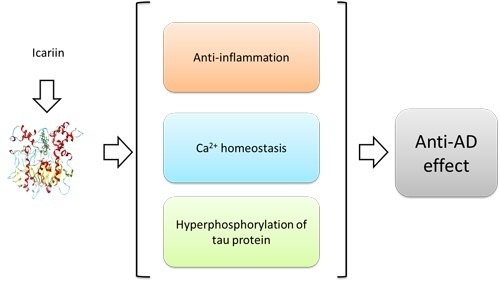
In Silico Insight into Potential Anti-Alzheimer’s Disease Mechanisms of Icariin
Abstract
:1. Introduction
2. Results
2.1. The Putative Protein Targets of Icariin

2.2. The Potential Targets Significantly Correlate with AD-Related Proteins
| Uniprot | Gene Symbol | PDB Code | Known Ligand | Icariin | |||
|---|---|---|---|---|---|---|---|
| Name | MM/GBVI (kcal/mol) | Affinity (pki) | MM/GBVI (kcal/mol) | Affinity (pki) | |||
| Q13464 | ROCK1 | 2ETK | Hydroxyfasudil | −21.03 | 8.10 | −33.03 | 14.34 |
| P00439 | PAH | 4PAH | Norepinephrine | −26.24 | 7.05 | −44.04 | 7.90 |
| Q9HAN9 | NMNAT1 | 1GZU | Nicotinamide Mononucleotide | −27.30 | 12.54 | −23.15 | 17.75 |
| Q9BW91 | NUDT9 | 1Q33 | β-d-Glucose | −24.64 | 10.70 | −34.00 | 13.26 |
| P50135 | HNMT | 2AOU | Amodiaquine | −26.05 | 6.91 | −22.14 | 6.10 |
| Q10588 | BST1 | 1ISG | Adenosine-5′-diphosphate Monothiophosphate | −14.33 | 9.41 | −24.31 | 10.99 |
| P06737 | PYGL | 1FA9 | Adenosine Monophosphate | −17.06 | 8.66 | −27.18 | 9.79 |
| P00750 | PLAT | 1PK2 | Aminocaproic Acid | −19.90 | 9.71 | −24.92 | 9.30 |
| O76074 | PDE5 | 2H42 | Sildenafil | −36.25 | 9.67 | −28.87 | 13.89 |
