Over-Expression of Rice CBS Domain Containing Protein, OsCBSX3, Confers Rice Resistance to Magnaporthe oryzae Inoculation
Abstract
:1. Introduction
2. Results
2.1. The Sequence Analysis of OsCBSX3 (Cystathionine β-Synthase (CBS) Domain Containing Protein 3)
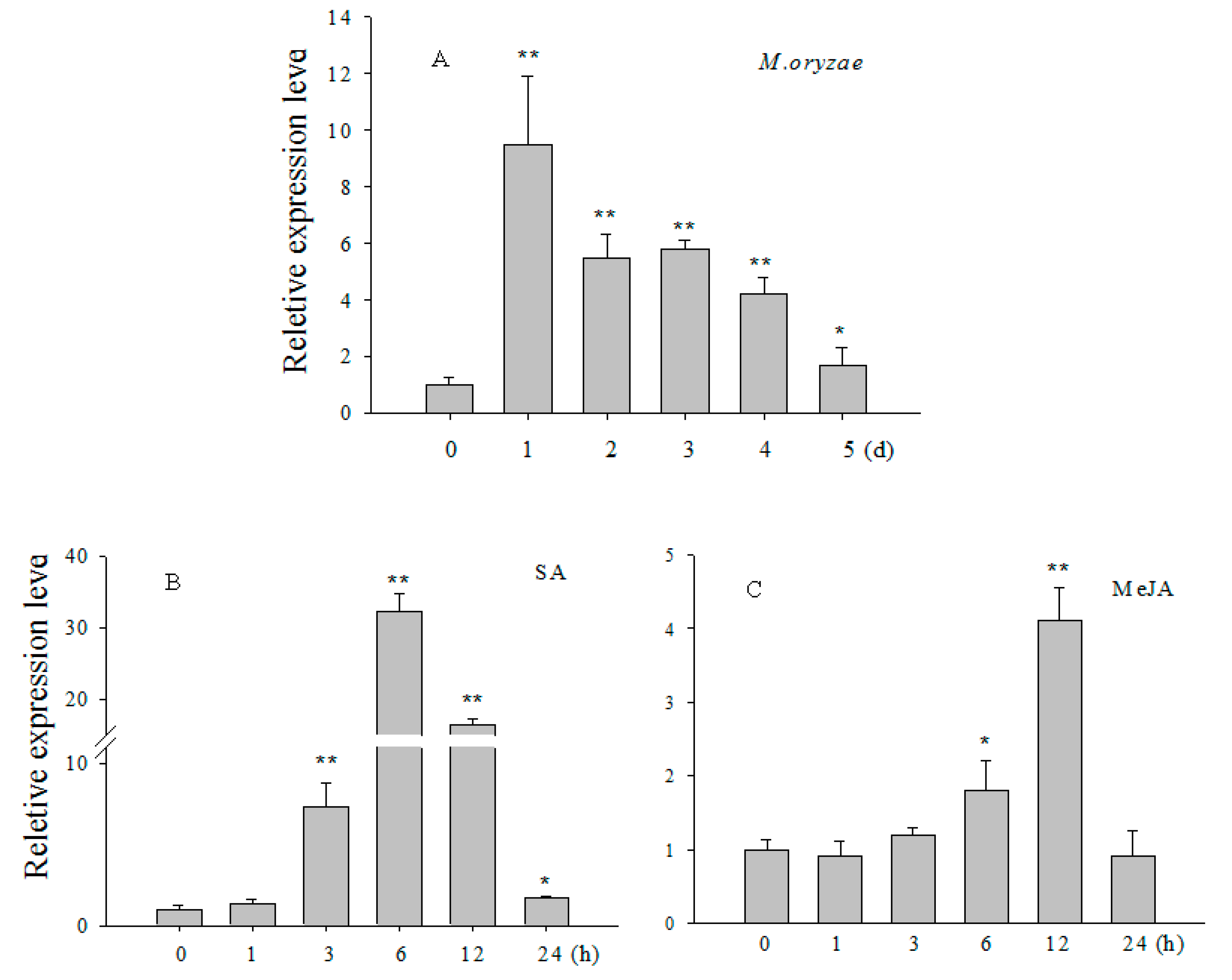
2.2. Transcript Levels of OsCBSX3 Enhanced in Response to M. oryzae Inoculation and Exogenous Application of Salicylic Acid (SA) and Methyl Jasmonate (MeJA)

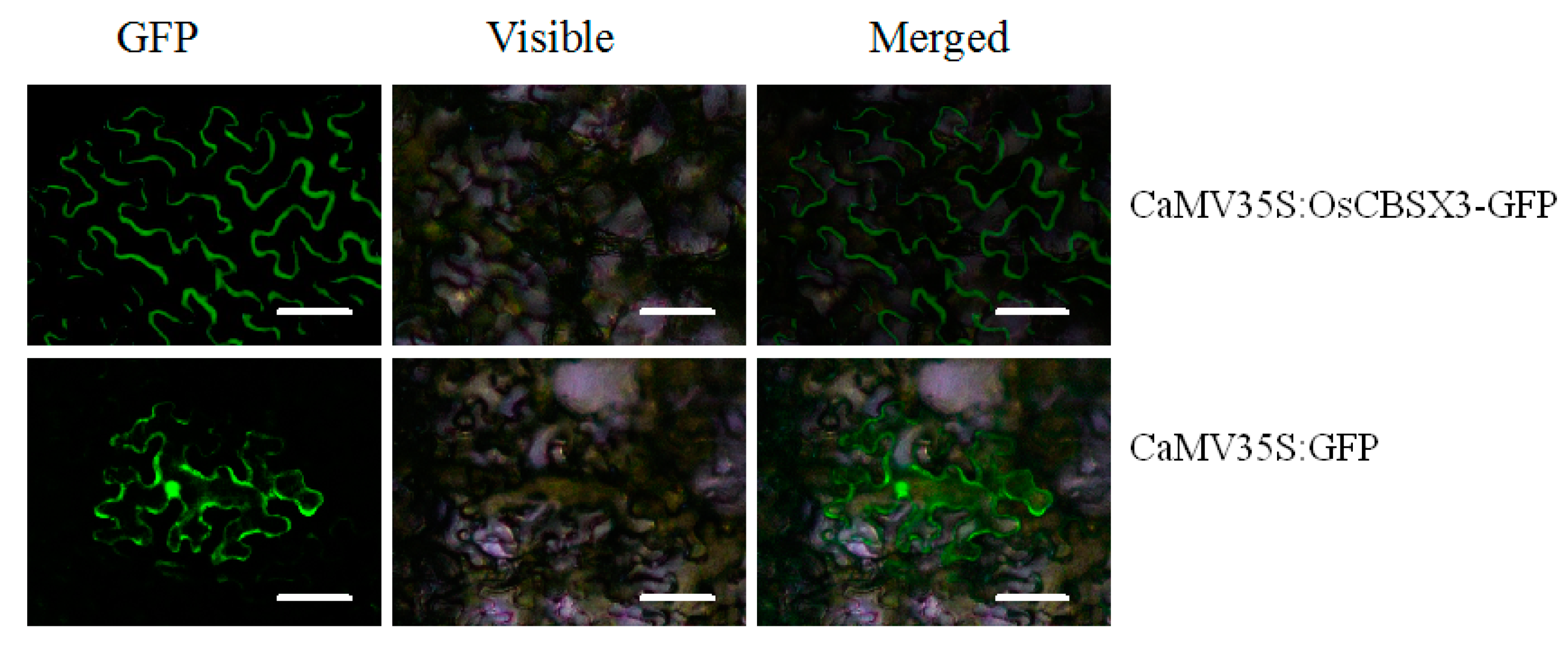
2.3. OsCBSX3 Is Localized in the Plasma Membrane
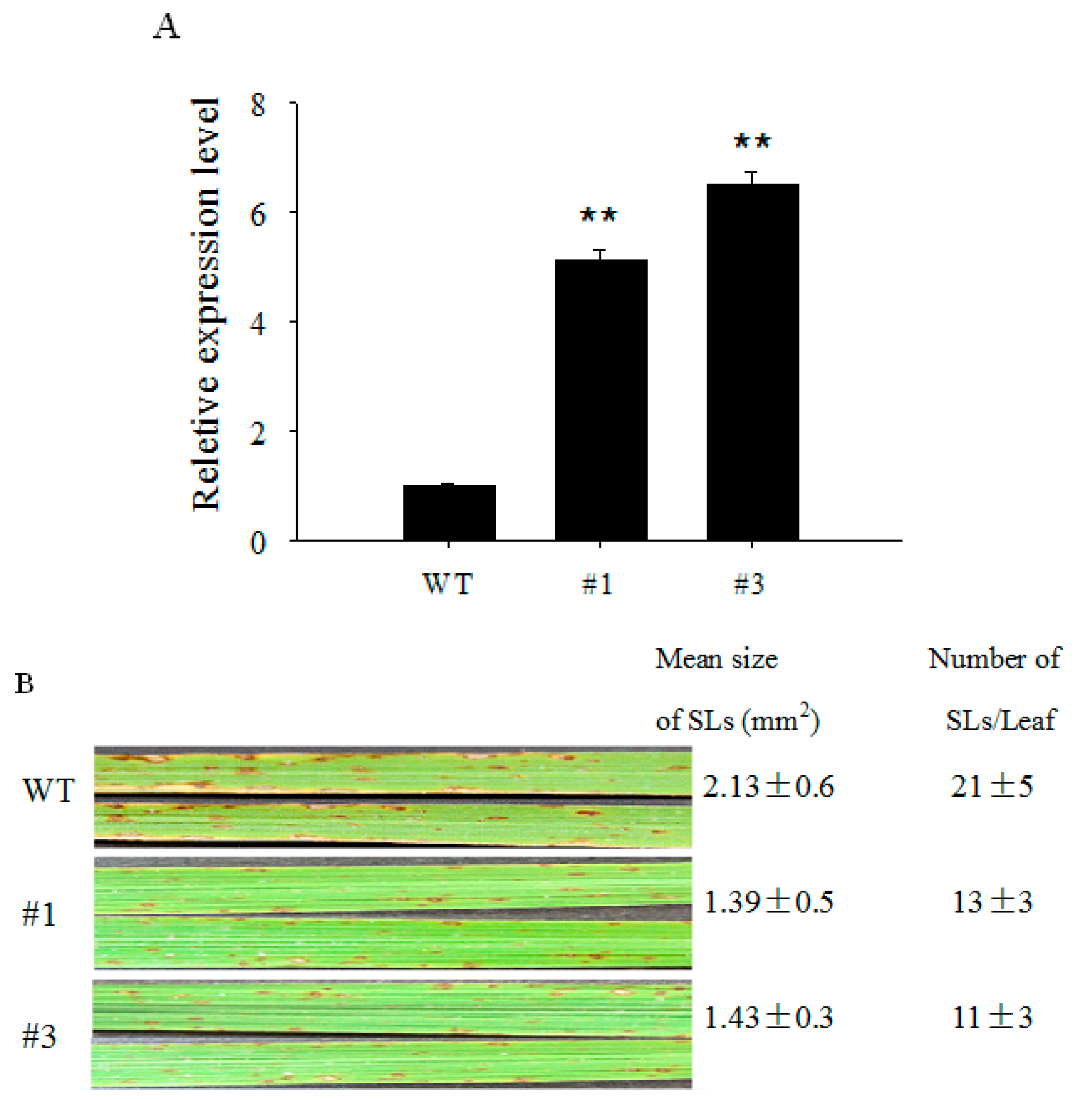
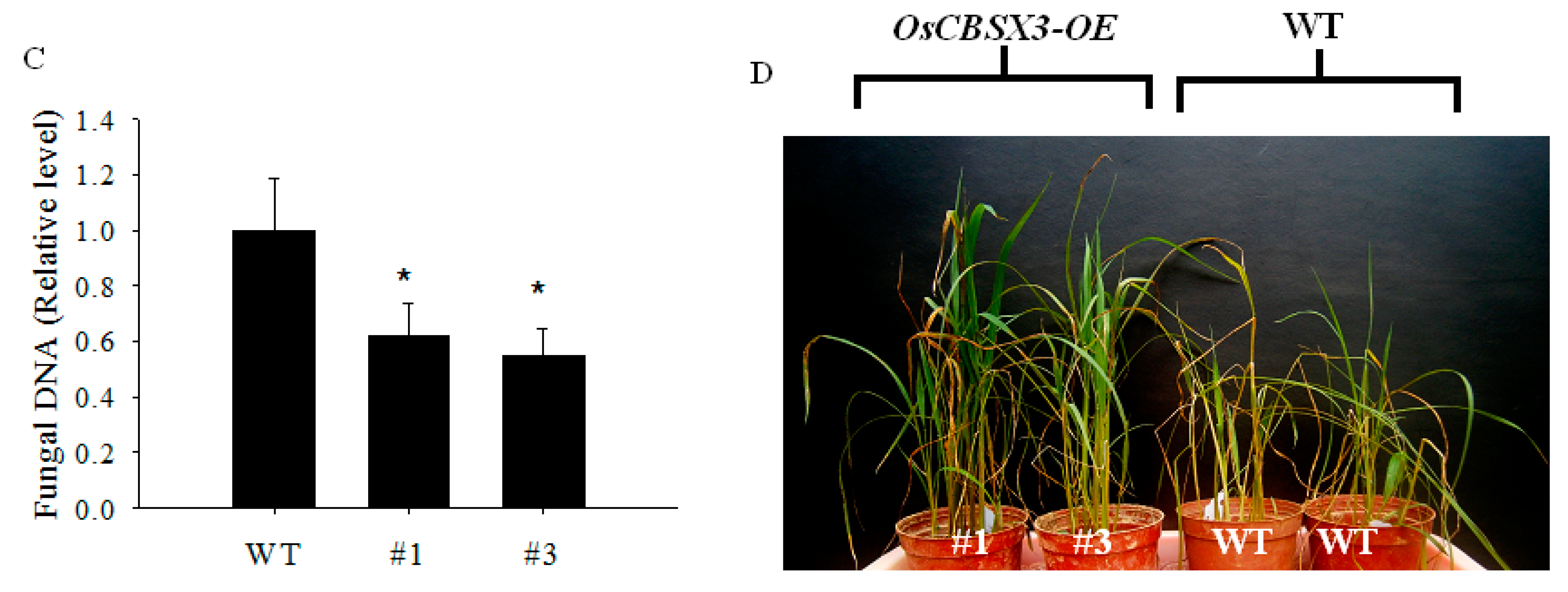
2.4. Over-Expression of OsCBSX3 in Transgenic Rice Plants Conferred Enhanced Resistance to M. oryzae Inoculation
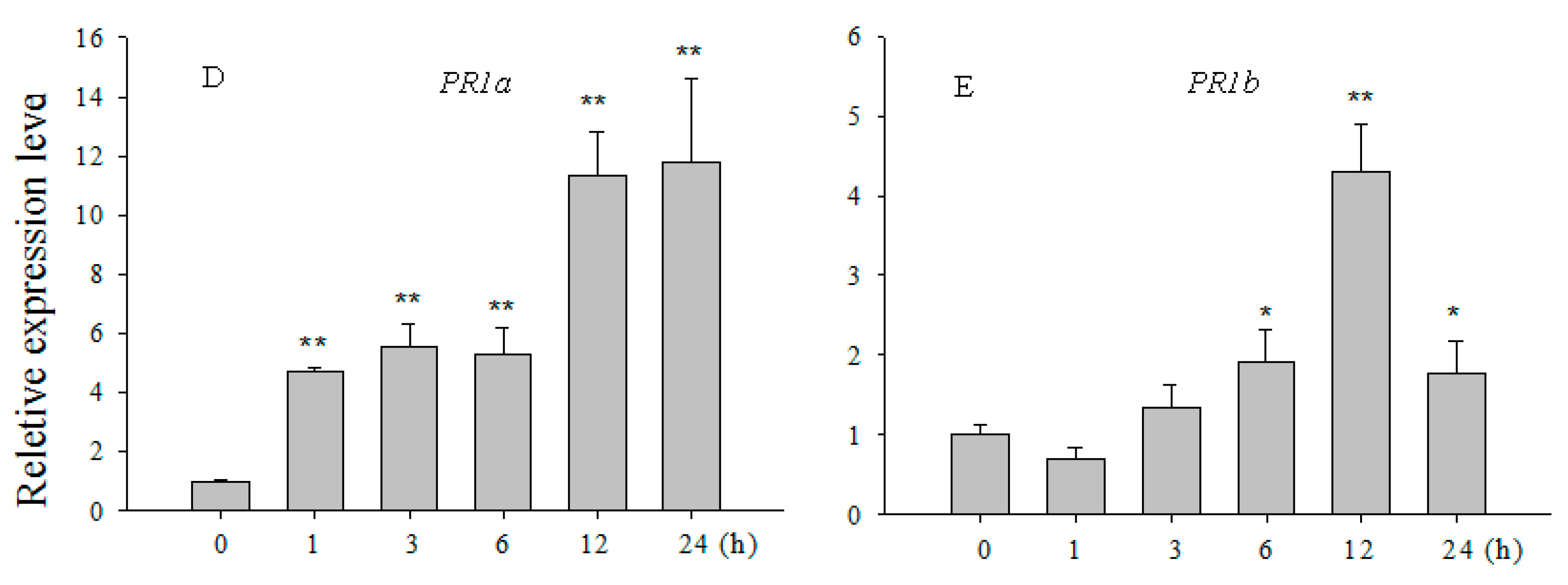
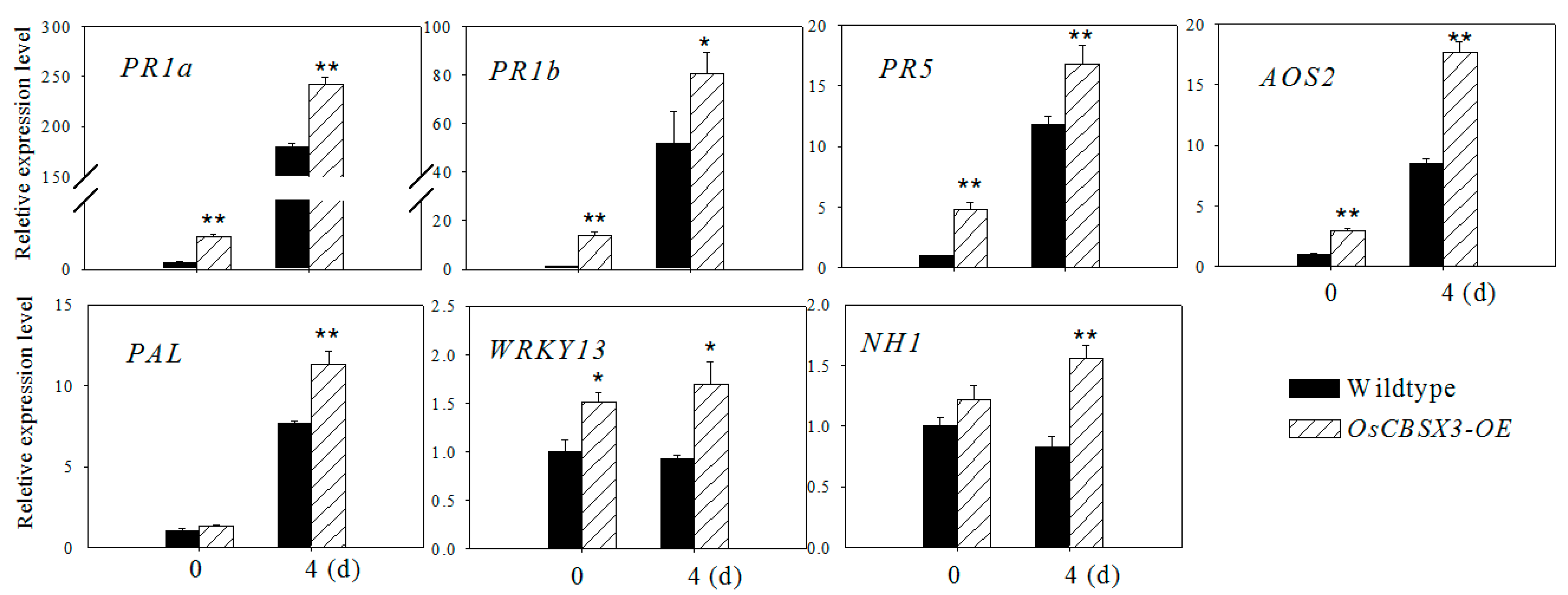
2.5. Over-Expression of OsCBSX3 Up-Regulated the Transcript Levels of Defense Marker Genes
3. Discussion
4. Experimental Section
4.1. Plant Growth and Treatments
4.2. Cloning, Plasmid Construction and Rice Transformation
4.3. RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
| Gene Name | Forward Primer Sequence (5′–3′) | Reverse Primer Sequence (5′–3′) |
|---|---|---|
| CBSX3 | GAGGAGGTTGAGTGCCACTTTG | GCCGCATCCATTACTGTTTTGTC |
| PR1a | CGTCTTCATCACCTGCAACTACTC | CATGCATAAACACGTAGCATAGCA |
| PR1b | GGCAACTTCGTCGGACAGA | CCGTGGACCTGTTTACATTTTCA |
| PR5 | CAACAGCAACTACCAAGTCGTCTT | CAAGGTGTCGTTTTATTCATCAACTTT |
| NH1 | CACGCCTAAGCCTCGGATTA | TCAGTGAGCAGCATCCTGACTAG |
| WRKY13 | TCAGTGGAGAAGCGGGTGGTG | GGGTGGTTGTGCTCGAAGGAG |
| AOS2 | CAATACGTGTACTGGTCGAATGG | AAGGTGTCGTACCGGAGGAA |
| PAL | AGCACATCTTGGAGGGAAGCT | GCGCGGATAACCTCAATTTG |
