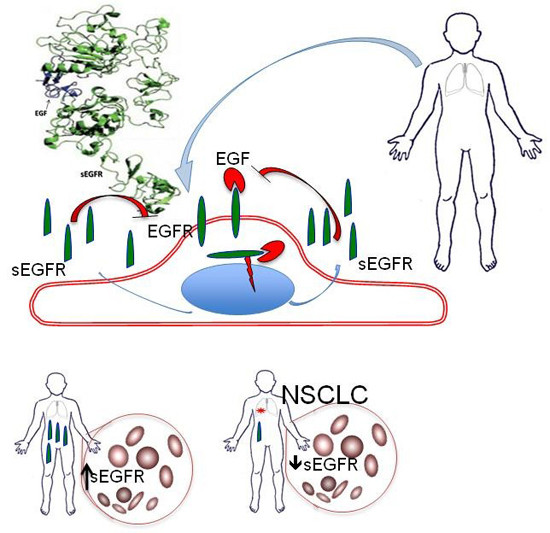
Preliminary Evidence on the Diagnostic and Molecular Role of Circulating Soluble EGFR in Non-Small Cell Lung Cancer
Abstract
:1. Introduction
2. Results
2.1. Plasma Levels of sEGFR Are Lower in NSCLC Patients than Healthy Controls
| sEGFR (ng/mL) | Groups | n | M ± SD | Me | Range | M-WU Text |
| P-group | 37 | 49.9 ± 9.3 | 48.6 | 24.7–69.7 | p = 0.0002 | |
| C-group | 54 | 58.2 ± 10.5 | 55.6 | 41.5–92.3 |

| Clinical-Pathological Variables | sEGFR (ng/mL) | p Value |
|---|---|---|
| Age | 0.306 | |
| Age ≤ 65 (n = 10) | 52.5 ± 13.3 | |
| Age > 65 (n = 27) | 48.9 ± 7.4 | |
| Gender | 0.512 | |
| Male (n = 26) | 49.2 ± 7.5 | |
| Female (n = 11) | 51.4 ± 12.9 | |
| Smoking Habits | 0.639 | |
| Smokers (n = 30) | 49.5 ± 8.1 | |
| Non-Smokers (n = 7) | 51.4 ± 14.0 | |
| Tumor Stage(TNM) | 0.588 | |
| pI (n = 21) | 49.4 ± 7.3 | |
| pII (n = 5) | 53.9 ± 9.8 | |
| pIII (n = 11) | 49.0 ± 12.4 | |
| Histologic cell type | 0.907 | |
| ADK (n = 27) | 49.7 ± 10.0 | |
| SCC (n = 10) | 50.2 ± 7.4 | |
| Grading | 0.082 | |
| G1 (n = 3) | 56.2 ± 9.7 | |
| G2 (n = 9) | 54.2 ± 10.7 | |
| G3 (n = 25) | 47.5 ± 8.1 |
2.2. Plasma Levels of sEGFR and Prognosis in NSCLC Patients
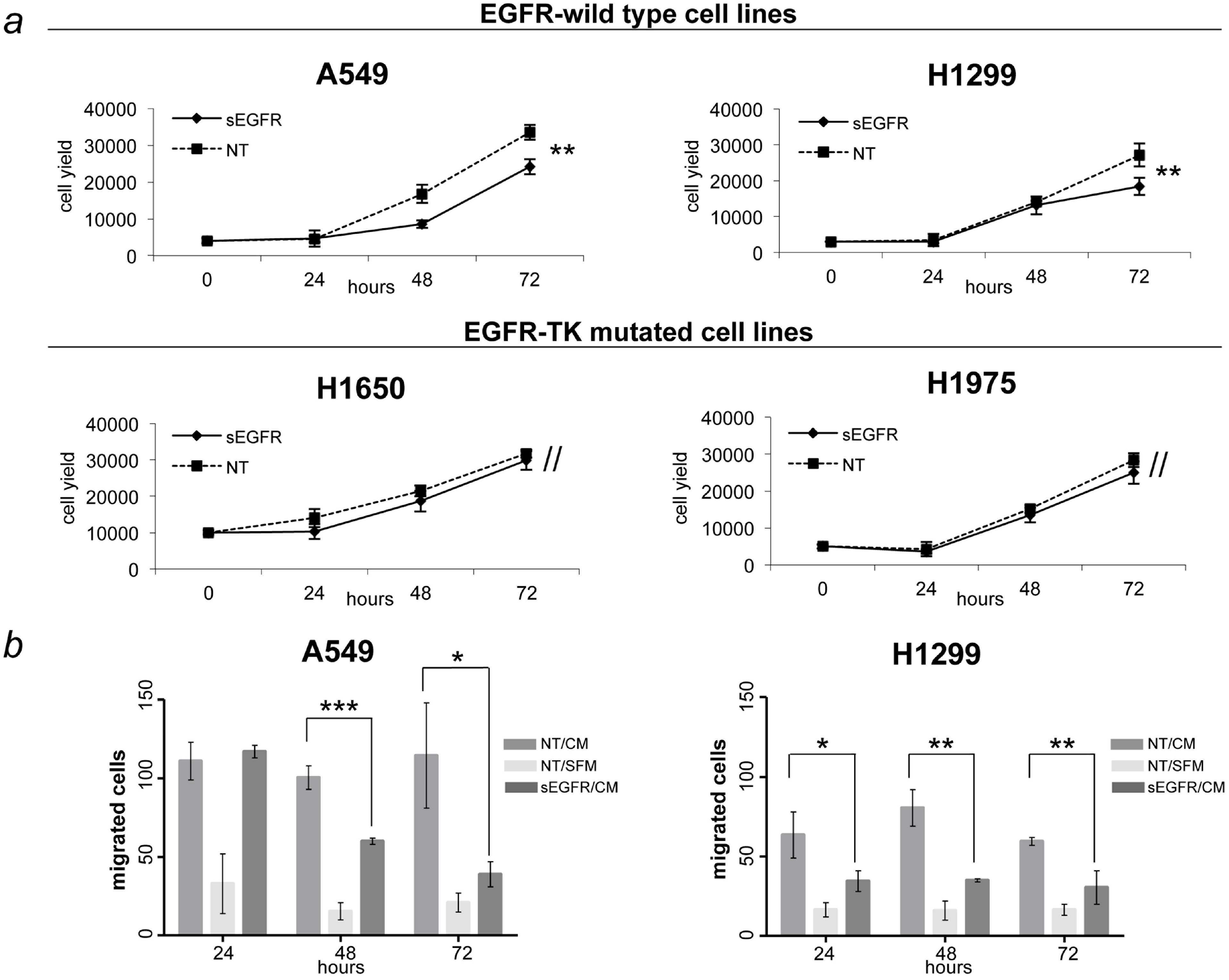
2.3. sEGFR Has Growth and Migration Inhibitory Effects in EGFR-Wild Type NSCLC Cell Lines
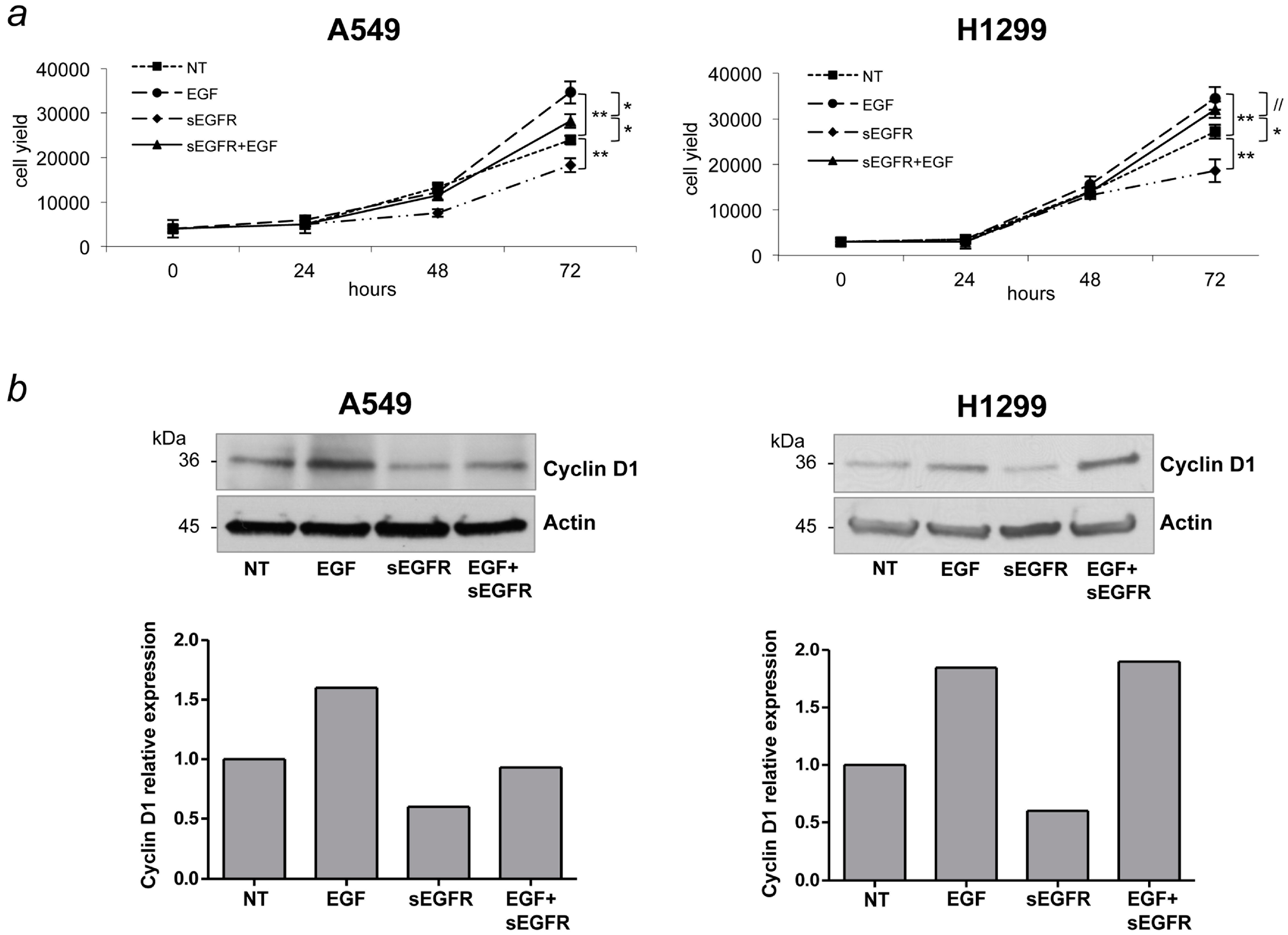
2.4. sEGFR Interferes with the EGF Signaling
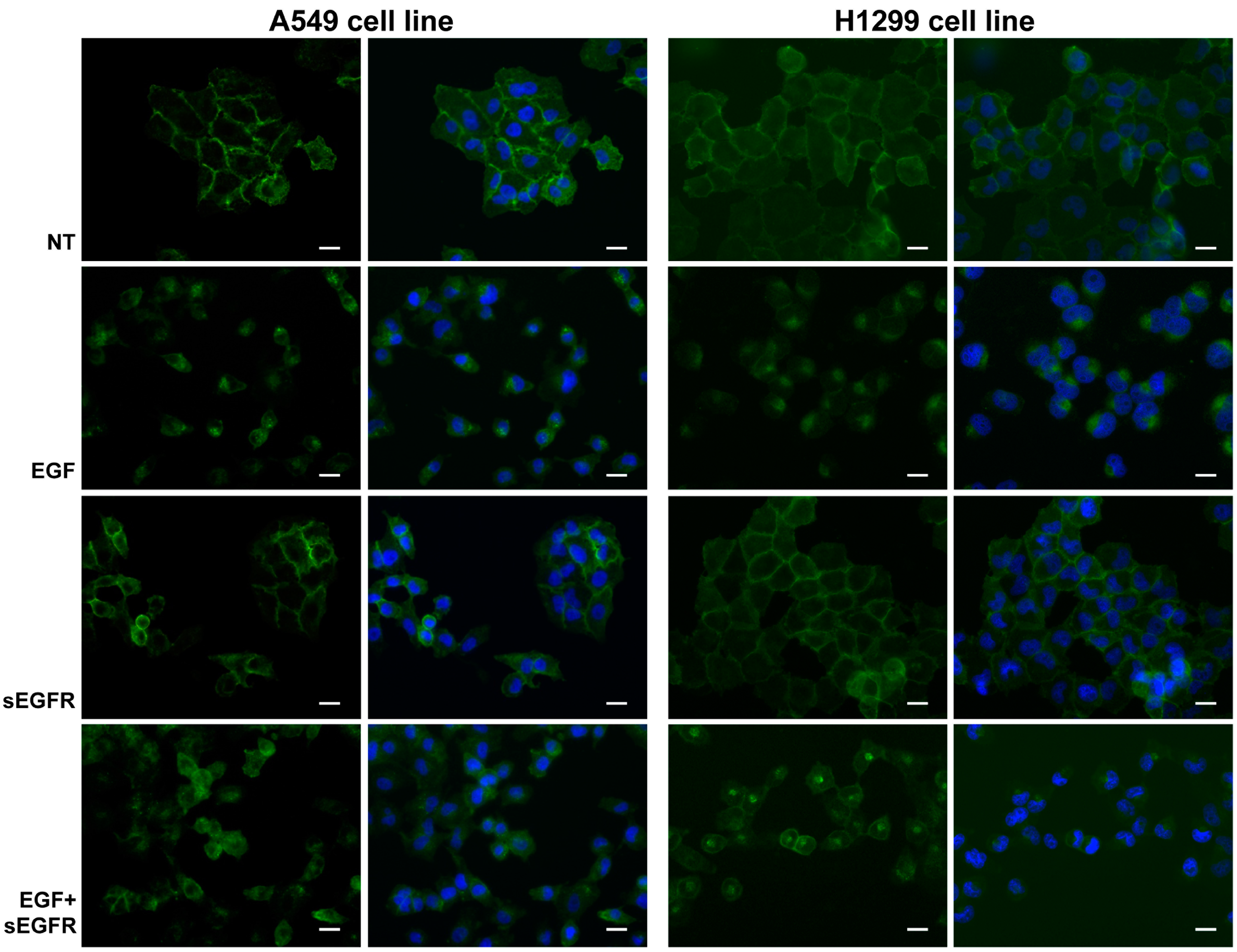