| P04062 | GBA | 2F61 | 2-(Acetylamino)-2-deoxy-a-d-glucopyranose | −16.41 | 5.57 | −21.80 | 11.17 |
| P15291 | B4GALT1 | 4EEG | N-Acetyl-d-glucosamine | −19.15 | 8.97 | −34.21 | 10.75 |
| P07737 | PFN1 | 1CJF | 7-Hydroxy-4-methyl-3-(2-hydroxy-ethyl)coumarin | −15.42 | 6.69 | −27.05 | 8.67 |
| P09012 | SNRPA | 1NU4 | Malonic Acid | −22.16 | 6.52 | −23.86 | 5.34 |
| P84077 | ARF1 | 1U81 | 1,3-Propandiol | −37.77 | 4.64 | −43.17 | 11.18 |
| Q08209 | PPP3CA | 4F0Z | Myristic Acid | −15.56 | 4.64 | −22.51 | 8.90 |
| P13569 | CFTR | 2BBO | Ibuprofen | −8.23 | 6.00 | −6.62 | 11.01 |
| P02774 | GC | 1J78 | Cholecalciferol | −10.99 | 5.05 | −21.56 | 6.43 |
| P11387 | TOP1 | 1TL8 | Irinotecan | −34.75 | 14.32 | −20.04 | 17.80 |
| P19883 | FST | 2B0U | d-Myo-inositol-hexasulphate | −9.54 | 7.03 | −24.81 | 9.78 |
| P27695 | APEX1 | 4QHE | Lucanthone | −16.70 | 4.25 | −17.73 | 4.56 |
| P22303 | AChE | 1F8U | Mefloquine | −11.44 | 6.54 | −34.86 | 7.97 |
2.3. An Integrated Network for Anti-AD Effects of Icariin
| KEGG Pathway | The Number of Icariin’s Targets | p-Value |
|---|---|---|
| Spliceosome | 5 | 2.26 × 10−6 |
| Vibrio cholerae infection | 2 | 3.17 × 10−2 |
| Carbohydrate digestion and absorption | 3 | 7.34 × 10−3 |
| Legionellosis | 3 | 4.59 × 10−2 |