| Actin | TGTATGCCAGTGGTCGTACCA | CCAGCAAGGTCGAGACGAA |
4.4. Subcellular Locatization
4.5. Quantification of M. oryzae DNA in Rice Leaves
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ali, A.; Alexandersson, E.; Sandin, M.; Resjo, S.; Lenman, M.; Hedley, P.; Levander, F.; Andreasson, E. Quantitative proteomics and transcriptomics of potato in response to Phytophthora infestans in compatible and incompatible interactions. BMC Genomics 2014, 15, 497. [Google Scholar] [CrossRef] [PubMed]
- Rosli, H.G.; Zheng, Y.; Pombo, M.A.; Zhong, S.; Bombarely, A.; Fei, Z.; Collmer, A.; Martin, G.B. Transcriptomics-based screen for genes induced by flagellin and repressed by pathogen effectors identifies a cell wall-associated kinase involved in plant immunity. Genome Biol. 2013, 14, R139. [Google Scholar] [CrossRef] [PubMed]
- Mathioni, S.M.; Patel, N.; Riddick, B.; Sweigard, J.A.; Czymmek, K.J.; Caplan, J.L.; Kunjeti, S.G.; Kunjeti, S.; Raman, V.; Hillman, B.I.; et al. Transcriptomics of the rice blast fungus Magnaporthe oryzae in response to the bacterial antagonist Lysobacter enzymogenes reveals candidate fungal defense response genes. PLoS ONE 2013, 8, e76487. [Google Scholar] [CrossRef] [PubMed]
- Rudd, J.J.; Kanyuka, K.; Hassani-Pak, K.; Derbyshire, M.; Andongabo, A.; Devonshire, J.; Lysenko, A.; Saqi, M.; Desai, N.M.; Powers, S.J.; et al. Transcriptome and metabolite profiling of the infection cycle of Zymoseptoria tritici on wheat (Triticum aestivum) reveals a biphasic interaction with plant immunity involving differential pathogen chromosomal contributions and a variation on the hemibiotrophic lifestyle definition. Plant Physiol. 2015, 167, 1158–1185. [Google Scholar] [PubMed]
