Extracellular Production of a Novel Endo-β-Agarase AgaA from Pseudomonas vesicularis MA103 that Cleaves Agarose into Neoagarotetraose and Neoagarohexaose
Abstract
:1. Introduction
2. Results and Discussion
2.1. Domain Prediction of AgaA from P. vesicularis MA103
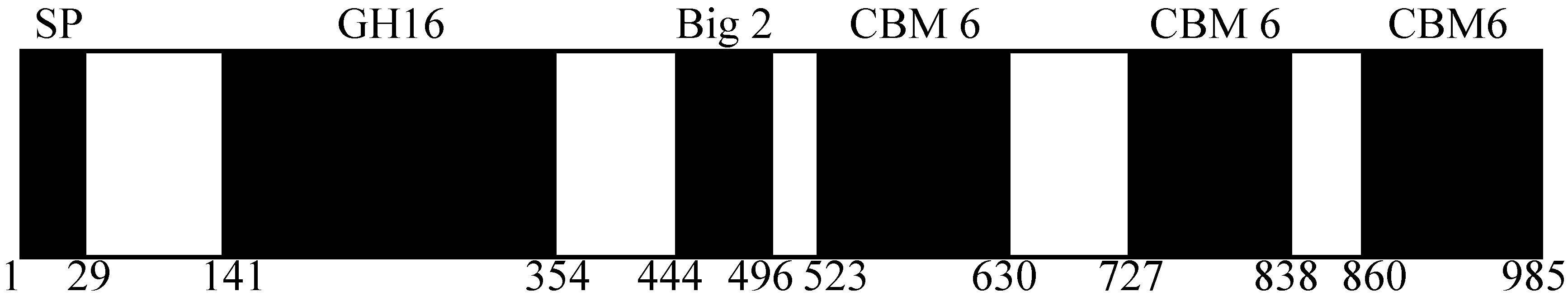
2.2. Cloning
2.3. Effect of Temperature on AgaA Overexpression
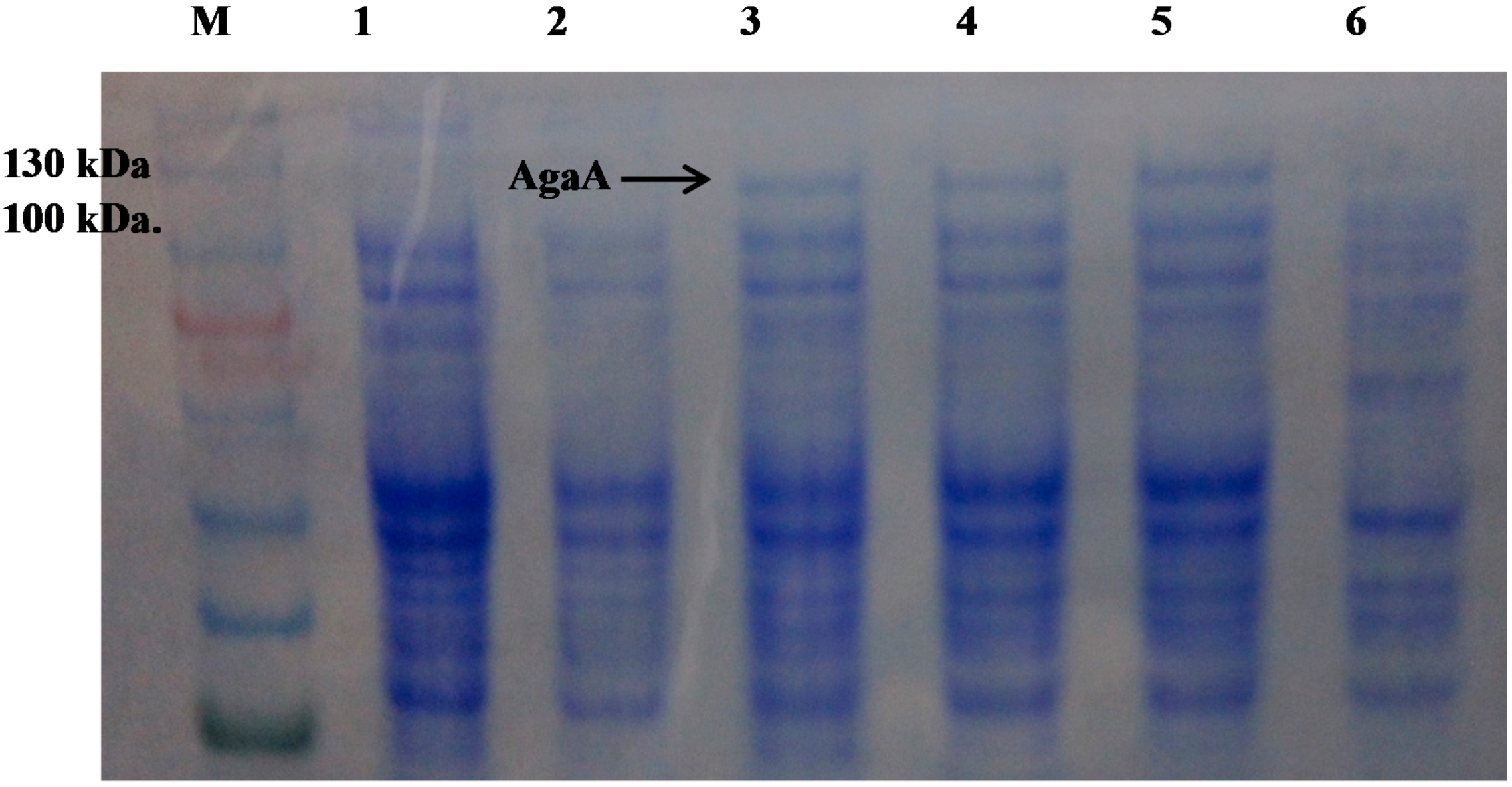
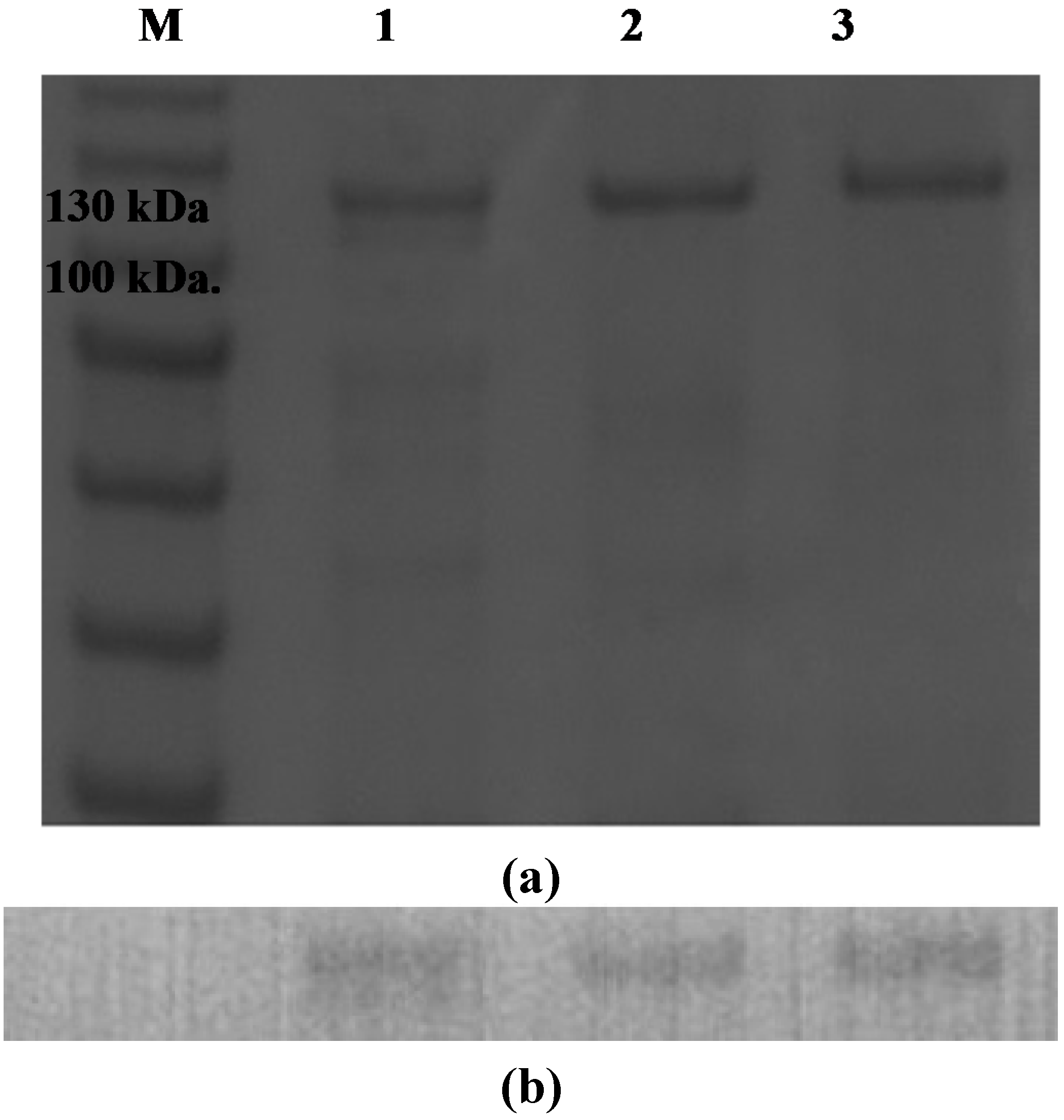
2.4. Effect of the N-Terminal Signal Peptide on the Overexpression of Functional AgaA
| Protein Source * | Total Protein (mg) | Total Activity (units) | Specific Activity (units/mg) |
|---|---|---|---|
| AgaA-FL | 12.2 | 5192.3 | 425.6 |
| AgaA-∆SP | 18.3 | 689.9 | 37.7 |
| AgaA-PelB | 6.2 | 816.8 | 132.6 |
2.5. AgaA from P. vesicularis Is an Endo-Type β-Agarase Yielding Neoagarotetraose and Neoagarohexaose Products
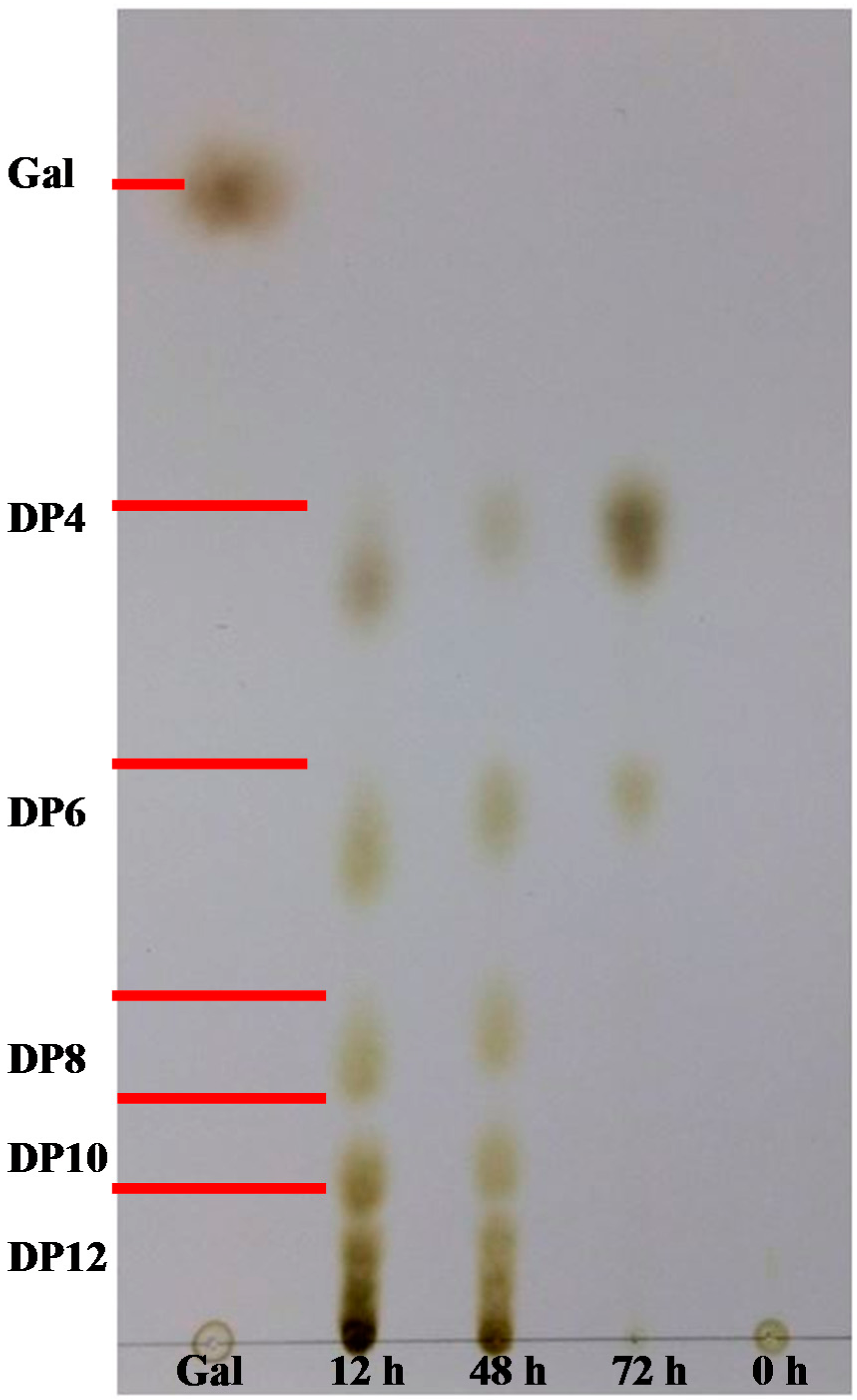
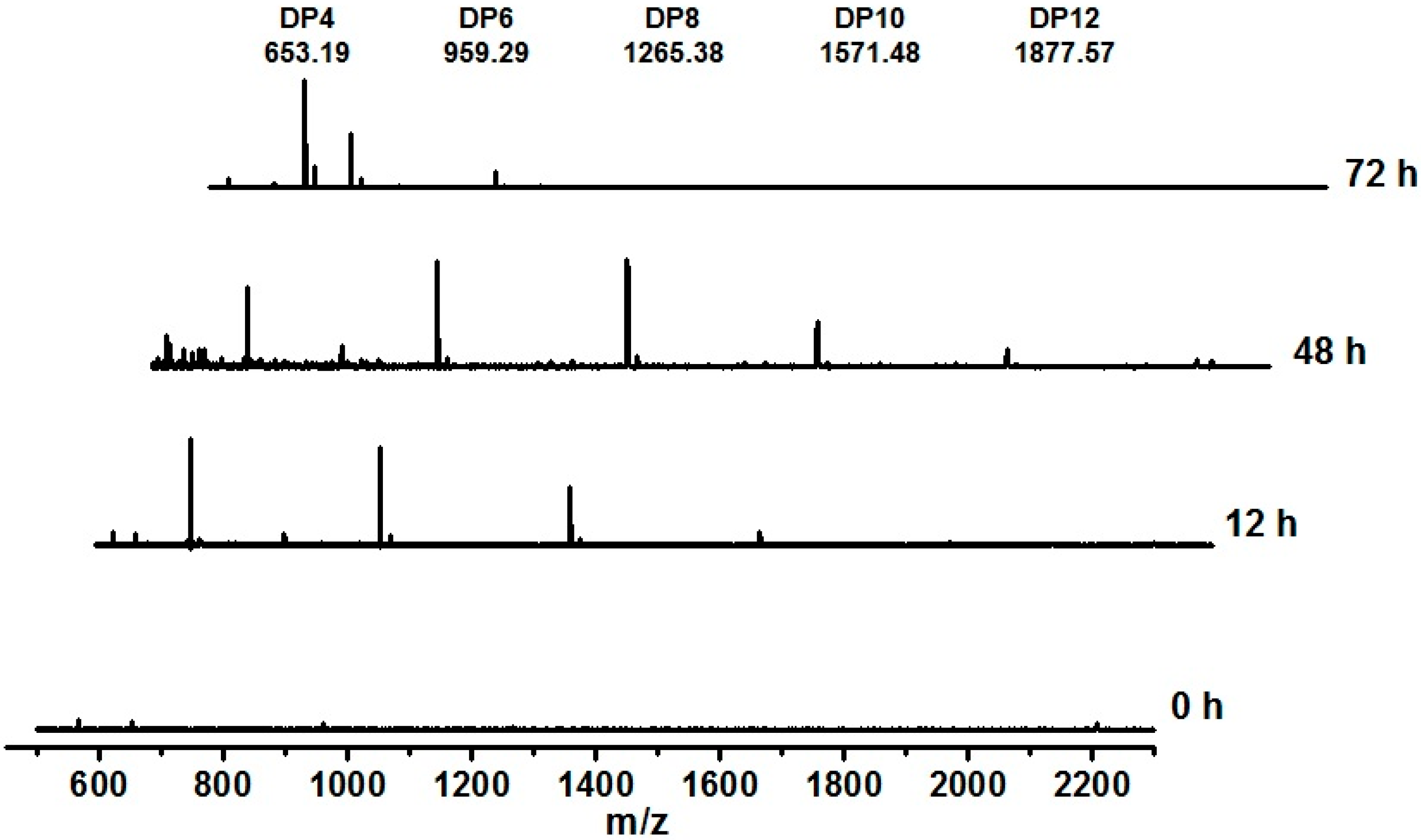
3. Experimental Section
3.1. Isolation of Pseudomonas vesicularis MA103
3.2. Cloning of AgaA
3.3. AgaA Overexpression and Purification
3.4. Western Blot
3.5. Enzyme Activity Assay
3.6. Time Course Hydrolysis of Agarose
3.7. Thin-Layer Chromatography (TLC)
3.8. Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS)
