Omics-Based Comparative Transcriptional Profiling of Two Contrasting Rice Genotypes during Early Infestation by Small Brown Planthopper
Abstract
:1. Introduction
2. Results
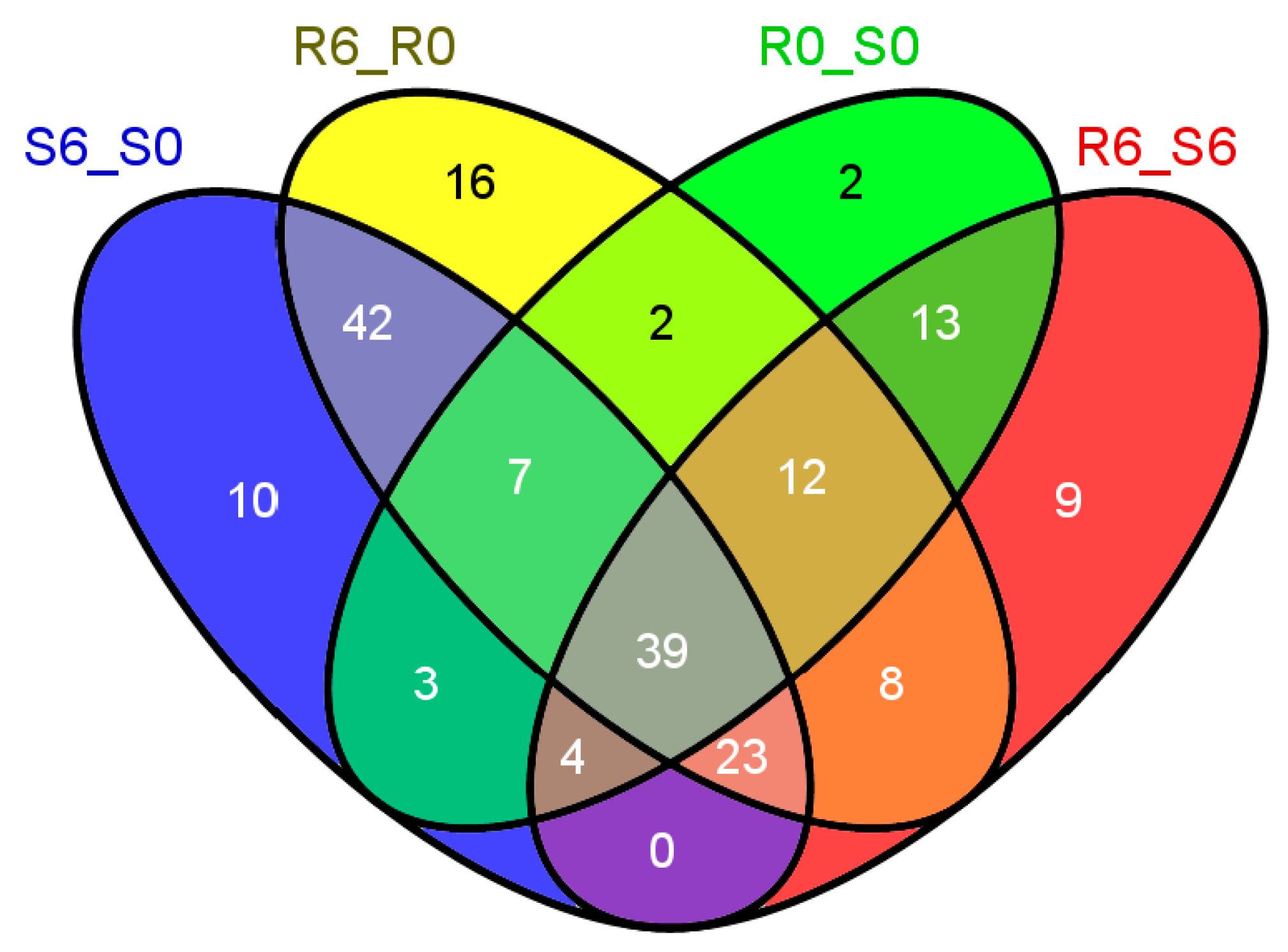
2.1. The Global Transcriptional Profiling of Rice In Response to Small Brown Planthopper (SBPH) Infestation
2.2. The Global Metabolic Map in Response to SBPH Infestation
2.3. An Overview of the Differentially Regulated Metabolic Pathways
2.4. The Global Batch Identification of Individual Metabolic Pathways by Pathway Tools Omics Viewer
| Category | R6_R0 | S6_S0 | R6_S6 | R0_S0 |
|---|---|---|---|---|
| Activation/Inactivation/Interconversion | ||||
| Inactivation | 2 | 2 | ||
| activation | 1 | |||
| Biosynthesis | ||||
| Secondary metabolites biosynthesis | 30 | 20 | 18 | 16 |
| Amino acids biosynthesis | 20 | 19 | 10 | 10 |
| Cofactors, prosthetic groups, electron carriers biosynthesis | 14 | 16 | 15 | 9 |
| Carbohydrates biosynthesis | 13 | 13 | 10 | 6 |
| Nucleosides and nucleotides biosynthesis | 9 | 9 | 5 | 5 |
| Cell Structures biosynthesis | 6 | 6 | 5 | 4 |
| Fatty acids and lipids biosynthesis | 6 | 8 | 7 | 7 |
| Hormones biosynthesis | 5 | 6 | 4 | 5 |
| Amines and polyamines biosynthesis | 4 | 2 | 2 | 2 |
| Aromatic compounds biosynthesis | 2 | 1 | 2 | 2 |
| Aminoacyl-tRNA charging | 1 | 1 | 1 | 1 |
| Degradation/Utilization/Assimilation | ||||
| Amino acids degradation | 14 | 16 | 15 | 8 |
| Carbohydrates degradation | 10 | 9 | 10 | 6 |
| Inorganic nutrients metabolism | 5 | 1 | 1 | |
| Fatty acid and lipids degradation | 4 | 2 | 4 | 4 |
| Detoxification | 3 | 3 | 1 | 2 |
| C1 compounds utilization and assimilation | 3 | 2 | 2 | 1 |
| Secondary metabolites degradation | 3 | 2 | 2 | 1 |
| Alcohols degradation | 2 | 3 | ||
| Nucleosides and nucleotides degradation | 2 | 1 | 1 | 2 |
| Hormones degradation | 1 | 1 | 1 | |
| Polymeric compounds degradation | 1 | 1 | 1 | |
| Amines and polyamines degradation | 1 | 1 | ||
| Aldehyde degradation | 1 | 1 | ||
| Degradation/Utilization/Assimilation-Other | 1 | |||
| Aromatic compounds degradation | 1 | |||
| Nicotine degradation | 1 | |||
| Generation of Precursor Metabolites and Energy | ||||
| Fermentation | 4 | 3 | 3 | 3 |
| Glycolysis | 2 | 2 | 1 | |
| Pentose phosphate pathways | 2 | 2 | 1 | |
