New Insights to Clathrin and Adaptor Protein 2 for the Design and Development of Therapeutic Strategies
Abstract
:1. Introduction
2. Results and Discussion
2.1. Y682G Mutation Prevents APP Binding to Clathrin and AP-2
| Peptide Name | Sequence |
|---|---|
| WT | Biotin-Ttsd linker-AAVTPEERHLSKMQQNGYENPTYKFFEQMQN |
| pWT | Biotin-Ttsd linker-AAVTPEERHLSKMQQNG-pY-ENPTYKFFEQMQN |
| YG | Biotin-Ttsd linker-AAVTPEERHLSKMQQNGGENPTYKFFEQMQN |
| Scr | Biotin-Ttsd linker-LVEYKQREQGFSQPHQTANPFNENAEYTMMK |
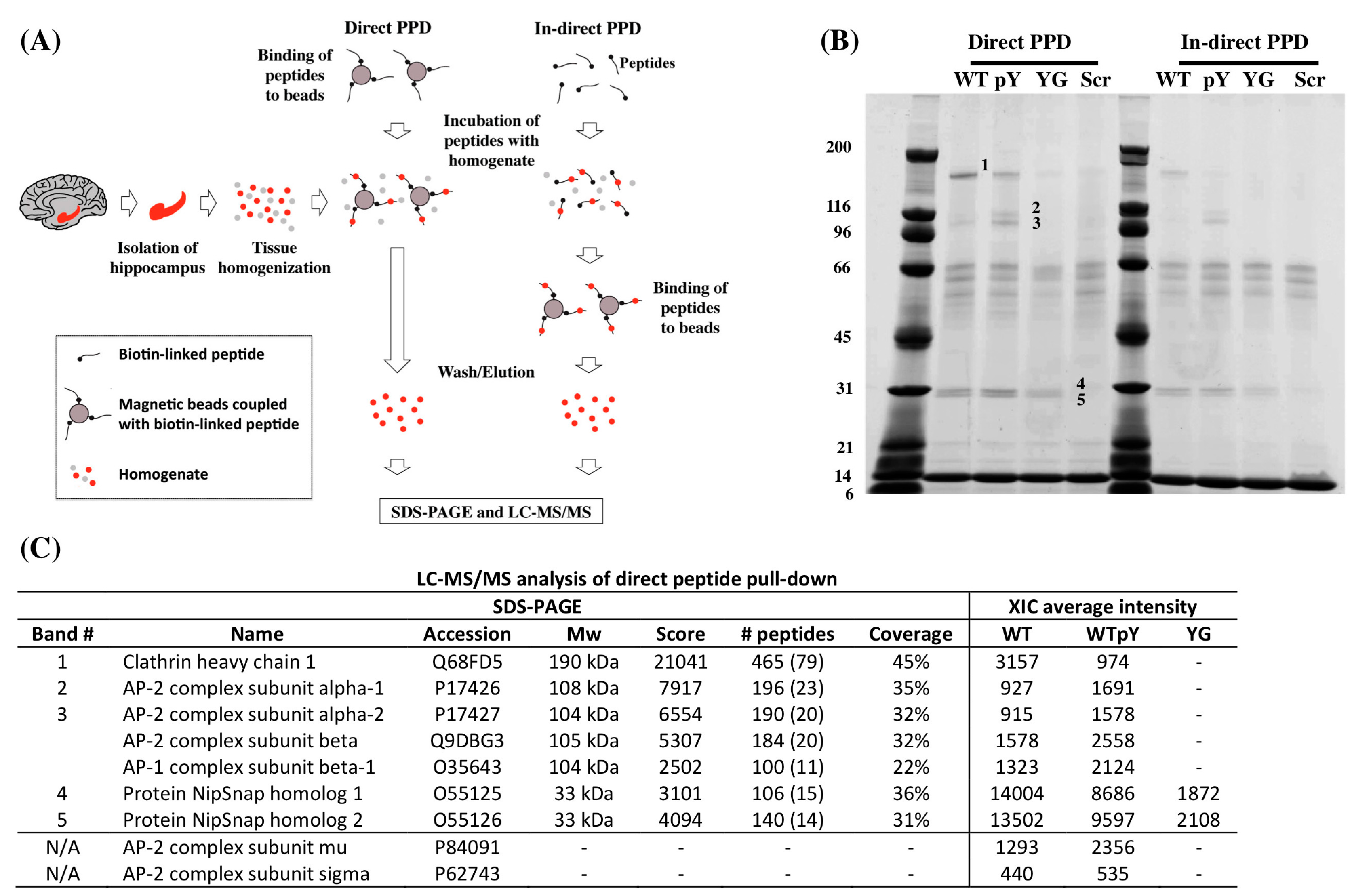
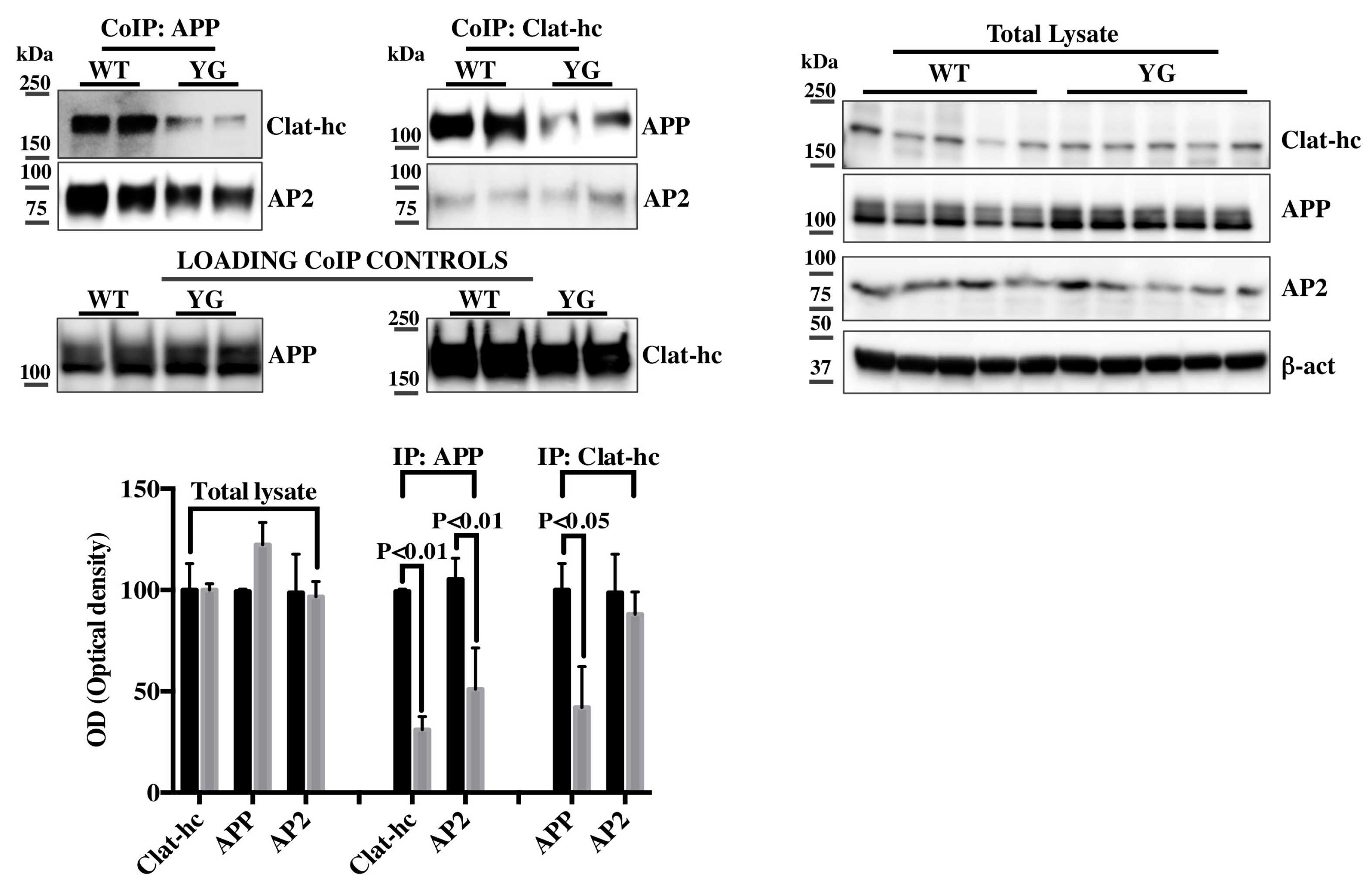
2.2. Clathrin and AP-2 Co-Immunoprecipitate with APP in WT but Not in Y682G Mice
3. Experimental Section
3.1. Tissue Homogenization
3.2. Biotinylated Peptides
3.3. Direct Peptide Pull-down
3.4. Indirect Peptide Pull-Down
3.5. SDS-PAGE
3.6. MS Sample Preparation for XIC Quantification
3.7. LC-MS/MS Analysis
3.8. Data Processing
3.9. Immunoprecipitation
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Basso, E.; Matrone, C. Ngf and app interplay: Focus on yenpty motif of amyloid precursor protein and y682 residue. Cell Biol.: Res. Ther. 2013, 2. [Google Scholar] [CrossRef]
- Saito, Y.; Akiyama, M.; Araki, Y.; Sumioka, A.; Shiono, M.; Taru, H.; Nakaya, T.; Yamamoto, T.; Suzuki, T. Intracellular trafficking of the amyloid beta-protein precursor (app) regulated by novel function of x11-like. PLoS ONE 2011, 6, e22108. [Google Scholar] [CrossRef] [PubMed]
- Treusch, S.; Hamamichi, S.; Goodman, J.L.; Matlack, K.E.; Chung, C.Y.; Baru, V.; Shulman, J.M.; Parrado, A.; Bevis, B.J.; Valastyan, J.S.; et al. Functional links between abeta toxicity, endocytic trafficking, and alzheimer's disease risk factors in yeast. Science 2011, 334, 1241–1245. [Google Scholar] [CrossRef] [PubMed]