- Douchkov, D.; Luck, S.; Johrde, A.; Nowara, D.; Himmelbach, A.; Rajaraman, J.; Stein, N.; Sharma, R.; Kilian, B.; Schweizer, P. Discovery of genes affecting resistance of barley to adapted and non-adapted powdery mildew fungi. Genome Biol. 2014, 15, 518. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Jiang, Z.; Peng, Y.L.; Zhang, Z. Revealing shared and distinct gene network organization in Arabidopsis immune responses by integrative analysis. Plant Physiol. 2015, 167, 1186–1203. [Google Scholar] [CrossRef] [PubMed]
- Azizi, P.; Rafii, M.Y.; Abdullah, S.N.; Nejat, N.; Maziah, M.; Hanafi, M.M.; Latif, M.A.; Sahebi, M. Toward understanding of rice innate immunity against Magnaporthe oryzae. Crit. Rev. Biotechnol. 2014, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, K.; Mine, A.; Bethke, G.; Igarashi, D.; Botanga, C.J.; Tsuda, Y.; Glazebrook, J.; Sato, M.; Katagiri, F. Dual regulation of gene expression mediated by extended MAPK activation and salicylic acid contributes to robust innate immunity in Arabidopsis thaliana. PLoS Genet. 2013, 9, e1004015. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wu, Y.; Gao, M.; Zhang, J.; Kong, Q.; Liu, Y.; Ba, H.; Zhou, J.; Zhang, Y. Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 2012, 11, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.; Nurnberger, T.; Joosten, M.H. Of PAMPs and effectors: The blurred PTI-ETI dichotomy. Plant Cell 2011, 23, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Hein, I.; Gilroy, E.M.; Armstrong, M.R.; Birch, P.R. The zig-zag-zig in oomycete–plant interactions. Mol. Plant Pathol. 2009, 10, 547–562. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem. Sci. 1997, 22, 12–13. [Google Scholar] [CrossRef]
