Functional Properties of the Catalytic Domain of Mouse Acidic Mammalian Chitinase Expressed in Escherichia coli
Abstract
:1. Introduction
2. Results
2.1. Recombinant N-Terminal Catalytic Domain (CatD) Expression Plasmids
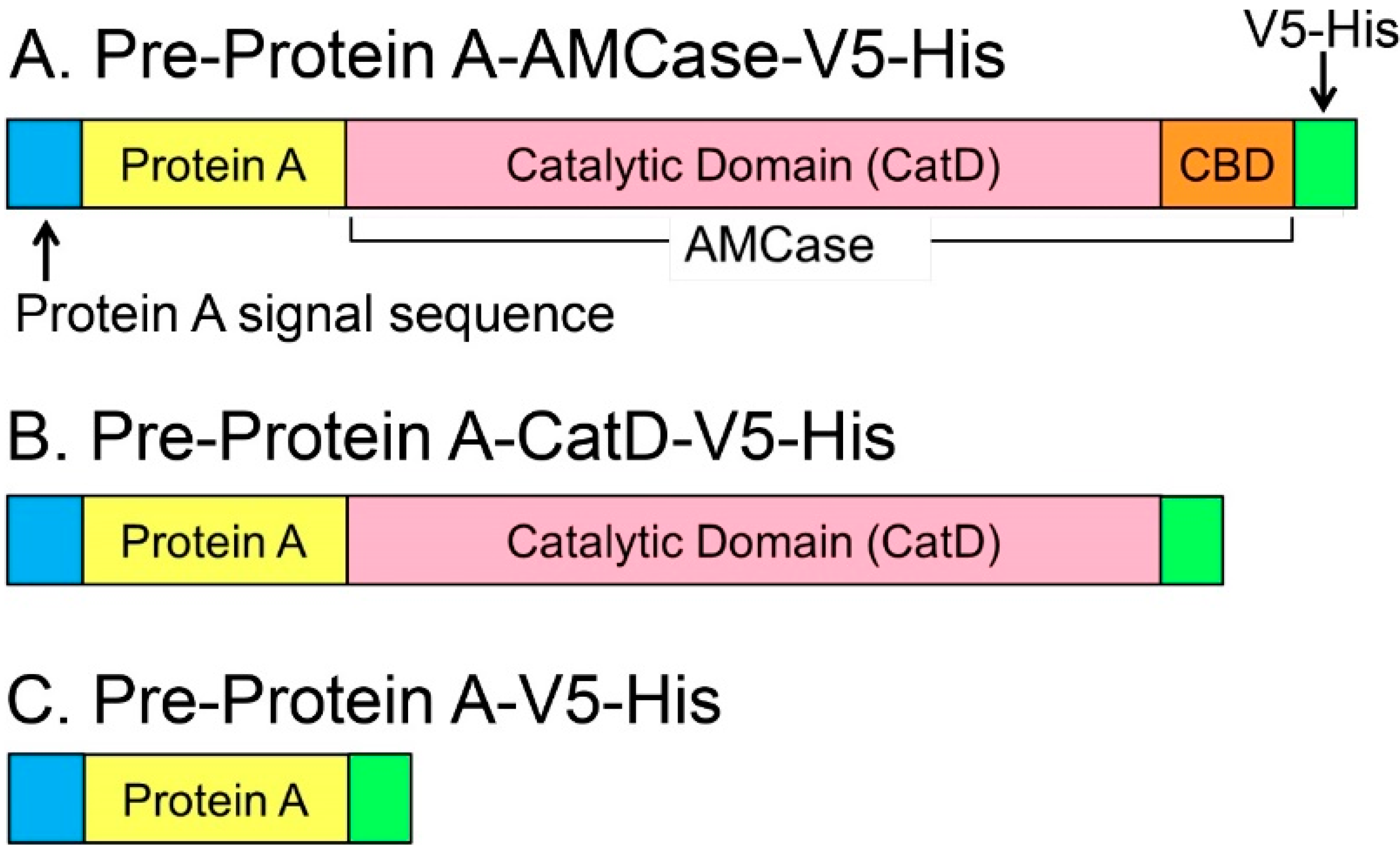
2.2. Purification of the Recombinant CatD from the Periplasmic Fraction of Escherichia coli
2.3. Recombinant CatD Has Chitinolytic Activity
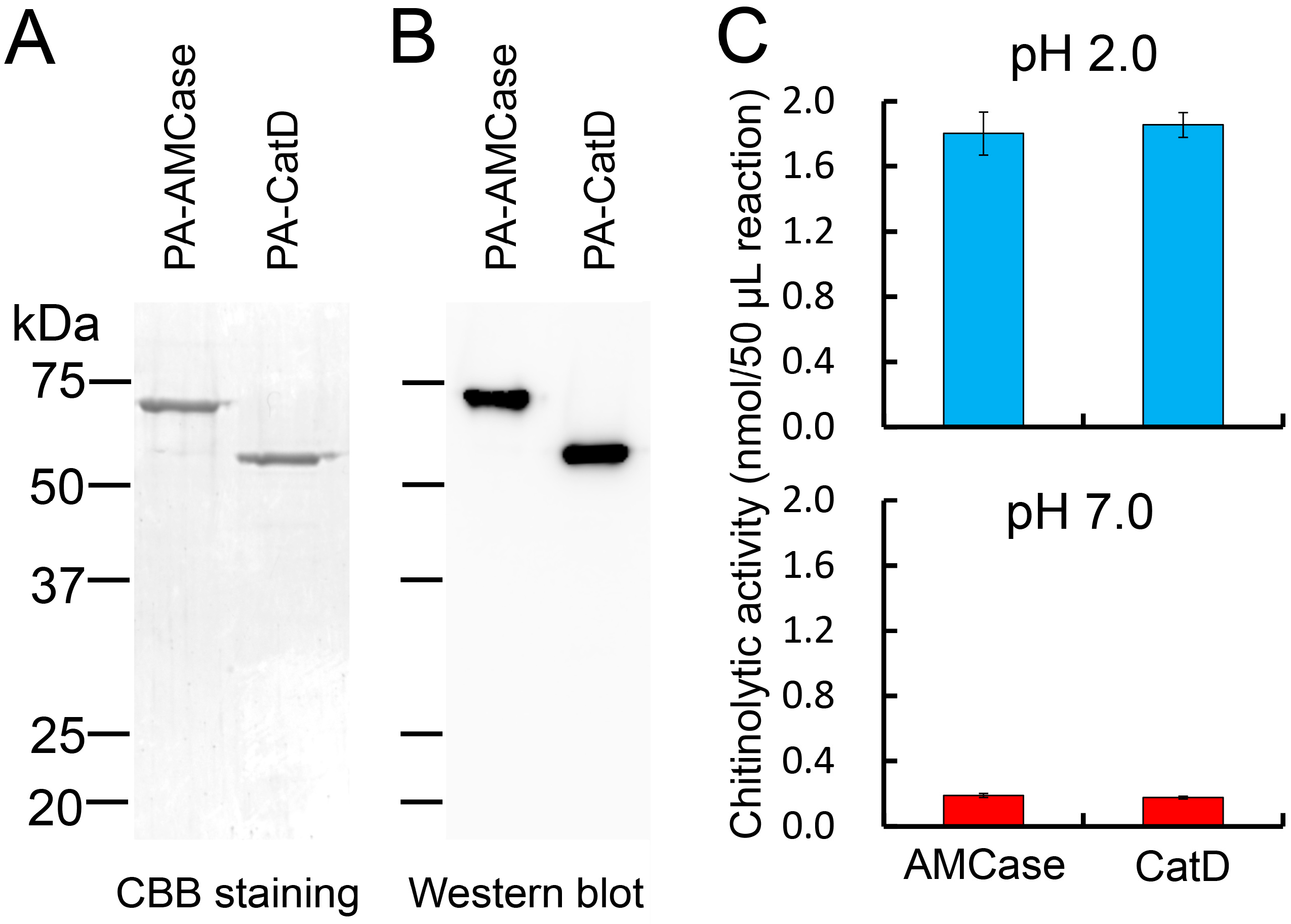
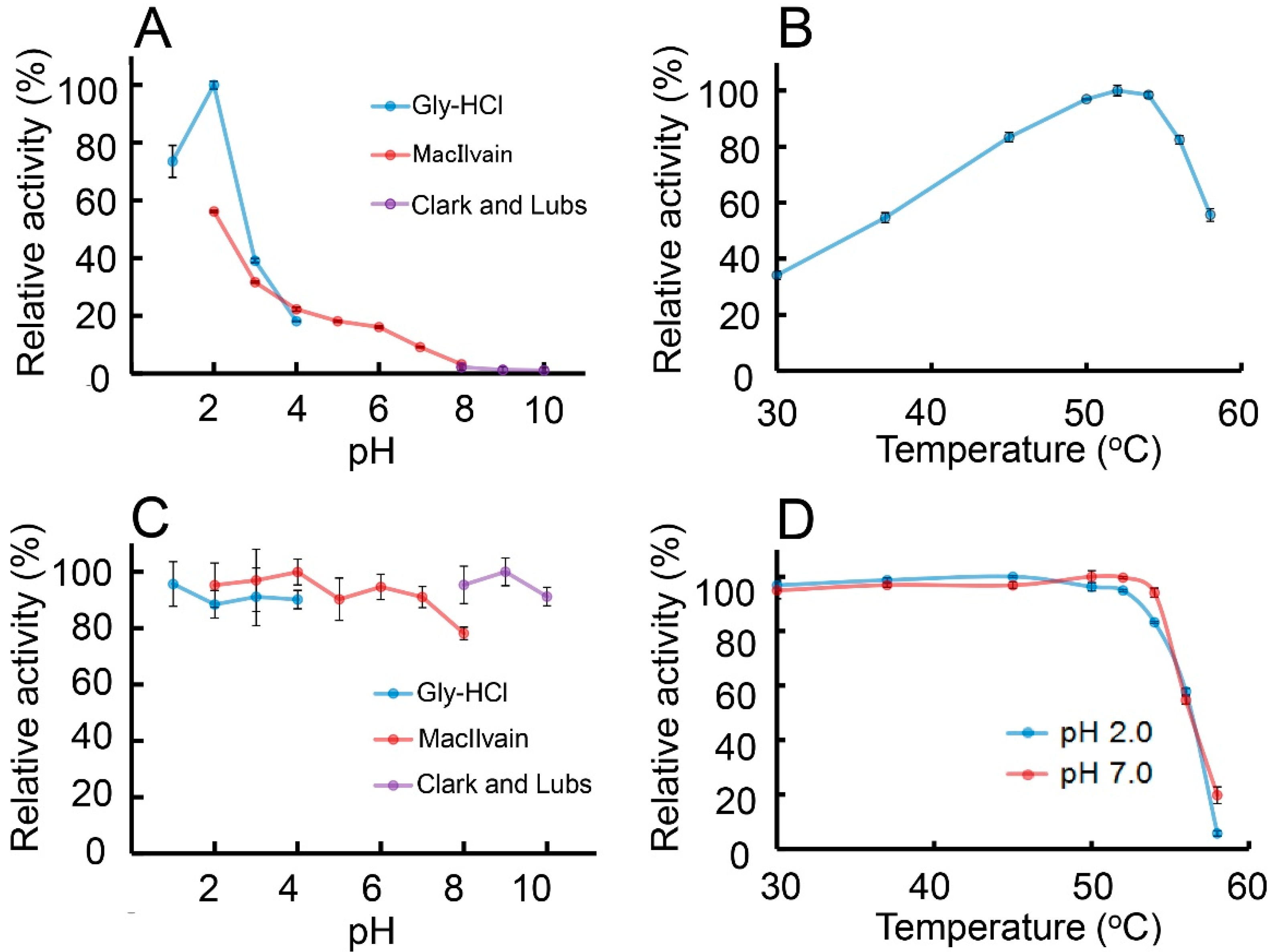
2.4. Characterization of the Chitinolytic Activity of Protein A-CatD-V5-His
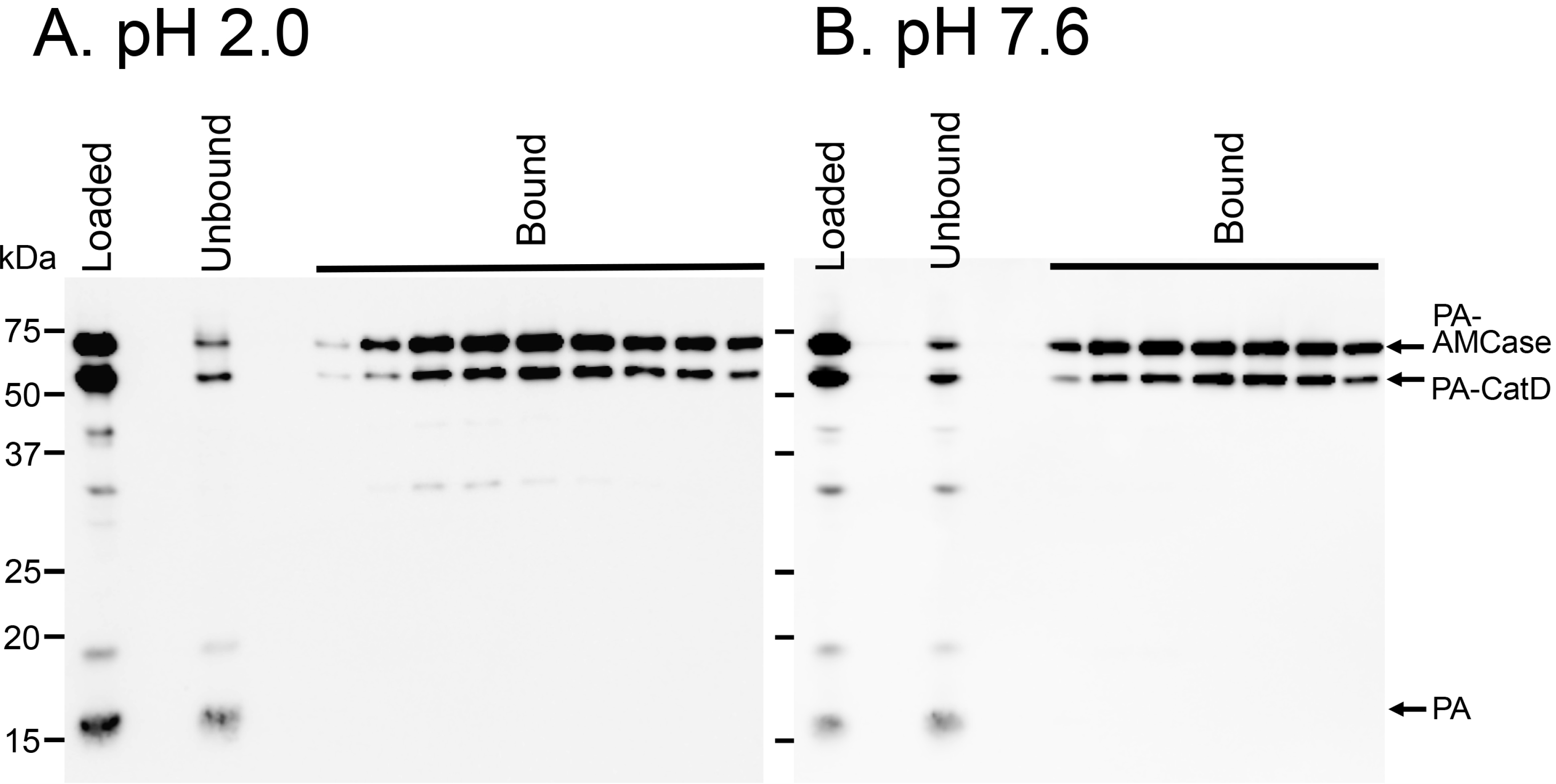
2.5. Protein A-CatD-V5-His Facilitates Chitin Binding
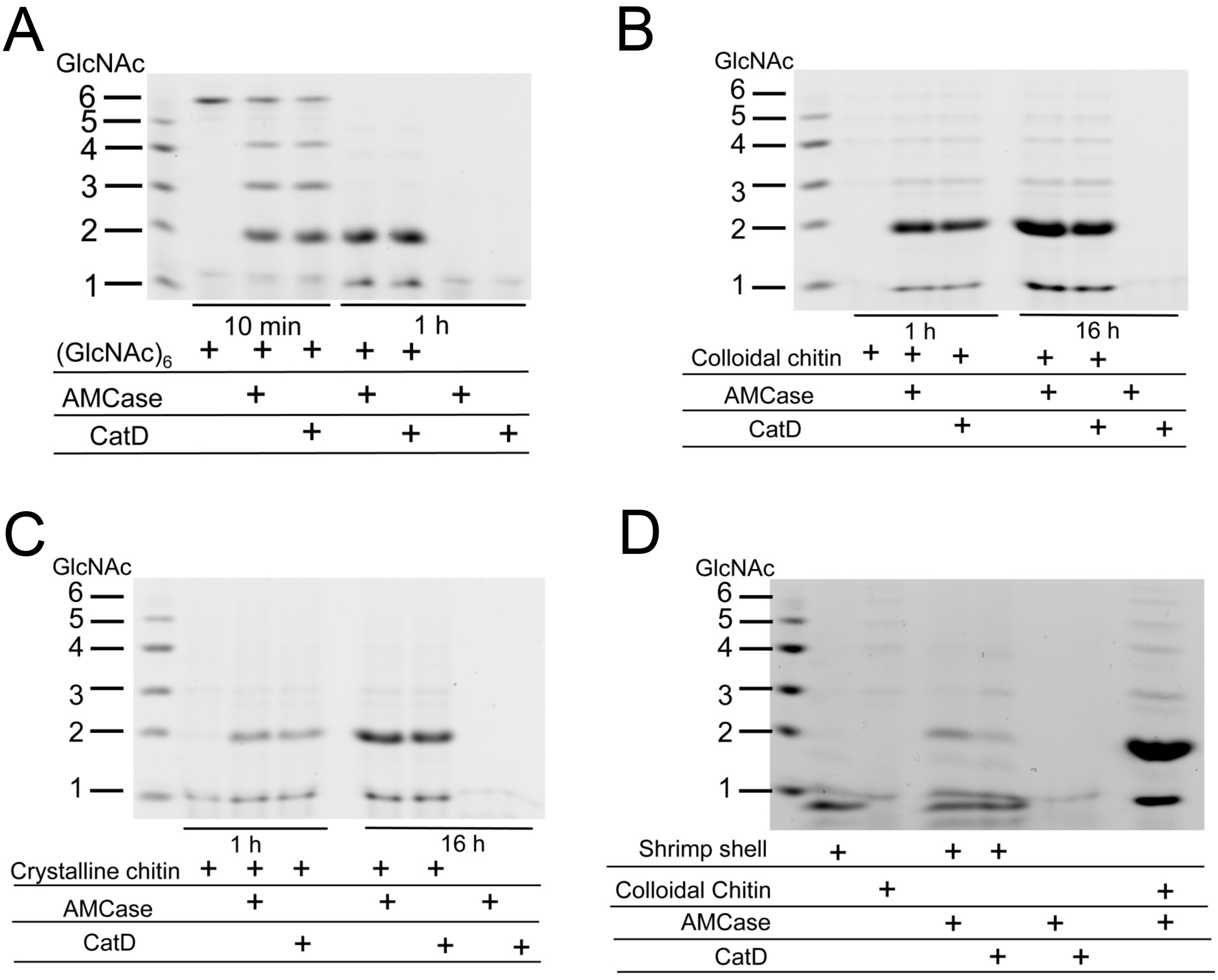
2.6. CatD Promotes Chitin Degradation
3. Discussion
4. Experimental Section
4.1. E. coli Expression Vector
4.2. Preparation of the Recombinant CatD Expressed in E. coli
4.3. SDS-Polyacrylamide Gel Electrophoresis and Western Blotting
4.4. Chitinase Enzymatic Assays
4.5. Chitin Binding Assay
4.6. Degradation of GlcNAc Hexamer, Colloidal and Crystalline Chitin and Shrimp Shell by AMCase and CatD
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Khoushab, F.; Yamabhai, M. Chitin research revisited. Mar. Drugs 2010, 8, 1988–2012. [Google Scholar] [CrossRef] [PubMed]
- Bueter, C.L.; Specht, C.A.; Levitz, S.M. Innate sensing of chitin and chitosan. PLoS Pathog. 2013, 9, e1003080. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.G.; da Silva, C.A.; Dela Cruz, C.S.; Ahangari, F.; Ma, B.; Kang, M.J.; He, C.H.; Takyar, S.; Elias, J.A. Role of chitin and chitinase/chitinase-like proteins in inflammation, tissue remodeling, and injury. Annu. Rev. Physiol. 2011, 73, 479–501. [Google Scholar] [CrossRef] [PubMed]