| Oxytocin signaling pathway | 5 | 4.25 × 10−2 |
| cGMP-PKG signaling pathway | 5 | 3.09 × 10−2 |
| Apoptosis | 5 | 1.14 × 10−2 |
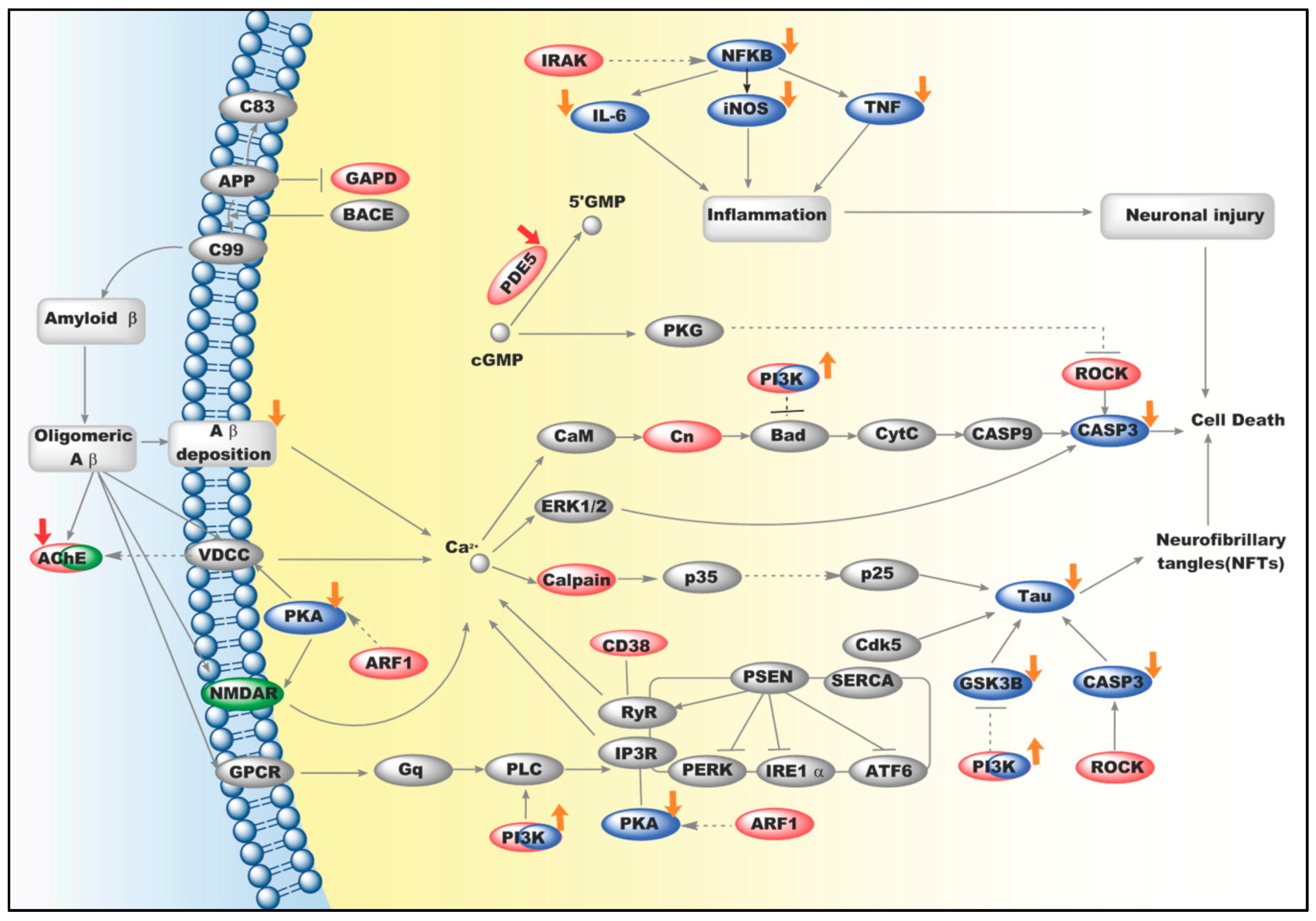
3. Discussion
4. Experimental Section
4.1. Identification of Putative Protein Targets
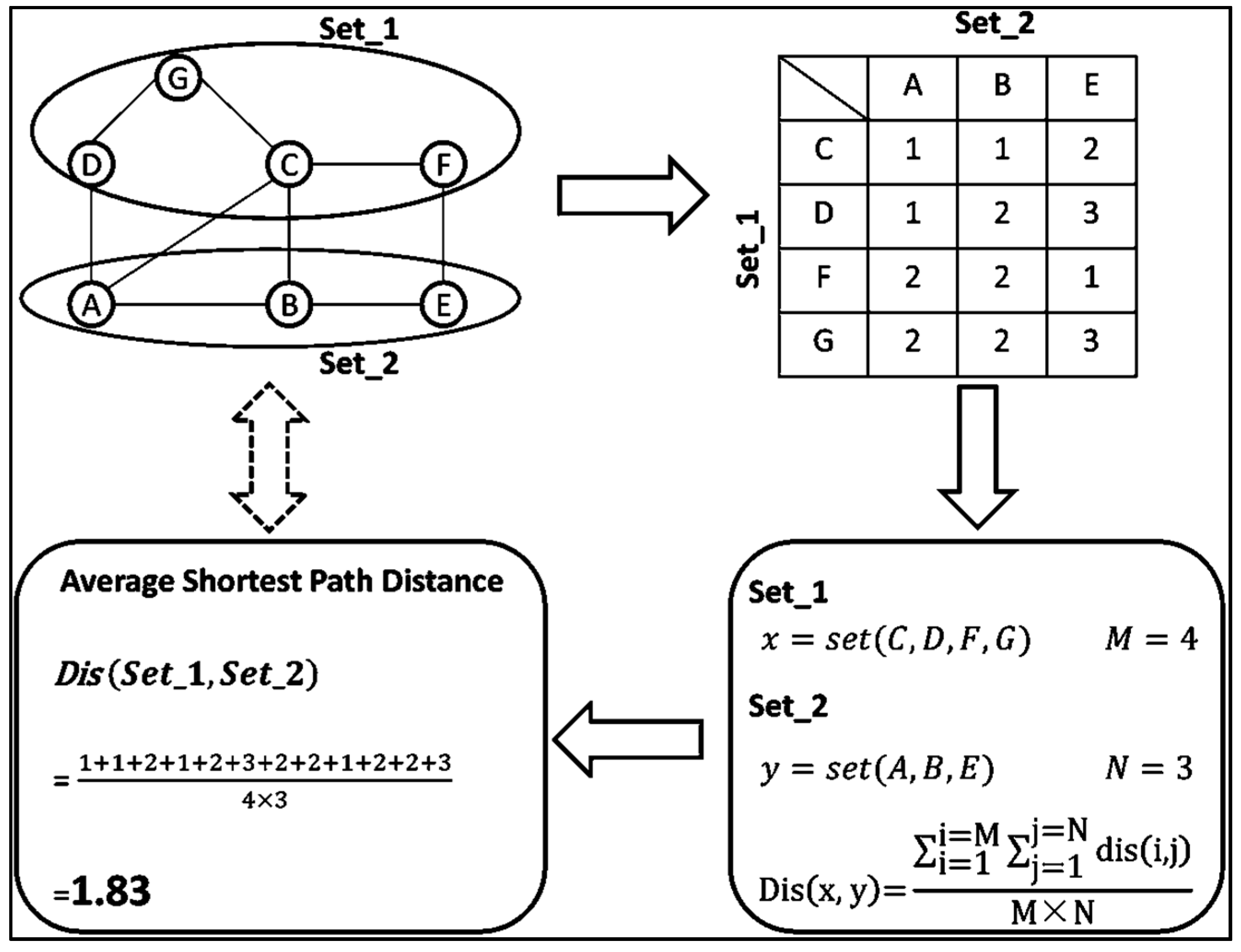
4.2. Average Shortest Path Calculation
4.3. The Semantic Similarity of Gene Ontology (GO) Profiles
4.4. Pathway Enrichment of Icariin’s Putative Targets
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Querfurth, H.W.; LaFerla, F.M. Alzheimer’s disease. N. Engl. J. Med. 2010, 362, 329–344. [Google Scholar] [CrossRef] [PubMed]
- Pohanka, M. Cholinesterases, a target of pharmacology and toxicology. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czechoslov. 2011, 155, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Reisberg, B.; Doody, R.; Stoffler, A.; Schmitt, F.; Ferris, S.; Mobius, H.J.; Memantine Study Group. Memantine in moderate-to-severe alzheimer’s disease. N. Engl. J. Med. 2003, 348, 1333–1341. [Google Scholar] [CrossRef] [PubMed]
- Drugs for alzheimer’s disease: Best avoided. No therapeutic advantage. Available online: http://www.ncbi.nlm.nih.gov/pubmed/22822592 (accessed on 28 April 2015).