2.5. sEGFR Prevents the Plasma Membrane-EGFR Holoreceptor Internalization
2.6. EGF Treatment Inhibits sEGFR Synthesis in A549 Cells
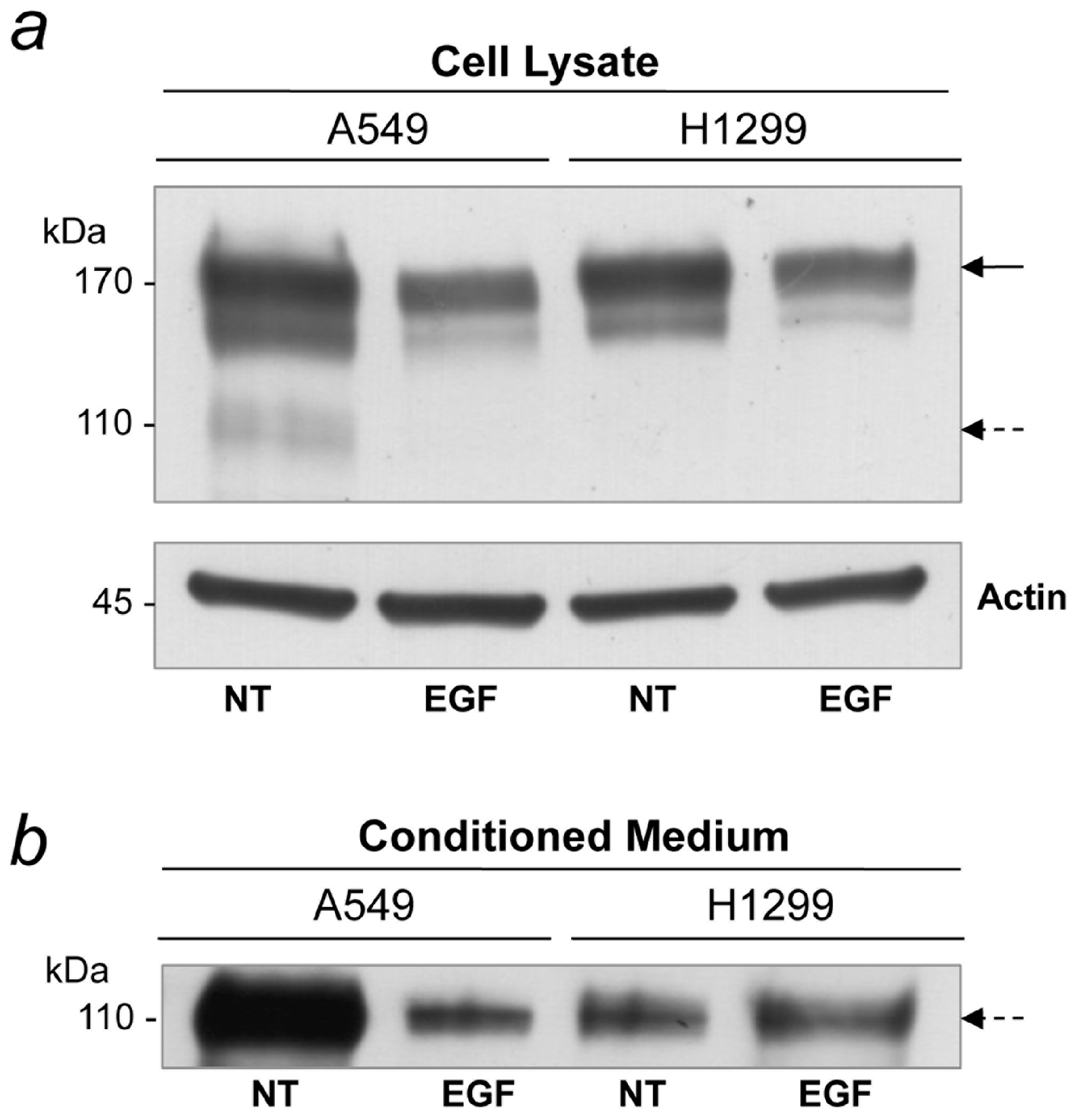
3. Discussion
4. Experimental Section
4.1. Patients and Control Subjects
4.2. Clinical Data Collection and Statistical Analysis
4.3. Enzyme-Linked Immunosorbent Assay (ELISA)
4.4. Fluorescent in Situ Hybridization (FISH)
4.5. Cell Culture Conditions, Cell Lysates and Conditioned Medium
4.6. Treatments
4.7. Cell Migration Assay
4.8. Western Blot Analysis
4.9. Immunofluorescence and Microscopy Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed]
- Infante, M.; Cavuto, S.; Lutman, F.R.; Passera, E.; Chiarenza, M.; Chiesa, G.; Brambilla, G.; Angeli, E.; Aranzulla, G.; Chiti, A. Long-term follow-up results of the DANTE trial, a randomized study of lung cancer screening with spiral computed tomography. Am. J. Respir. Crit. Care Med. 2015, 191, 1166–1175. [Google Scholar] [CrossRef] [PubMed]
- National Lung Screening Trial Research Team. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011, 365, 395–409. [Google Scholar]
- The International Early Lung Cancer Action Program Investigators. Survival of patients with stage I lung cancer detected on CT screening. N. Engl. J. Med. 2006, 355, 1763–1771. [Google Scholar]
- Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2000, 103, 211–225. [Google Scholar] [CrossRef]
- Peter, B.-J.; Tony, H. Oncogenic kinase signaling. Nature 2001, 411, 355–365. [Google Scholar]