- Haass, C.; Kaether, C.; Thinakaran, G.; Sisodia, S. Trafficking and proteolytic processing of app. Cold Spring Harb. Perspect. Med. 2012, 2, a006270. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Li, Y.; Zhang, X.; Bu, G.; Xu, H.; Zhang, Y.W. Trafficking regulation of proteins in Alzheimer’s disease. Mol. Neurodegener. 2014, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Matrone, C.; Barbagallo, A.P.; La Rosa, L.R.; Florenzano, F.; Ciotti, M.T.; Mercanti, D.; Chao, M.V.; Calissano, P.; D’Adamio, L. App is phosphorylated by trka and regulates ngf/trka signaling. J. Neurosci. 2011, 31, 11756–11761. [Google Scholar] [CrossRef] [PubMed]
- Matrone, C.; Luvisetto, S.; La Rosa, L.R.; Tamayev, R.; Pignataro, A.; Canu, N.; Yang, L.; Barbagallo, A.P.; Biundo, F.; Lombino, F.; et al. Tyr682 in the abeta-precursor protein intracellular domain regulates synaptic connectivity, cholinergic function, and cognitive performance. Aging Cell 2012, 11, 1084–1093. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, L.R.; Matrone, C.; Ferraina, C.; Panico, M.B.; Piccirilli, S.; Di Certo, M.G.; Strimpakos, G.; Mercuri, N.B.; Calissano, P.; D'Amelio, M.; et al. Age-related changes of hippocampal synaptic plasticity in abetapp-null mice are restored by ngf through p75ntr. J. Alzheimers Dis. 2013, 33, 265–272. [Google Scholar] [PubMed]
- Thomas, R.S.; Lelos, M.J.; Good, M.A.; Kidd, E.J. Clathrin-mediated endocytic proteins are upregulated in the cortex of the tg2576 mouse model of alzheimer's disease-like amyloid pathology. Biochem. Biophys. Res. Commun. 2011, 415, 656–661. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Matsuoka, Y.; Mattson, M.P.; Yao, P.J. The clathrin assembly protein ap180 regulates the generation of amyloid-beta peptide. Biochem. Biophys. Res. Commun. 2009, 385, 247–250. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Mattson, M.P.; Yao, P.J. Neuronal activity and the expression of clathrin-assembly protein ap180. Biochem. Biophys. Res. Commun. 2010, 402, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Yao, P.J. Clathrin-mediated endocytosis and alzheimer's disease: An update. Ageing Res. Rev. 2009, 8, 147–149. [Google Scholar] [CrossRef] [PubMed]