- Birks, J.; Harvey, R.J. Donepezil for dementia due to alzheimer’s disease. Cochrane Database Syst. Rev. 2006. [Google Scholar] [CrossRef]
- Sha, D.; Li, L.; Ye, L.; Liu, R.; Xu, Y. Icariin inhibits neurotoxicity of beta-amyloid by upregulating cocaine-regulated and amphetamine-regulated transcripts. Neuroreport 2009, 20, 1564–1567. [Google Scholar] [CrossRef] [PubMed]
- Zeng, K.W.; Ko, H.; Yang, H.O.; Wang, X.M. Icariin attenuates β-amyloid-induced neurotoxicity by inhibition of tau protein hyperphosphorylation in pc12 cells. Neuropharmacology 2010, 59, 542–550. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wang, Z.; Sheng, C.; Peng, W.; Hui, S.; Gong, W.; Chen, S. Icariin prevents amyloid β-induced apoptosis via the pi3k/akt pathway in PC-12 cells. Evid. Based Complement. Alternat. Med. 2015. [Google Scholar] [CrossRef] [PubMed]
- Li, W.W.; Gao, X.M.; Wang, X.M.; Guo, H.; Zhang, B.L. Icariin inhibits hydrogen peroxide-induced toxicity through inhibition of phosphorylation of jnk/p38 mapk and p53 activity. Mutat. Res. 2011, 708, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Tsai, H.J.; Li, L.; Wang, X.M. Icariin inhibits the increased inward calcium currents induced by amyloid-beta(25–35) peptide in ca1 pyramidal neurons of neonatal rat hippocampal slice. Am. J. Chin. Med. 2010, 38, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Nie, J.; Luo, Y.; Huang, X.N.; Gong, Q.H.; Wu, Q.; Shi, J.S. Icariin inhibits β-amyloid peptide segment 25–35 induced expression of β-secretase in rat hippocampus. Eur. J. Pharmacol. 2010, 626, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Urano, T.; Tohda, C. Icariin improves memory impairment in alzheimer’s disease model mice (5xfad) and attenuates amyloid β-induced neurite atrophy. Phytother. Res.: PTR 2010, 24, 1658–1663. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.; Gong, Q.H.; Xu, Y.S.; Wang, L.N.; Jin, H.; Li, F.; Li, L.S.; Ma, Y.M.; Shi, J.S. Icariin, a phosphodiesterase-5 inhibitor, improves learning and memory in app/ps1 transgenic mice by stimulation of no/cgmp signalling. Int. J. Neuropsychopharmacol. 2014, 17, 871–881. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.D.; Cai, Y.N.; Zhang, Q.; Qi, Z.L.; Gao, Q.Q. Inhibitory effect of icariin on acetylcholinesterase. Yao Xue Xue Bao 2012, 47, 1141–1146. [Google Scholar] [PubMed]
- Cao, Y.F.; He, R.R.; Cao, J.; Chen, J.X.; Huang, T.; Liu, Y. Drug-drug interactions potential of icariin and its intestinal metabolites via inhibition of intestinal udp-glucuronosyltransferases. Evid. Based Complement. Altern. Med. 2012, 2012, 395912. [Google Scholar] [CrossRef] [PubMed]
- Xin, Z.C.; Kim, E.K.; Lin, C.S.; Liu, W.J.; Tian, L.; Yuan, Y.M.; Fu, J. Effects of icariin on cgmp-specific pde5 and camp-specific pde4 activities. Asian J. Androl. 2003, 5, 15–18. [Google Scholar] [PubMed]
- Sun, Y.; Zhu, R.; Ye, H.; Tang, K.; Zhao, J.; Chen, Y.; Liu, Q.; Cao, Z. Towards a bioinformatics analysis of anti-alzheimer’s herbal medicines from a target network perspective. Brief. Bioinform. 2013, 14, 327–343. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Z.; Zhi, D.G. Ligand-protein inverse docking and its potential use in the computer search of protein targets of a small molecule. Proteins 2001, 43, 217–226. [Google Scholar] [CrossRef]
- Zhang, H.P.; Pan, J.B.; Zhang, C.; Ji, N.; Wang, H.; Ji, Z.L. Network understanding of herb medicine via rapid identification of ingredient-target interactions. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Ulrich-Merzenich, G.; Panek, D.; Zeitler, H.; Vetter, H.; Wagner, H. Drug development from natural products: Exploiting synergistic effects. Indian J. Exp. Biol. 2010, 48, 208–219. [Google Scholar] [PubMed]