3.9. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chi, W.J.; Chang, Y.K.; Hong, S.K. Agar degradation by microorganisms and agar-degrading enzymes. Appl. Microbiol. Biotechnol. 2012, 94, 917–930. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.T.; Kim, S.M. Agarase: Review of major sources, categories, purification method, enzyme characteristics and applications. Mar. Drugs 2010, 8, 200–218. [Google Scholar] [CrossRef] [PubMed]
- Ohta, Y.; Hatada, Y.; Nogi, Y.; Miyazaki, M.; Li, Z.; Akita, M.; Hidaka, Y.; Goda, S.; Ito, S.; Horikoshi, K. Enzymatic properties and nucleotide and amino acid sequences of a thermostable β-agarase from a novel species of deep-sea Microbulbifer. Appl. Microbiol. Biotechnol. 2004, 64, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.G.; Jang, M.K.; Lee, O.H.; Kim, N.Y.; Ju, S.A.; Lee, S.H. Over-production of a glycoside hydrolase family 50 β-agarase from Agarivorans sp. JA-1 in Bacillus subtilis and the whitening effect of its product. Biotechnol. Lett. 2008, 30, 911–918. [Google Scholar]
- Yun, E.J.; Lee, S.; Kim, J.H.; Kim, B.B.; Kim, H.T.; Lee, S.H.; Pelton, J.G.; Kang, N.J.; Choi, I.G.; Kim, K.H. Enzymatic production of 3,6-anhydro-l-galactose from agarose and its purification and in vitro skin whitening and anti-inflammatory activities. Appl. Microbiol. Biotechnol. 2013, 97, 2961–2970. [Google Scholar] [CrossRef] [PubMed]
- Giordano, A.; Andreotti, G.; Tramice, A.; Trincone, A. Marine glycosyl hydrolases in the hydrolysis and synthesis of oligosaccharides. Biotechnol. J. 2006, 1, 511–530. [Google Scholar] [CrossRef] [PubMed]