- Yarden, Y.; Shilo, B.Z. SnapShot: EGFR signaling pathway. Cell 2007, 131. [Google Scholar] [CrossRef] [PubMed]
- Marianela, P.-T.; Valle, B.L.; Maihle, N.J.; Maihle, N.J.; Lisandra, N.-V.; Rene, N.-A.; Elsa, M.C. Shedding of epidermal growth factor receptor is a regulated process that occurs with overexpression in malignant cells. Exp. Cell Res. 2008, 314, 2907–2918. [Google Scholar]
- Baron, A.T.; Cora, E.M.; Lafky, J.M.; Boardman, C.H.; Buenafe, M.C.; Rademaker, A.; Liu, D.; Fishman, D.A.; Podratz, K.C.; Maihle, N.J. Soluble epidermal growth factor receptor (sEGFR/sErbB1) as a potential risk, screening, and diagnostic serum biomarker of epithelial ovarian cancer. Cancer Epidemiol. Biomark. Prev. 2003, 12, 103–113. [Google Scholar]
- Reiter, J.L.; Threadgill, D.W.; Eley, G.D.; Strunk, K.E.; Danielsen, A.J.; Sinclair, C.S.; Pearsall, R.S.; Green, P.J.; Yee, D.; Lampland, A.L.; et al. Comparative genomic sequence analysis and isolation of human and mouse alternative EGFR transcripts encoding truncated receptor isoforms. Genomics 2001, 71, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Reiter, J.L.; Maihle, N.J. Characterization and expression of novel 60 kDa and 110 kDa EGFR isoforms in human placenta. Ann. N. Y. Acad. Sci. 2003, 995, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Lafky, J.M.; Wilken, J.A.; Baron, A.T.; Maihle, N.J. Clinical implications of the ErbB/epidermal growth factor (EGF) receptor family and its ligands in ovarian cancer. Biochim. Biophys. Acta 2008, 1785, 232–265. [Google Scholar] [CrossRef] [PubMed]