- Kelly, B.T.; Graham, S.C.; Liska, N.; Dannhauser, P.N.; Honing, S.; Ungewickell, E.J.; Owen, D.J. Clathrin adaptors. Ap2 controls clathrin polymerization with a membrane-activated switch. Science 2014, 345, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.; Pan, W.; Jones, S.A.; Zhang, Y.; Zhuang, X.; Wu, D. Clathrin and ap2 are required for ptdins(4,5)p2-mediated formation of lrp6 signalosomes. J. Cell Biol. 2013, 200, 419–428. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, L.R.; Perrone, L.; Nielsen, M.S.; Calissano, P.; Andersen, O.M.; Matrone, C. Y682g mutation of amyloid precursor protein promotes endo-lysosomal dysfunction by disrupting app-sorla interaction. Front. Cell. Neurosci. 2015, 9, 109. [Google Scholar] [CrossRef] [PubMed]
- Matrone, C. A new molecular explanation for age-related neurodegeneration: The tyr682 residue of amyloid precursor protein. Bioessays 2013, 35, 847–852. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Chang, J.C.; Fan, E.Y.; Flajolet, M.; Greengard, P. Adaptor complex ap2/picalm, through interaction with lc3, targets alzheimer's app-ctf for terminal degradation via autophagy. Proc. Natl. Acad. Sci. USA 2013, 110, 17071–17076. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.H.; Kao, P.Y.; Fan, Y.H.; Ho, D.T.; Chan, C.S.; Yik, P.Y.; Ha, J.C.; Chu, L.W.; Song, Y.Q. Polymorphisms of cr1, clu and picalm confer susceptibility of alzheimer's disease in a southern chinese population. Neurobiol. Aging 2012, 33. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zhang, S.; Cai, Z.; Ma, G.; Zhang, L.; Jiang, Y.; Feng, R.; Liao, M.; Chen, Z.; Zhao, B.; et al. Picalm gene rs3851179 polymorphism contributes to alzheimer's disease in an asian population. Neuromol. Med. 2013, 15, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Parikh, I.; Medway, C.; Younkin, S.; Fardo, D.W.; Estus, S. An intronic picalm polymorphism, rs588076, is associated with allelic expression of a picalm isoform. Mol. Neurodegener. 2014, 9, 32. [Google Scholar] [CrossRef] [PubMed]