- Sugano, Y.; Terada, I.; Arita, M.; Noma, M.; Matsumoto, T. Purification and characterization of a new agarase from a marine bacterium, Vibrio sp. strain JT0107. Appl. Environ. Microbiol. 1993, 59, 1549–1554. [Google Scholar]
- Minegishi, H.; Shimane, Y.; Echigo, A.; Ohta, Y.; Hatada, Y.; Kamekura, M.; Maruyama, T.; Usami, R. Thermophilic and halophilic β-agarase from a halophilic archaeon Halococcus sp. 197A. Extremophiles 2013, 17, 931–939. [Google Scholar] [CrossRef]
- Kim, B.J.; Kim, H.J.; Ha, S.D.; Hwang, S.H.; Byun, D.S.; Lee, T.H.; Kong, J.K. Purification and characterization of β-agarase from marine bacterium Bacillus cereus ASK202. Biotechnol. Lett. 1999, 21, 1011–1015. [Google Scholar] [CrossRef]
- Song, T.; Cao, Y.; Xu, H.; Zhang, W.J.; Fei, B.J.; Qiao, D.R.; Cao, Y. Purification and characterization of a novel β-agarase of Paenibacillus sp. SSG-1 isolated from soil. J. Biosci. Bioeng. 2014, 118, 125–129. [Google Scholar] [CrossRef]
- Feng, Z.H.; Li, M.Y. Purification and characterization of agarase from Rhodococcus sp. Q5, a novel agarolytic bacterium isolated from printing and dyeing wastewater. Aquaculture 2013, 372, 74–79. [Google Scholar]
- Parro, V.; Mellado, R.P.; Harwood, C.R. Effects of phosphate limitation on agarase production by Streptomyces lividans TK21. FEMS Microbiol. Lett. 1998, 158, 107–113. [Google Scholar]
- Lakshmikanth, M.; Manohar, S.; Souche, Y.; Lalitha, J. Extracellular β-agarase LSL-1 producing neoagarobiose from a newly isolated agar-liquefying soil bacterium, Acinetobacter sp., AG LSL-1. World J. Microb. Biot. 2006, 22, 1087–1094. [Google Scholar] [CrossRef]