- Baron, A.T.; Boardman, C.H.; Lafky, J.M.; Rademaker, A.; Liu, D.; Fishman, D.A.; Podratz, K.C.; Maihle, N.J. Soluble epidermal growth factor receptor (sEGFR) and cancer antigen 125 (CA125) as screening and diagnostic tests for epithelial ovarian cancer. Cancer Epidemiol. Biomark. Prev. 2005, 14, 306–318. [Google Scholar] [CrossRef] [PubMed]
- Müller, V.; Witzel, I.; Pantel, K.; Krenkel, S.; Lück, H.J.; Neumann, R.; Keller, T.; Dittmer, J.; Jänicke, F.; Thomssen, C. Prognostic and predictive impact of soluble epidermal growth factor receptor (sEGFR) protein in the serum of patients treated with chemotherapy for metastatic breast cancer. Anticancer Res. 2006, 26, 1479–1487. [Google Scholar] [PubMed]
- Asgeirsson, K.S.; Agrawal, A.; Allen, C.; Hitch, A.; Ellis, I.O.; Chapman, C.; Cheung, K.L.; Robertson, J.F. Serum epidermal growth factor receptor and HER2 expression in primary and metastatic breast cancer patients. Breast Cancer Res. 2007, 9. [Google Scholar] [CrossRef] [PubMed]
- Jantus-Lewintre, E.; Sirera, R.; Cabrera, A.; Blasco, A.; Caballero, C.; Iranzo, V.; Rosell, R.; Camps, C. Analysis of the prognostic value of soluble epidermal growth factor receptor plasma concentration in advanced non-small-cell lung cancer patients. Clin. Lung Cancer 2011, 12, 320–327. [Google Scholar] [CrossRef] [PubMed]