- Ignoul, S.; Eggermont, J. CBS domains: Structure, function, and pathology in human proteins. Am. J. Physiol. Cell Physiol. 2005, 289, C1369–C1378. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, H.R.; Singh, A.K.; Sopory, S.K.; Singla-Pareek, S.L.; Pareek, A. Genome wide expression analysis of CBS domain containing proteins in Arabidopsis thaliana (L.) Heynh and Oryza sativa L. reveals their developmental and stress regulation. BMC Genomics 2009, 10, 200. [Google Scholar] [CrossRef] [PubMed]
- Gissot, L.; Polge, C.; Jossier, M.; Girin, T.; Bouly, J.P.; Kreis, M.; Thomas, M. AKINβγ contributes to SnRK1 heterotrimeric complexes and interacts with two proteins implicated in plant pathogen resistance through its KIS/GBD sequence. Plant Physiol. 2006, 142, 931–944. [Google Scholar] [CrossRef] [PubMed]
- Yoo, K.S.; Ok, S.H.; Jeong, B.C.; Jung, K.W.; Cui, M.H.; Hyoung, S.; Lee, M.R.; Song, H.K.; Shin, J.S. Single cystathionine β-synthase domain-containing proteins modulate development by regulating the thioredoxin system in Arabidopsis. Plant Cell 2011, 23, 3577–3594. [Google Scholar] [CrossRef] [PubMed]
- Ramon, M.; Ruelens, P.; Li, Y.; Sheen, J.; Geuten, K.; Rolland, F. The hybrid four-CBS-domain KINβγ subunit functions as the canonical gamma subunit of the plant energy sensor SnRK1. Plant J. 2013, 75, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Kumar, R.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Overexpression of rice CBS domain containing protein improves salinity, oxidative, and heavy metal tolerance in transgenic tobacco. Mol. Biotechnol. 2012, 52, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Ren, X.; Zhu, L.L.; He, G.C. OsBi1, a rice gene, encodes a novel protein with a CBS-like domain and its expression is induced in responses to herbivore feeding. Plant Sci. 2004, 166, 1581–1588. [Google Scholar] [CrossRef]