- Hamid, R.; Khan, M.A.; Ahmad, M.; Ahmad, M.M.; Abdin, M.Z.; Musarrat, J.; Javed, S. Chitinases: An update. J. Pharm. Bioallied Sci. 2013, 5, 21–29. [Google Scholar] [PubMed]
- Bussink, A.P.; Speijer, D.; Aerts, J.M.; Boot, R.G. Evolution of mammalian chitinase(-like) members of family 18 glycosyl hydrolases. Genetics 2007, 177, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Kawada, M.; Hachiya, Y.; Arihiro, A.; Mizoguchi, E. Role of mammalian chitinases in inflammatory conditions. Keio J. Med. 2007, 56, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Renkema, G.H.; Boot, R.G.; Muijsers, A.O.; Donker-Koopman, W.E.; Aerts, J.M. Purification and characterization of human chitotriosidase, a novel member of the chitinase family of proteins. J. Biol. Chem. 1995, 270, 2198–2202. [Google Scholar] [CrossRef] [PubMed]
- Boot, R.G.; Renkema, G.H.; Strijland, A.; van Zonneveld, A.J.; Aerts, J.M. Cloning of a cDNA encoding chitotriosidase, a human chitinase produced by macrophages. J. Biol. Chem. 1995, 270, 26252–26256. [Google Scholar] [CrossRef] [PubMed]
- Boot, R.G.; Blommaart, E.F.; Swart, E.; Ghauharali-van der Vlugt, K.; Bijl, N.; Moe, C.; Place, A.; Aerts, J.M. Identification of a novel acidic mammalian chitinase distinct from chitotriosidase. J. Biol. Chem. 2001, 276, 6770–6778. [Google Scholar] [CrossRef] [PubMed]