- Gregorc, V.; Ceresoli, G.L.; Floriani, I.; Spreafico, A.; Bencardino, K.B.; Ludovini, V.; Pistola, L.; Mihaylova, Z.; Tofanetti, F.R.; Ferraldeschi, M.; et al. Effects of Gefitinib on serum epidermal growth factor receptor and HER2 in patients with advanced non-small cell lung cancer. Clin. Cancer Res. 2004, 10, 6006–6012. [Google Scholar] [CrossRef] [PubMed]
- Carney, W.P. Circulating oncoproteins HER2/neu, EGFR and CAIX (MN) as novel cancer biomarkers. Expert Rev. Mol. Diagn. 2007, 7, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Lemos-González, Y.; Rodríguez-Berrocal, F.J.; Cordero, O.J.; Gómez, C.; Páez de la Cadena, M. Alteration of the serum levels of the epidermal growth factor receptor and its ligands in patients with non-small cell lung cancer and head and neck carcinoma. Br. J. Cancer 2007, 96, 1569–1578. [Google Scholar] [CrossRef] [PubMed]
- Maramotti, S.; Paci, M.; Miccichè, F.; Ciarrocchi, A.; Cavazza, A.; de Bortoli, M.; Vaghi, E.; Formisano, D.; Canovi, L.; Sgarbi, G.; et al. Soluble epidermal growth factor receptor isoforms in non-small cell lung cancer tissue and in blood. Lung Cancer 2012, 76, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Heaney, M.L.; Golde, D.W. Soluble receptors in human disease. J. Leukoc. Biol. 1998, 64, 135–146. [Google Scholar] [PubMed]
- Aberle, D.R.; Adams, A.M.; Berg, C.D.; Black, W.C.; Clapp, J.D.; Fagerstrom, R.M.; Gareen, I.F.; Gatsonis, C.; Marcus, P.M.; Sicks, J.D. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011, 365, 395–409. [Google Scholar] [PubMed]