- Espinoza, C.; Medina, C.; Somerville, S.; Arce-Johnson, P. Senescence-associated genes induced during compatible viral interactions with grapevine and Arabidopsis. J. Exp. Bot. 2007, 58, 3197–3212. [Google Scholar] [CrossRef] [PubMed]
- Fabro, G.; di Rienzo, J.A.; Voigt, C.A.; Savchenko, T.; Dehesh, K.; Somerville, S.; Alvarez, M.E. Genome-wide expression profiling Arabidopsis at the stage of Golovinomyces cichoracearum haustorium formation. Plant Physiol. 2008, 146, 1421–1439. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Sabharwal, V.P.; Kushwaha, H.R.; Sopory, S.K.; Singla-Pareek, S.L.; Pareek, A. Transcriptome map for seedling stage specific salinity stress response indicates a specific set of genes as candidate for saline tolerance in Oryza sativa L. Funct. Integr. Genomics 2009, 9, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Sahu, B.B.; Shaw, B.P. Isolation, identification and expression analysis of salt-induced genes in Suaeda maritima, a natural halophyte, using PCR-based suppression subtractive hybridization. BMC Plant Biol. 2009, 9, 69. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lai, T.; Qin, G.; Tian, S. Response of jujube fruits to exogenous oxalic acid treatment based on proteomic analysis. Plant Cell Physiol. 2009, 50, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.A.; Talbot, N.J.; Ebbole, D.J.; Farman, M.L.; Mitchell, T.K.; Orbach, M.J.; Thon, M.; Kulkarni, R.; Xu, J.R.; Pan, H.; et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 2005, 434, 980–986. [Google Scholar] [CrossRef] [PubMed]