- Boot, R.G.; Bussink, A.P.; Verhoek, M.; de Boer, P.A.; Moorman, A.F.; Aerts, J.M. Marked differences in tissue-specific expression of chitinases in mouse and man. J. Histochem. Cytochem. 2005, 53, 1283–1292. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Zheng, T.; Homer, R.J.; Kim, Y.K.; Chen, N.Y.; Cohn, L.; Hamid, Q.; Elias, J.A. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science 2004, 304, 1678–1682. [Google Scholar] [CrossRef] [PubMed]
- Bierbaum, S.; Nickel, R.; Koch, A.; Lau, S.; Deichmann, K.A.; Wahn, U.; Superti-Furga, A.; Heinzmann, A. Polymorphisms and haplotypes of acid mammalian chitinase are associated with bronchial asthma. Am. J. Respir. Crit. Care Med. 2005, 172, 1505–1509. [Google Scholar] [CrossRef] [PubMed]
- Seibold, M.A.; Reese, T.A.; Choudhry, S.; Salam, M.T.; Beckman, K.; Eng, C.; Atakilit, A.; Meade, K.; Lenoir, M.; Watson, H.G.; et al. Differential enzymatic activity of common haplotypic versions of the human acidic mammalian chitinase protein. J. Biol. Chem. 2009, 284, 19650–19658. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, T.E.; Andersen, O.A.; Betou, M.; Eggleston, I.M.; Maizels, R.M.; van Aalten, D.; Allen, J.E. Analyzing airway inflammation with chemical biology: Dissection of acidic mammalian chitinase function with a selective drug-like inhibitor. Chem. Biol. 2011, 18, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.J.; Liu, Y.K.; Liu, C.L.; Shen, C.N.; Kuo, M.L.; Su, C.C.; Tseng, C.P.; Yen, T.C.; Shen, C.R. Inhibition of acidic mammalian chitinase by rna interference suppresses ovalbumin-sensitized allergic asthma. Hum. Gene Ther. 2009, 20, 1597–1606. [Google Scholar] [CrossRef] [PubMed]