- Ohta, Y.; Hatada, Y.; Ito, S.; Horikoshi, K. High-level expression of a neoagarobiose-producing β-agarase gene from Agarivorans sp. JAMB-AII in Bacillus subtilis and enzymic properties of the recombinant enzyme. Biotechnol. Appl. Biochem. 2005, 41, 183–191. [Google Scholar]
- Lee, D.G.; Park, G.T.; Kim, N.Y.; Lee, E.J.; Jang, M.K.; Shin, Y.G.; Park, G.S.; Kim, T.M.; Lee, J.H.; Lee, J.H.; et al. Cloning, expression, and characterization of a glycoside hydrolase family 50 β-agarase from a marine Agarivorans isolate. Biotechnol. Lett. 2006, 28, 1925–1932. [Google Scholar]
- Potin, P.; Richard, C.; Rochas, C.; Kloareg, B. Purification and characterization of the α-agarase from Alteromonas agarlyticus (Cataldi) comb. nov., strain GJ1B. Eur. J. Biochem. 1993, 214, 599–607. [Google Scholar] [CrossRef]
- Shi, X.C.; Yu, M.; Yan, S.L.; Dong, S.J.; Zhang, X.H. Genome sequence of the thermostable-agarase-producing marine bacterium Catenovulum agarivorans YM01(T), which reveals the presence of a series of agarase-encoding genes. J. Bacteriol. 2012, 194, 5484–5484. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.L.; Chen, L.C.; Shih, Y.Y.; Hsieh, C.; Chen, C.Y.; Chen, W.M.; Chen, C.C. Cloning and characterization of β-agarase AgaYT from Flammeovirga yaeyamensis strain YT. J. Biosci. Bioeng. 2011, 112, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.L.; Lu, X.Z.; Yu, W.G. A new β-agarase from marine bacterium Janthinobacterium sp. SY12. World J. Microb. Biot. 2008, 24, 2659–2664. [Google Scholar] [CrossRef]