| Photosynthesis | 2 | 2 | 1 | |
| Respiration | 1 | 1 | 2 | |
| TCA cycle | 1 | 1 | 2 | 2 |
| Acetyl-CoA biosynthesis | 1 | |||
| Methanogenesis | 1 | |||
2.5. The Metabolic Pathways Potentially Related to SBPH Resistance in Rice
| Pathway | S6_S0 | R6_R0 | R0_S0 | R6_S6 |
|---|---|---|---|---|
| Class I | ||||
| Betanidin degradation | down | lower | ||
| Cytokinins degradation | down | lower | ||
| Glutamate degradation III | down | lower | ||
| IAA conjugate biosynthesis I | up | higher | ||
| IAA conjugate biosynthesis II | up | higher | ||
| Spermine biosynthesis | up | higher | ||
| Very long chain fatty acid biosynthesis | up | higher | ||
| Momilactone biosynthesis | up | higher | ||
| Class II | ||||
| Flavonoid biosynthesis | down | higher | higher | |
| Mixed acid fermentation | down | higher | higher | |
| Pinobanksin biosynthesis | down | higher | higher | |
| Aminopropanol biosynthesis | down | lower | lower | |
| Salicylate biosynthesis | down | lower | lower | |
| Serine biosynthesis | down | lower | lower | |
| Threonine degradation II | down | lower | lower | |
| Threonine degradation III (to methylglyoxal) | down | lower | lower | |
| Reductive TCA cycle I | up | higher | higher | |
| 13-LOX and 13-HPL pathway | up | lower | lower | |
| Divinyl ether biosynthesis II (13-LOX) | up | lower | lower | |
| Class III | ||||
| Ureide biosynthesis | up | down | lower | lower |
| phenylalanine degradation III | up | down | lower | |
2.6. Construction the Network of the Metabolic Pathways Potentially Related to SBPH Resistance in the SBPH-Resistant Rice Plant

2.7. The Metabolic Pathways Potentially Related to SBPH Susceptibility in Rice
| Pathway | S6_S0 | R6_R0 | R0_S0 | R6_S6 |
|---|---|---|---|---|
| Class II | ||||
| methylerythritol phosphate pathway | down | lower | lower | |
| thiamine biosynthesis | down | lower | lower | |
| lysine degradation I | up | higher | higher | |
| phospholipid biosynthesis II | up | lower | lower | |
| Class III | ||||
| phenylalanine degradation III | up | down | lower | |
| ureide biosynthesis | up | down | lower | lower |
2.8. The Changes of Free Amino Acid Levels in Response to SBPH Infestation
| Free Amino Acid | Concentration of Free Amino Acids (µg per g FW) x | Percent Change of Concentration | ||||
|---|---|---|---|---|---|---|