- Li, R.Y.; Wu, X.M.; Yin, X.H.; Liang, J.N.; Li, M. The natural product citral can cause significant damage to the hyphal cell walls of Magnaporthe grisea. Molecules 2014, 19, 10279–10290. [Google Scholar] [CrossRef] [PubMed]
- Ahn, I.P. Glufosinate ammonium-induced pathogen inhibition and defense responses culminate in disease protection in bar-transgenic rice. Plant Physiol. 2008, 146, 213–227. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Liu, H.; Qiu, D.; Zhou, Y.; Li, X.; Xu, C.; Wang, S. A pair of allelic WRKY genes play opposite roles in rice–bacteria interactions. Plant Physiol. 2009, 151, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Mou, S.; Liu, Z.; Guan, D.; Qiu, A.; Lai, Y.; He, S. Functional analysis and expressional characterization of rice ankyrin repeat-containing protein, OsPIANK1, in basal defense against Magnaporthe oryzae attack. PLoS ONE 2013, 8, e59699. [Google Scholar] [CrossRef] [PubMed]
- Qiu, D.; Xiao, J.; Ding, X.; Xiong, M.; Cai, M.; Cao, Y.; Li, X.; Xu, C.; Wang, S. OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol. Plant Microbe Interact. 2007, 20, 492–499. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.F.; Liu, H.B.; Li, Y.; Li, X.H.; Xu, C.G.; Long, M.Y.; Wang, S.P. A rice gene of de novo origin negatively regulates pathogen-induced defense response. PLoS ONE 2009, 4, e4603. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Liu, H.; Yuan, B.; Li, X.; Xu, C.; Wang, S. OsEDR1 negatively regulates rice bacterial resistance via activation of ethylene biosynthesis. Plant Cell Environ. 