- Pavelic, K.; Banjac, Z.; Pavelic, J.; Spaventi, S. Evidence for a role of EGF receptor in the progression of human lung-carcinoma. Anticancer Res. 1993, 13, 1133–1138. [Google Scholar] [PubMed]
- Hirsch, F.R.; Varella-Garcia, M.; Bunn, P.A.; di Maria, M.V.; Veve, R.; Bremmes, R.M.; Barón, A.E.; Zeng, C.; Franklin, W.A. Epidermal growth factor receptor in non-small-cell lung carcinomas: Correlation between gene copy number and protein expression and impact on prognosis. J. Clin. Oncol. 2003, 21, 3798–3807. [Google Scholar] [CrossRef] [PubMed]
- Domingo, G.; Perez, C.A.; Velez, M.; Cudris, J.; Raez, L.E.; Santos, E.S. EGF receptor in lung cancer: A successful story of targeted therapy. Expert Rev. Anticancer Ther. 2010, 10, 1577–1587. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.H.; Obuchowski, N.; McClish, D. Statistical Methods in Diagnostic Medicine, 2nd ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2011. [Google Scholar]
- Carney, W.P.; Burrell, M.; Morris, L.D.; Hamer, P.J. Normal levels of serum EGFr and decreases in several cancers. Proc. AACR 2002, 43, 47. [Google Scholar]
- Riquet, M.; Manac’h, D.; Le Pimpec Barthes, F.; Dujon, A.; Debrosse, D.; Debesse, B. Prognostic value of T and N in non-small cell lung cancer three centimeters or less in diameter. Eur. J. Cardiothorac. Surg. 1997, 11, 440–443. [Google Scholar] [CrossRef]
- Siegelin, M.D.; Borczuk, A.C. Epidermal growth factor receptor mutations in lung adenocarcinoma. Lab. Investig. 2014, 94, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Citri, A.; Yarden, Y. EGF–ERBB signalling: Towards the systems level. Nat. Rev. Mol. Cell Biol. 2006, 7, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J. D-Type Cyclins. Trends Biochem. Sci. 1995, 20, 187–190. [Google Scholar] [CrossRef]