- Reese, T.A.; Liang, H.E.; Tager, A.M.; Luster, A.D.; van Rooijen, N.; Voehringer, D.; Locksley, R.M. Chitin induces accumulation in tissue of innate immune cells associated with allergy. Nature 2007, 447, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Bucolo, C.; Musumeci, M.; Maltese, A.; Drago, F.; Musumeci, S. Effect of chitinase inhibitors on endotoxin-induced uveitis (EIU) in rabbits. Pharmacol. Res. 2008, 57, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, M.; Aragona, P.; Bellin, M.; Maugeri, F.; Rania, L.; Bucolo, C.; Musumeci, S. Acidic mammalian chitinase in dry eye conditions. Cornea 2009, 28, 667–672. [Google Scholar] [CrossRef] [PubMed]
- Bucolo, C.; Musumeci, M.; Musumeci, S.; Drago, F. Acidic mammalian chitinase and the eye: Implications for ocular inflammatory diseases. Front. Pharmacol. 2011, 2, 43. [Google Scholar] [CrossRef] [PubMed]
- Cozzarini, E.; Bellin, M.; Norberto, L.; Polese, L.; Musumeci, S.; Lanfranchi, G.; Paoletti, M.G. CHIT1 and amcase expression in human gastric mucosa: Correlation with inflammation and Helicobacter pylori infection. Eur. J. Gastroenterol. Hepatol. 2009, 21, 1119–1126. [Google Scholar] [CrossRef] [PubMed]
- Nookaew, I.; Thorell, K.; Worah, K.; Wang, S.; Hibberd, M.L.; Sjovall, H.; Pettersson, S.; Nielsen, J.; Lundin, S.B. Transcriptome signatures in Helicobacter pylori-infected mucosa identifies acidic mammalian chitinase loss as a corpus atrophy marker. BMC Med. Genomics 2013, 6, 41. [Google Scholar] [CrossRef] [PubMed]