2011, 34, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Qiu, D.; Xiao, J.; Xie, W.; Liu, H.; Li, X.; Xiong, L.; Wang, S. Rice gene network inferred from expression profiling of plants overexpressing OsWRKY13, a positive regulator of disease resistance. Mol. Plant 2008, 1, 538–551. [Google Scholar] [CrossRef] [PubMed]
- Mei, C.S.; Qi, M.; Sheng, G.Y.; Yang, Y.N. Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol. Plant Microbe Interact. 2006, 19, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Cecchini, N.M.; Jung, H.W.; Engle, N.; Tschaplinski, T.J.; Greenberg, J. ALD1 regulates basal immune components and early inducible defense responses in Arabidopsis. Mol. Plant Microbe Interact. 2015, 28, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Djami-Tchatchou, A.T.; Maake, M.P.; Piater, L.A.; Dubery, I.A. Isonitrosoacetophenone drives transcriptional reprogramming in Nicotiana tabacum cells in support of innate immunity and defense. PLoS ONE 2015, 10, e0117377. [Google Scholar] [CrossRef] [PubMed]
- Beckers, G.J.; Spoel, S.H. Fine-tuning plant defence signalling: Salicylate versus jasmonate. Plant Biol. (Stuttg.) 2006, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, K.; Sato, M.; Stoddard, T.; Glazebrook, J.; Katagiri, F. Network properties of robust immunity in plants. PLoS Genet. 2009, 5, e1000772. [Google Scholar] [CrossRef] [PubMed]
- Proietti, S.; Bertini, L.; Timperio, A.M.; Zolla, L.; Caporale, C.; Caruso, C. Crosstalk between salicylic acid and jasmonate in Arabidopsis investigated by an integrated proteomic and transcriptomic approach. Mol. Biosyst. 2013, 9, 1169–1187. [Google Scholar] [CrossRef] [PubMed]