- Lin, S.Y.; Makino, K.; Xia, W.; Matin, A.; Wen, Y.; Kwong, K.Y.; Bourguignon, L.; Hung, M.C. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell Biol. 2001, 3, 802–808. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.C.; Hung, M.C. Nuclear translocation of the epidermal growth factor receptor family membrane tyrosine kinase receptors. Clin. Cancer Res. 2009, 15, 6484–6489. [Google Scholar] [CrossRef] [PubMed]
- Varella-Garcia, M.; Diebold, J.; Eberhard, D.A.; Geenen, K.; Hirschmann, A.; Kockx, M.; Nagelmeier, I.; Rüschoff, J.; Schmitt, M.; Arbogast, S.; et al. EGFR fluorescence in situ hybridisation assay: Guidelines for application to non-small-cell lung cancer. J. Clin. Pathol. 2009, 62, 970–977. [Google Scholar] [CrossRef] [PubMed]
- Lynch, T.J.; Bell, D.W.; Sordella, R.; Gurubhagavatula, S.; Okimoto, R.A.; Brannigan, B.W.; Harris, P.L.; Haserlat, S.M.; Supko, J.G.; Haluska, F.G.; et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small cell lung cancer to gefitinib. N. Engl. J. Med. 2004, 350, 2129–2139. [Google Scholar] [CrossRef] [PubMed]
