Isolation of DNA from plants with large amounts of secondary metabolites. Methods Enzymol. 2005, 395, 1–12. [Google Scholar]
- Katterman, F.R.; Shattuck, V.I. An effective method of DNA isolation from the mature leaves of Gossypium species that contain large amounts of phenolic terpenoids and tannins. Prep. Biochem. 1983, 13, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.N.; Adams, R.P.; Flournoy, L.E. Inhibition of random amplified polymorphic DNAs (RAPDs) by plant polysaccharides. Plant Mol. Biol. Rep. 1996, 14, 17–22. [Google Scholar] [CrossRef]
- Stange, C.; Prehn, D.; Arce-Johnson, P. Isolation of Pinus radiata genomic DNA suitable for RAPD analysis. Plant Mol. Biol. Rep. 1998, 16, 1–8. [Google Scholar] [CrossRef]
- Kim, C.S.; Lee, C.H.; Shin, J.S.; Chung, Y.S.; Hyung, N.I. A simple and rapid method for isolation of high quality genomic DNA from fruit trees and conifers using PVP. Nucleic Acids Res. 1997, 25, 1085–1086. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, B.; Richards, S.; Dierig, D.A. DNA isolation method for high polysaccharide Lesquerella species. Ind. Crop. Prod. 1999, 9, 111–114. [Google Scholar] [CrossRef]
- Michiels, A.; van den Ende, W.; Tucker, M.; van Riet, L.; van Laere, A. Extraction of high-quality genomic DNA from latex-containing plants. Anal. Biochem. 2003, 315, 85–89. [Google Scholar] [CrossRef]
- Echevarría-Machado, I.; Sánchez-Cach, L.A.; Hernández-Zepeda, C.; Rivera-Madrid, R.; Moreno-Valenzuela, O.A. A simple and efficient method for isolation of DNA in high mucilaginous plant tissues. Mol. Biotechnol. 2005, 31, 129–135. [Google Scholar] [CrossRef]
- Moyo, M.; Amoo, S.O.; Bairu, M.W.; Finnie, J.F.; van Staden, J. Optimising DNA isolation for medicinal plants. S. Afr. J. Bot. 2008, 74, 771–775. [Google Scholar] [CrossRef]
- Nagori, R.; Sharma, P.; Habibi, N.; Purohit, S.D. An efficient genomic DNA extraction protocol for molecular analysis in Annona reticulata. Natl. Acad. Sci. Lett. 2014, 37, 137–140. [Google Scholar] [CrossRef]
- Fang, G.; Hammar, S.; Grumet, R. A quick and inexpensive method for removing polysaccharides from plant genomic DNA. Biotechniques 1992, 13, 52–56. [Google Scholar] [PubMed]
- Das, S.; Tiwari, K.L.; Sen, S.; Singh, A. Rapid one step DNA extraction method from (Gastrimargus musicus) through formaldehyde. J. Pharm. Biomed. Sci. 2012, 20, 4. [Google Scholar]
- Grevelding, C.G.; Kampkötter, A.; Hollmann, M.; Schäfer, U.; Kunz, W. Direct PCR on fruitflies and blood flukes without prior DNA isolation. Nucleic Acids Res. 1996, 24, 4100–4101. [Google Scholar] [CrossRef] [PubMed]
- Bu, Y.; Huang, H.; Zhou, G. Direct polymerase chain reaction (PCR) from human whole blood and filter-paper-dried blood by using a PCR buffer with a higher pH. Anal. Biochem. 2008, 375, 370–372. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xu, H.; Zhao, C.; Sulaiman, Y.; Wu, C. A PCR amplification method without DNA extraction. Electrophoresis 2011, 32, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Virdi, A.S.; Singh, P. A novel method for whole blood PCR without pretreatment. Gene 2012, 501, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.G.; Kim, J.Y.; Soh, M.S.; Kim, D.S. A simple and rapid gene amplification from Arabidopsis leaves using AnyDirect system. J. Biochem. Mol. Biol. 2007, 40, 444–447. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.H.; Yang, S.Y.; Lin, C.L.; Wang, C.H.; Li, P.C.; Chen, T.Y.; Jan, F.J.; Lee, G.B. Detection of viruses directly from the fresh leaves of a Phalaenopsis orchid using a microfluidic system. Nanomedicine 2013, 9, 1274–1282. [Google Scholar] [CrossRef] [PubMed]
- Bellstedt, D.U.; Pirie, M.D.; Visser, J.C.; de Villiers, M.J.; Gehrke, B. A rapid and inexpensive method for the direct PCR amplification of DNA from plants. Am. J. Bot. 2010, 97, e65–e68. [Google Scholar] [CrossRef] [PubMed]