- Toki, S.; Hara, N.; Ono, K.; Onodera, H.; Tagiri, A.; Oka, S.; Tanaka, H. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 2006, 47, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Curtis, M.D.; Grossniklaus, U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003, 133, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Yang, Y. Quantification of Magnaporthe grisea during infection of rice plants using real-time polymerase chain reaction and northern blot/phosphoimaging analyses. Phytopathology 2002, 92, 870–876. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mou, S.; Shi, L.; Lin, W.; Liu, Y.; Shen, L.; Guan, D.; He, S. Over-Expression of Rice CBS Domain Containing Protein, OsCBSX3, Confers Rice Resistance to Magnaporthe oryzae Inoculation. Int. J. Mol. Sci. 2015, 16, 15903-15917. https://doi.org/10.3390/ijms160715903
Mou S, Shi L, Lin W, Liu Y, Shen L, Guan D, He S. Over-Expression of Rice CBS Domain Containing Protein, OsCBSX3, Confers Rice Resistance to Magnaporthe oryzae Inoculation. International Journal of Molecular Sciences. 2015; 16(7):15903-15917. https://doi.org/10.3390/ijms160715903
Chicago/Turabian StyleMou, Shaoliang, Lanping Shi, Wei Lin, Yanyan Liu, Lei Shen, Deyi Guan, and Shuilin He. 2015. "Over-Expression of Rice CBS Domain Containing Protein, OsCBSX3, Confers Rice Resistance to Magnaporthe oryzae Inoculation" International Journal of Molecular Sciences 16, no. 7: 15903-15917. https://doi.org/10.3390/ijms160715903
APA StyleMou, S., Shi, L., Lin, W., Liu, Y., Shen, L., Guan, D., & He, S. (2015). Over-Expression of Rice CBS Domain Containing Protein, OsCBSX3, Confers Rice Resistance to Magnaporthe oryzae Inoculation. International Journal of Molecular Sciences, 16(7), 15903-15917. https://doi.org/10.3390/ijms160715903