- Ohta, Y.; Hatada, Y.; Nogi, Y.; Li, Z.J.; Ito, S.; Horikoshi, K. Cloning, expression, and characterization of a glycoside hydrolase family 86 β-agarase from a deep-sea Microbulbifer-like isolate. Appl. Microbiol. Biotechnol. 2004, 66, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Ha, J.C.; Kim, G.T.; Kim, S.K.; Oh, T.K.; Yu, J.H.; Kong, I.S. β-agarase from Pseudomonas sp. W7: Purification of the recombinant enzyme from Escherichia coli and the effects of salt on its activity. Biotechnol. Appl. Biochem. 1997, 26, 1–6. [Google Scholar]
- Vera, J.; Alvarez, R.; Murano, E.; Slebe, J.C.; Leon, O. Identification of a marine agarolytic Pseudoalteromonas isolate and characterization of its extracellular agarase. Appl. Environ. Microbiol. 1998, 64, 4378–4383. [Google Scholar] [PubMed]
- Kim, H.T.; Lee, S.; Lee, D.; Kim, H.S.; Bang, W.G.; Kim, K.H.; Choi, I.G. Overexpression and molecular characterization of Aga50D from Saccharophagus degradans 2–40: An exo-type β-agarase producing neoagarobiose. Appl. Microbiol. Biotechnol. 2010, 86, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Ohta, Y.; Hatada, Y.; Miyazaki, M.; Nogi, Y.; Ito, S.; Horikoshi, K. Purification and characterization of a novel α-agarase from a Thalassomonas sp. Curr. Microbiol. 2005, 50, 212–216. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.H.; Hashikawa, S.; Konishi, T.; Tamaru, Y.; Araki, T. Cloning of the novel gene encoding β-agarase C from a marine bacterium, Vibrio sp. strain PO-303, and characterization of the gene product. Appl. Environ. Microbiol. 2006, 72, 6399–6401. [Google Scholar]
- Jang, M.K.; Lee, S.W.; Lee, D.G.; Kim, N.Y.; Yu, K.H.; Jang, H.J.; Kim, S.; Kim, A.; Lee, S.H. Enhancement of the thermostability of a recombinant β-agarase, AgaB, from Zobellia galactanivorans by random mutagenesis. Biotechnol. Lett. 2010, 32, 943–949. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Michel, G.; Nyval-Collen, P.; Barbeyron, T.; Czjzek, M.; Helbert, W. Bioconversion of red seaweed galactans: A focus on bacterial agarases and carrageenases. Appl. Microbiol. Biotechnol. 2006, 71, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.H.; Tamaru, Y.; Araki, T. A unique β-agarase, AgaA, from a marine bacterium, Vibrio sp. strain PO-303. Appl. Microbiol. Biotechnol. 2007, 74, 1248–1255. [Google Scholar] [CrossRef]
- Ma, C.P.; Lu, X.Z.; Shi, C.; Li, J.B.; Gu, Y.C.; Ma, Y.M.; Chu, Y.; Han, F.; Gong, Q.H.; Yu, W.G. Molecular cloning and characterization of a novel β-agarase, AgaB, from marine Pseudoalteromonas sp. CY24. J. Biol. Chem. 2007, 282, 3747–3754. [Google Scholar] [CrossRef]
- Wu, S.C.; Pan, C.L. Preparation of algal-oligosaccharide mixtures by bacterial agarases and their antioxidative properties. Fish. Sci. 2004, 70, 1164–1173. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef] [PubMed]