- Ohno, M.; Tsuda, K.; Sakaguchi, M.; Sugahara, Y.; Oyama, F. Chitinase mrna levels by quantitative PCR using the single standard DNA: Acidic mammalian chitinase is a major transcript in the mouse stomach. PLoS One 2012, 7, e50381. [Google Scholar] [CrossRef] [PubMed]
- Ohno, M.; Togashi, Y.; Tsuda, K.; Okawa, K.; Kamaya, M.; Sakaguchi, M.; Sugahara, Y.; Oyama, F. Quantification of chitinase mRNA levels in human and mouse tissues by real-time PCR: Species-specific expression of acidic mammalian chitinase in stomach tissues. PLoS One 2013, 8, e67399. [Google Scholar] [CrossRef] [PubMed]
- Kashimura, A.; Okawa, K.; Ishikawa, K.; Kida, Y.; Iwabuchi, K.; Matsushima, Y.; Sakaguchi, M.; Sugahara, Y.; Oyama, F. Protein A-mouse acidic mammalian chitinase-v5-his expressed in periplasmic space of Escherichia coli possesses chitinase functions comparable to CHO-expressed protein. PLoS One 2013, 8, e78669. [Google Scholar] [CrossRef] [PubMed]
- Tjoelker, L.W.; Gosting, L.; Frey, S.; Hunter, C.L.; Trong, H.L.; Steiner, B.; Brammer, H.; Gray, P.W. Structural and functional definition of the human chitinase chitin-binding domain. J. Biol. Chem. 2000, 275, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Jackson, P. The use of polyacrylamide-gel electrophoresis for the high-resolution separation of reducing saccharides labelled with the fluorophore 8-aminonaphthalene-1,3,6-trisulphonic acid. Detection of picomolar quantities by an imaging system based on a cooled charge-coupled device. Biochem. J. 1990, 270, 705–713. [Google Scholar] [PubMed]