- Gong, C.X.; Iqbal, K. Hyperphosphorylation of microtubule-associated protein tau: A promising therapeutic target for alzheimer disease. Curr. Med. Chem. 2008, 15, 2321–2328. [Google Scholar] [CrossRef] [PubMed]
- Baki, L.; Shioi, J.; Wen, P.; Shao, Z.; Schwarzman, A.; Gama-Sosa, M.; Neve, R.; Robakis, N.K. Ps1 activates pi3k thus inhibiting gsk-3 activity and tau overphosphorylation: Effects of fad mutations. EMBO J. 2004, 23, 2586–2596. [Google Scholar] [CrossRef] [PubMed]
- Jarero-Basulto, J.J.; Luna-Munoz, J.; Mena, R.; Kristofikova, Z.; Ripova, D.; Perry, G.; Binder, L.I.; Garcia-Sierra, F. Proteolytic cleavage of polymeric tau protein by caspase-3: Implications for alzheimer disease. J. Neuropathol. Exp. Neurol. 2013, 72, 1145–1161. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Zhao, J.; Zhang, X.; Li, H.; Zhou, Y. Icariin induces osteoblast proliferation, differentiation and mineralization through estrogen receptor-mediated erk and jnk signal activation. Eur. J. Pharmacol. 2013, 714, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, H.; Barger, S.; Barnum, S.; Bradt, B.; Bauer, J.; Cole, G.M.; Cooper, N.R.; Eikelenboom, P.; Emmerling, M.; Fiebich, B.L.; et al. Inflammation and alzheimer’s disease. Neurobiol. Aging 2000, 21, 383–421. [Google Scholar] [CrossRef]
- Heneka, M.T.; O’Banion, M.K.; Terwel, D.; Kummer, M.P. Neuroinflammatory processes in alzheimer’s disease. J. Neural Transm. 2010, 117, 919–947. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Sun, T.; Wu, J.; Kalionis, B.; Zhang, C.; Yuan, D.; Huang, J.; Cai, W.; Fang, H.; Xia, S. Icariin intervenes in cardiac inflammaging through upregulation of sirt6 enzyme activity and inhibition of the nf-kappa b pathway. BioMed Res. Int. 2015, 2015, 895976. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.R.; Xu, X.Z.; Wang, Y.H.; Chen, J.W.; Xu, S.W.; Gu, L.Q.; Liu, P.Q. Icariin derivative inhibits inflammation through suppression of p38 mitogen-activated protein kinase and nuclear factor-κB pathways. Biol. Pharm. Bull. 2010, 33, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.Q.; Liu, B.J.; Wu, J.F.; Xu, Y.C.; Duan, X.H.; Cao, Y.X.; Dong, J.C. Icariin attenuates lps-induced acute inflammatory responses: Involvement of pi3k/akt and nf-κB signaling pathway. Eur. J. Pharm. 2010, 642, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.T.; Chang, R.C.; Tan, L. Calcium dysregulation in alzheimer’s disease: From mechanisms to therapeutic opportunities. Prog. Neurobiol. 2009, 89, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Dechant, R.; Saad, S.; Ibanez, A.J.; Peter, M. Cytosolic ph regulates cell growth through distinct gtpases, arf1 and gtr1, to promote ras/pka and torc1 activity. Mol. Cell 2014, 55, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Skeberdis, V.A.; Chevaleyre, V.; Lau, C.G.; Goldberg, J.H.; Pettit, D.L.; Suadicani, S.O.; Lin, Y.; Bennett, M.V.; Yuste, R.; Castillo, P.E.; et al. Protein kinase a regulates calcium permeability of nmda receptors. Nat. Neurosci. 2006, 9, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhang, J.; Fu, L.; Deng, W.; Wu, A.; Wang, X. Icariin inhibits the apoptosis of cemx174 triggered by simian immuno deficiency virus infection in vitro. Chin. Pharmacol. Bull. 2008, 24, 684–687. [Google Scholar]