- Allouch, J.; Jam, M.; Helbert, W.; Barbeyron, T.; Kloareg, B.; Henrissat, B.; Czjzek, M. The three-dimensional structures of two β-agarases. J. Biol. Chem. 2003, 278, 47171–47180. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.Y.; Dong, S.J.; Shi, X.C.; Zhao, X.; Zhang, X.H. Overexpression and characterization of a novel thermostable β-agarase YM01-3, from marine bacterium Catenovulum agarivorans YM01(T). Mar. Drugs 2014, 12, 2731–2747. [Google Scholar] [CrossRef] [PubMed]
- Bruser, T. The twin-arginine translocation system and its capability for protein secretion in biotechnological protein production. Appl. Microbiol. Biotechnol. 2007, 76, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Weiss, J.B.; Ray, P.H.; Bassford, P.J., Jr. Purified secB protein of Escherichia coli retards folding and promotes membrane translocation of the maltose-binding protein in vitro. Proc. Natl. Acad. Sci. USA 1988, 85, 8978–8982. [Google Scholar] [CrossRef] [PubMed]
- Manting, E.H.; Driessen, A.J. Escherichia coli translocase: The unravelling of a molecular machine. Mol. Microbiol. 2000, 37, 226–238. [Google Scholar] [CrossRef] [PubMed]
- Berks, B.C.; Palmer, T.; Sargent, F. The Tat protein translocation pathway and its role in microbial physiology. Adv. Microb. Physiol. 2003, 47, 187–254. [Google Scholar] [PubMed]