- Bury, R.W.; Mashford, M.L. Use of a drug-screening service in an inner-city teaching hospital. Med. J. Aust. 1981, 1, 132–133. [Google Scholar] [PubMed]
- Dyrlund, T.F.; Poulsen, E.T.; Scavenius, C.; Sanggaard, K.W.; Enghild, J.J. Ms data miner: A web-based software tool to analyze, compare, and share mass spectrometry protein identifications. Proteomics 2012, 12, 2792–2796. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poulsen, E.T.; Larsen, A.; Zollo, A.; Jørgensen, A.L.; Sanggaard, K.W.; Enghild, J.J.; Matrone, C. New Insights to Clathrin and Adaptor Protein 2 for the Design and Development of Therapeutic Strategies. Int. J. Mol. Sci. 2015, 16, 29446-29453. https://doi.org/10.3390/ijms161226181
Poulsen ET, Larsen A, Zollo A, Jørgensen AL, Sanggaard KW, Enghild JJ, Matrone C. New Insights to Clathrin and Adaptor Protein 2 for the Design and Development of Therapeutic Strategies. International Journal of Molecular Sciences. 2015; 16(12):29446-29453. https://doi.org/10.3390/ijms161226181
Chicago/Turabian StylePoulsen, Ebbe Toftgaard, Agnete Larsen, Alen Zollo, Arne L. Jørgensen, Kristian W. Sanggaard, Jan J. Enghild, and Carmela Matrone. 2015. "New Insights to Clathrin and Adaptor Protein 2 for the Design and Development of Therapeutic Strategies" International Journal of Molecular Sciences 16, no. 12: 29446-29453. https://doi.org/10.3390/ijms161226181
APA StylePoulsen, E. T., Larsen, A., Zollo, A., Jørgensen, A. L., Sanggaard, K. W., Enghild, J. J., & Matrone, C. (2015). New Insights to Clathrin and Adaptor Protein 2 for the Design and Development of Therapeutic Strategies. International Journal of Molecular Sciences, 16(12), 29446-29453. https://doi.org/10.3390/ijms161226181