- Lowenadler, B.; Jansson, B.; Paleus, S.; Holmgren, E.; Nilsson, B.; Moks, T.; Palm, G.; Josephson, S.; Philipson, L.; Uhlen, M. A gene fusion system for generating antibodies against short peptides. Gene 1987, 58, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Oyama, F.; Hashino, K.; Oyama, R.; Kato, I.; Titani, K. Improved method for expression of kunitz-type serine proteinase inhibitor domain of beta-amyloid protein precursor in Escherichia coli and characterization of disulfide bonds of the product. J. Biochem. 1993, 114, 813–819. [Google Scholar] [PubMed]
- Tiwary, E.; Gupta, R. Extracellular expression of keratinase from bacillus licheniformis ER-15 in Escherichia coli. J. Agric. Food Chem. 2010, 58, 8380–8385. [Google Scholar] [CrossRef] [PubMed]
- Kumari, A.; Gupta, R. Extracellular expression and characterization of thermostable lipases, lip8, lip14 and lip18, from yarrowia lipolytica. Biotechnol. Lett. 2012, 34, 1733–1739. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, M.; Matsushima, Y.; Nankumo, T.; Seino, J.; Miyakawa, S.; Honda, S.; Sugahara, Y.; Oyama, F.; Kawakita, M. Glucoamylase of Caulobacter crescentus CB15: Cloning and expression in Escherichia coli and functional identification. AMB Express 2014, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Olland, A.M.; Strand, J.; Presman, E.; Czerwinski, R.; Joseph-McCarthy, D.; Krykbaev, R.; Schlingmann, G.; Chopra, R.; Lin, L.; Fleming, M.; et al. Triad of polar residues implicated in pH specificity of acidic mammalian chitinase. Protein Sci. 2009, 18, 569–578. [Google Scholar] [PubMed]