- Zhang, W.W.; Sun, L. Cloning, characterization, and molecular application of a β-agarase gene from Vibrio sp. strain V134. Appl. Environ. Microbiol. 2007, 73, 2825–2831. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsu, P.-H.; Wei, C.-H.; Lu, W.-J.; Shen, F.; Pan, C.-L.; Lin, H.-T.V. Extracellular Production of a Novel Endo-β-Agarase AgaA from Pseudomonas vesicularis MA103 that Cleaves Agarose into Neoagarotetraose and Neoagarohexaose. Int. J. Mol. Sci. 2015, 16, 5590-5603. https://doi.org/10.3390/ijms16035590
Hsu P-H, Wei C-H, Lu W-J, Shen F, Pan C-L, Lin H-TV. Extracellular Production of a Novel Endo-β-Agarase AgaA from Pseudomonas vesicularis MA103 that Cleaves Agarose into Neoagarotetraose and Neoagarohexaose. International Journal of Molecular Sciences. 2015; 16(3):5590-5603. https://doi.org/10.3390/ijms16035590
Chicago/Turabian StyleHsu, Pang-Hung, Chien-Han Wei, Wen-Jung Lu, Fen Shen, Chorng-Liang Pan, and Hong-Ting Victor Lin. 2015. "Extracellular Production of a Novel Endo-β-Agarase AgaA from Pseudomonas vesicularis MA103 that Cleaves Agarose into Neoagarotetraose and Neoagarohexaose" International Journal of Molecular Sciences 16, no. 3: 5590-5603. https://doi.org/10.3390/ijms16035590
APA StyleHsu, P.-H., Wei, C.-H., Lu, W.-J., Shen, F., Pan, C.-L., & Lin, H.-T. V. (2015). Extracellular Production of a Novel Endo-β-Agarase AgaA from Pseudomonas vesicularis MA103 that Cleaves Agarose into Neoagarotetraose and Neoagarohexaose. International Journal of Molecular Sciences, 16(3), 5590-5603. https://doi.org/10.3390/ijms16035590