- Albini, A.; Benelli, R. The chemoinvasion assay: A method to assess tumor and endothelial cell invasion and its modulation. Nat. Protoc. 2007, 2, 504–511. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lococo, F.; Paci, M.; Rapicetta, C.; Rossi, T.; Sancisi, V.; Braglia, L.; Cavuto, S.; Bisagni, A.; Bongarzone, I.; Noonan, D.M.; et al. Preliminary Evidence on the Diagnostic and Molecular Role of Circulating Soluble EGFR in Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2015, 16, 19612-19630. https://doi.org/10.3390/ijms160819612
Lococo F, Paci M, Rapicetta C, Rossi T, Sancisi V, Braglia L, Cavuto S, Bisagni A, Bongarzone I, Noonan DM, et al. Preliminary Evidence on the Diagnostic and Molecular Role of Circulating Soluble EGFR in Non-Small Cell Lung Cancer. International Journal of Molecular Sciences. 2015; 16(8):19612-19630. https://doi.org/10.3390/ijms160819612
Chicago/Turabian StyleLococo, Filippo, Massimiliano Paci, Cristian Rapicetta, Teresa Rossi, Valentina Sancisi, Luca Braglia, Silvio Cavuto, Alessandra Bisagni, Italia Bongarzone, Douglas M. Noonan, and et al. 2015. "Preliminary Evidence on the Diagnostic and Molecular Role of Circulating Soluble EGFR in Non-Small Cell Lung Cancer" International Journal of Molecular Sciences 16, no. 8: 19612-19630. https://doi.org/10.3390/ijms160819612
APA StyleLococo, F., Paci, M., Rapicetta, C., Rossi, T., Sancisi, V., Braglia, L., Cavuto, S., Bisagni, A., Bongarzone, I., Noonan, D. M., Albini, A., & Maramotti, S. (2015). Preliminary Evidence on the Diagnostic and Molecular Role of Circulating Soluble EGFR in Non-Small Cell Lung Cancer. International Journal of Molecular Sciences, 16(8), 19612-19630. https://doi.org/10.3390/ijms160819612