- Watanabe, T.; Kimura, K.; Sumiya, T.; Nikaidou, N.; Suzuki, K.; Suzuki, M.; Taiyoji, M.; Ferrer, S.; Regue, M. Genetic analysis of the chitinase system of serratia marcescens 2170. J. Bacteriol. 1997, 179, 7111–7117. [Google Scholar] [PubMed]
- Suzuki, K.; Taiyoji, M.; Sugawara, N.; Nikaidou, N.; Henrissat, B.; Watanabe, T. The third chitinase gene (chiC) of Serratia marcescens 2170 and the relationship of its product to other bacterial chitinases. Biochem. J. 1999, 343, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Ohno, M.; Kida, Y.; Sakaguchi, M.; Sugahara, Y.; Oyama, F. Establishment of a quantitative PCR system for discriminating chitinase-like proteins: Catalytically inactive breast regression protein-39 and Ym1 are constitutive genes in mouse lung. BMC Mol. Biol. 2014, 15, 23. [Google Scholar] [CrossRef] [PubMed]
- Richter, C.; Tanaka, T.; Yada, R.Y. Mechanism of activation of the gastric aspartic proteinases: Pepsinogen, progastricsin and prochymosin. Biochem. J. 1998, 335, 481–490. [Google Scholar] [PubMed]
- Kageyama, T. Pepsinogens, progastricsins, and prochymosins: Structure, function, evolution, and development. Cell Mol. Life Sci. 2002, 59, 288–306. [Google Scholar] [CrossRef] [PubMed]
- Renkema, G.H.; Boot, R.G.; Au, F.L.; Donker-Koopman, W.E.; Strijland, A.; Muijsers, A.O.; Hrebicek, M.; Aerts, J.M. Chitotriosidase, a chitinase, and the 39-kDa human cartilage glycoprotein, a chitin-binding lectin, are homologues of family 18 glycosyl hydrolases secreted by human macrophages. Eur. J. Biochem. 