- Ming, R.; Hou, S.; Feng, Y.; Yu, Q.; Dionne-Laporte, A.; Saw, J.H.; Senin, P.; Wang, W.; Ly, B.V.; Lewis, K.L.T. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 2008, 452, 991–996. [Google Scholar] [CrossRef] [PubMed]
- Rocha, A.J.; de Souza Miranda, R.; da Silva Cunha, R.M. Assessment of DNA polymerases in microsatellite amplification assays through PowerPlex® 16 BIO System. Biochem. Biotechnol. Rep. 2014, 3, 1–8. [Google Scholar] [CrossRef]
- Rohland, N.; Hofreiter, M. Ancient DNA extraction from bones and teeth. Nat. Protoc. 2007, 2, 1756–1762. [Google Scholar] [CrossRef] [PubMed]
- John, M.E. An efficient method for isolation of RNA and DNA from plants containing polyphenolics. Nucleic Acids Res. 1992, 20, 2381. [Google Scholar] [CrossRef] [PubMed]
- Chum, P.Y.; Haimes, J.D.; André, C.P.; Kuusisto, P.K.; Kelley, M.L. Genotyping of plant and animal samples without prior DNA purification. J. Vis. Exp. 2014, 67. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.M.; Lu, S.H.; Tong, Q.B.; Lou, D.; Chen, R.; Zheng, B.; Kumagai, T.; Wen, L.Y.; Ohta, N.; Zhou, X.N. Loop-mediated isothermal amplification (LAMP): Early detection of Toxoplasma gondii infection in mice. Parasites Vectors 2012, 5. [Google Scholar] [CrossRef] [PubMed]
- Ming, R.; Yu, Q.Y.; Moore, P.H. Sex determination in papaya. Semin. Cell Dev. Biol. 2007, 18, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Eiken Chemical Co., Ltd. Primer Explorer Version 4 Software. 2016. Available online: http://primerexplorer.jp/v5_manual/index.html (accessed on 18 August 2016).
- Tsai, C.C.; Huang, S.C.; Chen, C.H.; Tseng, Y.H.; Huang, P.L.; Tsai, S.H.; Chou, C.H. Genetic relationship of Rhododendron (Ericaceae) in Taiwan based on the sequence of the internal transcribed spacer of ribosomal DNA. J. Hortic. Sci. Biotechnol. 2003, 78, 234–240. [Google Scholar] [CrossRef]


| Primer Name | Sequence |
|---|---|
| papaya-F3 | 5′-GTGGCATTAATGCAACGC-3′ |
| papaya-B3 | 5′-TGTCACCATGAGCACTAG-3′ |
| papaya-FIP | 5′-CAGAGGAAGAGTGGGTGTTTTGCGGGTCCTACGAACCTAG-3′ |
| papaya-BIP | 5′-GCTTGCCGAACATAGAGGCTTGAGCACGATCCACGAGAT-3′ |
| papaya-LF | 5′-GTGGTGTTGTAGGCATCAAATGTT-3′ |
| papaya-LB | 5′-TCTCCCCTCACACCCAATCC-3′ |
| Papaya Sample No. | Varieties Name | Sex Type |
|---|---|---|
| 1 | Line Ph13047-1 | hermaphrodite |
| 2 | Inbred line Y5043 | Females |
| 3 | Inbred line Y5043 | Females |
| 4 | Inbred line Y5043 | Males |
| 5 | Line Ph13047-2 | hermaphrodite |
| 6 | Inbred line Y5043 | Males |
| Papaya Sample No. | Varieties Name | Source |
|---|---|---|
| 1 | Tainung No. 1 | Taiwan |
| 2 | Tainung No. 2 | Taiwan |
| 3 | Red Lady | Taiwan |
| 4 | Holland | Thailand |
| 5 | Golden | Brazil |
| 6 | Sunrise | Brazil |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsai, C.-C.; Shih, H.-C.; Ko, Y.-Z.; Wang, R.-H.; Li, S.-J.; Chiang, Y.-C. Direct LAMP Assay without Prior DNA Purification for Sex Determination of Papaya. Int. J. Mol. Sci. 2016, 17, 1630. https://doi.org/10.3390/ijms17101630
Tsai C-C, Shih H-C, Ko Y-Z, Wang R-H, Li S-J, Chiang Y-C. Direct LAMP Assay without Prior DNA Purification for Sex Determination of Papaya. International Journal of Molecular Sciences. 2016; 17(10):1630. https://doi.org/10.3390/ijms17101630
Chicago/Turabian StyleTsai, Chi-Chu, Huei-Chuan Shih, Ya-Zhu Ko, Ren-Huang Wang, Shu-Ju Li, and Yu-Chung Chiang. 2016. "Direct LAMP Assay without Prior DNA Purification for Sex Determination of Papaya" International Journal of Molecular Sciences 17, no. 10: 1630. https://doi.org/10.3390/ijms17101630
APA StyleTsai, C.-C., Shih, H.-C., Ko, Y.-Z., Wang, R.-H., Li, S.-J., & Chiang, Y.-C. (2016). Direct LAMP Assay without Prior DNA Purification for Sex Determination of Papaya. International Journal of Molecular Sciences, 17(10), 1630. https://doi.org/10.3390/ijms17101630