- Zhu, H.; Gao, W.; Jiang, H.; Jin, Q.H.; Shi, Y.F.; Tsim, K.W.K.; Zhang, X.J. Regulation of acetylcholinesterase expression by calcium signaling during calcium ionophore a23187-and thapsigargin-induced apoptosis. Int. J. Biochem. Cell Biol. 2007, 39, 93–108. [Google Scholar] [CrossRef] [PubMed]
- Bellera, C.L.; Balcazar, D.E.; Alberca, L.; Labriola, C.A.; Talevi, A.; Carrillo, C. Application of computer-aided drug repurposing in the search of new cruzipain inhibitors: Discovery of amiodarone and bromocriptine inhibitory effects. J. Chem. Inf. Model. 2013, 53, 2402–2408. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Z.; Ung, C.Y. Prediction of potential toxicity and side effect protein targets of a small molecule by a ligand-protein inverse docking approach. J. Mol. Graph. Model. 2001, 20, 199–218. [Google Scholar] [CrossRef]
- Davis, A.P.; Murphy, C.G.; Johnson, R.; Lay, J.M.; Lennon-Hopkins, K.; Saraceni-Richards, C.; Sciaky, D.; King, B.L.; Rosenstein, M.C.; Wiegers, T.C.; et al. The comparative toxicogenomics database: Update 2013. Nucleic Acids Res. 2013, 41, D1104–D1114. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ji, Z.L.; Zhi, D.G.; Chen, Y.Z. Clibe: A database of computed ligand binding energy for ligand-receptor complexes. Comput. Chem. 2002, 26, 661–666. [Google Scholar] [CrossRef]
- Fronczak, A.; Fronczak, P.; Hołyst, J.A. Average path length in random networks. Phys. Rev. E 2004, 70, 056110. [Google Scholar] [CrossRef] [PubMed]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. Int. J. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Keshava Prasad, T.S.; Goel, R.; Kandasamy, K.; Keerthikumar, S.; Kumar, S.; Mathivanan, S.; Telikicherla, D.; Raju, R.; Shafreen, B.; Venugopal, A.; et al. Human protein reference database—2009 update. Nucleic Acids Res. 2009, 37, D767–D772. [Google Scholar] [CrossRef] [PubMed]
- Lord, P.W.; Stevens, R.D.; Brass, A.; Goble, C.A. Investigating semantic similarity measures across the gene ontology: The relationship between sequence and annotation. Bioinformatics 2003, 19, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Du, L.; Zhou, Y. Evaluation of go-based functional similarity measures using S. cerevisiae protein interaction and expression profile data. BMC Bioinform. 2008, 9, 472. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.Z.; Du, Z.; Payattakool, R.; Yu, P.S.; Chen, C.F. A new method to measure the semantic similarity of go terms. Bioinformatics 2007, 23, 1274–1281. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Li, F.; Qin, Y.; Bo, X.; Wu, Y.; Wang, S. Gosemsim: An R package for measuring semantic similarity among go terms and gene products. Bioinformatics 2010, 26, 976–978. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, Z.; Sheng, Z.; Yan, X.; Cao, Z.; Tang, K. In Silico Insight into Potential Anti-Alzheimer’s Disease Mechanisms of Icariin. Int. J. Mol. Sci. 2016, 17, 113. https://doi.org/10.3390/ijms17010113
Cui Z, Sheng Z, Yan X, Cao Z, Tang K. In Silico Insight into Potential Anti-Alzheimer’s Disease Mechanisms of Icariin. International Journal of Molecular Sciences. 2016; 17(1):113. https://doi.org/10.3390/ijms17010113
Chicago/Turabian StyleCui, Zhijie, Zhen Sheng, Xinmiao Yan, Zhiwei Cao, and Kailin Tang. 2016. "In Silico Insight into Potential Anti-Alzheimer’s Disease Mechanisms of Icariin" International Journal of Molecular Sciences 17, no. 1: 113. https://doi.org/10.3390/ijms17010113
APA StyleCui, Z., Sheng, Z., Yan, X., Cao, Z., & Tang, K. (2016). In Silico Insight into Potential Anti-Alzheimer’s Disease Mechanisms of Icariin. International Journal of Molecular Sciences, 17(1), 113. https://doi.org/10.3390/ijms17010113