1998, 251, 504–509. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kashimura, A.; Kimura, M.; Okawa, K.; Suzuki, H.; Ukita, A.; Wakita, S.; Okazaki, K.; Ohno, M.; Bauer, P.O.; Sakaguchi, M.; et al. Functional Properties of the Catalytic Domain of Mouse Acidic Mammalian Chitinase Expressed in Escherichia coli. Int. J. Mol. Sci. 2015, 16, 4028-4042. https://doi.org/10.3390/ijms16024028
Kashimura A, Kimura M, Okawa K, Suzuki H, Ukita A, Wakita S, Okazaki K, Ohno M, Bauer PO, Sakaguchi M, et al. Functional Properties of the Catalytic Domain of Mouse Acidic Mammalian Chitinase Expressed in Escherichia coli. International Journal of Molecular Sciences. 2015; 16(2):4028-4042. https://doi.org/10.3390/ijms16024028
Chicago/Turabian StyleKashimura, Akinori, Masahiro Kimura, Kazuaki Okawa, Hirotaka Suzuki, Atsushi Ukita, Satoshi Wakita, Kana Okazaki, Misa Ohno, Peter O. Bauer, Masayoshi Sakaguchi, and et al. 2015. "Functional Properties of the Catalytic Domain of Mouse Acidic Mammalian Chitinase Expressed in Escherichia coli" International Journal of Molecular Sciences 16, no. 2: 4028-4042. https://doi.org/10.3390/ijms16024028
APA StyleKashimura, A., Kimura, M., Okawa, K., Suzuki, H., Ukita, A., Wakita, S., Okazaki, K., Ohno, M., Bauer, P. O., Sakaguchi, M., Sugahara, Y., & Oyama, F. (2015). Functional Properties of the Catalytic Domain of Mouse Acidic Mammalian Chitinase Expressed in Escherichia coli. International Journal of Molecular Sciences, 16(2), 4028-4042. https://doi.org/10.3390/ijms16024028