| S0 | R0 | S6 | R6 | S6_S0 | R6_R0 | |
| Isoleucine | 27.7 ± 2.7 | 43.5 ± 4.2 | 1.9 ± 0.4 | 12.5 ± 1.4 | −93.1 | −71.3 |
| Citrulline | 12 ± 1.3 | 13 ± 1.4 | 2.4 ± 0.3 | 7 ± 0.8 | −80.0 | −46.2 |
| Lysine | 27 ± 2.8 | 31.1 ± 3.4 | 6.3 ± 0.6 | 9.8 ± 0.9 | −76.7 | −68.5 |
| Methionine | 11.80 ± 0.59 | 3.60 ± 0.28 | 11.03 ± 0.67 | 0.42 ± 0.04 | −69.5 | −96.2 |
| Cystine | 11.44 ± 0.99 | 5.97 ± 0.45 | 24.47 ± 1.79 | 10.14 ± 0.91 | −47.8 | −58.6 |
| Valine | 23.4 ± 1.9 | 23.1 ± 2.5 | 13.6 ± 1.5 | 16.8 ± 1.3 | −41.9 | −27.3 |
| Aspartic acid | 26.95 ± 3.59 | 15.68 ± 1.23 | 23.70 ± 2.25 | 21.65 ± 1.94 | −41.8 | −8.6 |
| Cystathionine | 2.73 ± 0.25 | 1.74 ± 0.13 | 4.58 ± 0.68 | 0.39 ± 0.03 | −36.3 | −91.5 |
| Ornithine | 2.1 ± 0.1 | 3.6 ± 0.1 | 1.4 ± 0.1 | 2.9 ± 0.2 | −33.3 | −19.4 |
| β-Aminoisobutyric Acid | 3.6 ± 0.3 | 5 ± 0.4 | 2.7 ± 0.2 | 1.8 ± 0.2 | −25.0 | −64.0 |
| Ethanolamine | 11 ± 1.3 | 9.1 ± 1 | 8.7 ± 0.9 | 7.5 ± 0.8 | −20.9 | −17.6 |
| Phenylalanine | 18.5 ± 1.8 | 7.5 ± 0.3 | 3.2 ± 0.3 | 7.9 ± 0.5 | −82.7 | 5.3 |
| Arginine | 90.4 ± 10.9 | 30 ± 3.4 | 18.6 ± 1.8 | 46.7 ± 4.8 | −79.4 | 55.7 |
| β-Alanine | 6.4 ± 0.3 | 5.5 ± 0.4 | 3.9 ± 0.4 | 8.1 ± 0.6 | −39.1 | 47.3 |
| Serine | 7.19 ± 0.71 | 4.43 ± 0.31 | 2.87 ± 0.26 | 5.46 ± 0.45 | −38.4 | 90.2 |
| Glycine | 7.7 ± 1 | 9.3 ± 1.6 | 5.2 ± 0.4 | 11.1 ± 1.2 | −32.5 | 19.4 |
| Leucine | 2.2 ± 0.1 | 6.3 ± 0.2 | 1.5 ± 0.1 | 11.3 ± 1.3 | −31.8 | 79.4 |
| Glutamic Acid | 114.3 ± 8.8 | 143.8 ± 14.2 | 81.9 ± 8.9 | 160.4 ± 15.4 | −28.3 | 11.5 |
| Threonine | 2.9 ± 0.3 | 1.9 ± 0.2 | 2.1 ± 0.2 | 3.6 ± 0.3 | −27.6 | 89.5 |
| Alanine | 10 ± 1.1 | 10.9 ± 1.2 | 8.9 ± 0.4 | 20 ± 0.8 | −11.0 | 83.5 |
| α-Amino-n-butyric Acid | 2.5 ± 0.2 | 3.6 ± 0.2 | 4 ± 0.2 | 2.9 ± 0.2 | 60.0 | −19.4 |
| Tyrosine | 4.7 ± 0.3 | 3.7 ± 0.3 | 11.3 ± 1.2 | 14.7 ± 1.2 | 140.4 | 297.3 |
| α-Aminoadipic Acid | 7.9 ± 0.6 | 11.5 ± 1.4 | 14.3 ± 0.8 | 28.4 ± 1.2 | 81.0 | 147.0 |
| γ-Amino-n-butyric Acid | 4.4 ± 0.4 | 2.3 ± 0.2 | 6.2 ± 0.5 | 22.9 ± 2 | 40.9 | 895.7 |
| Proline | 5 ± 0.3 | 2.6 ± 0.2 | 5.7 ± 0.3 | 5.3 ± 0.4 | 14.0 | 103.8 |
| Total | 443.7 ± 40.7 | 434.1 ± 40.9 | 235.3 ± 21.1 | 439.6 ± 37.9 | −47.0 | 1.3 |
3. Discussion
4. Materials and Methods
4.1. Plant and Insect Materials
4.2. SBPH Infestation Experiments
4.3. Affymetrix GeneChip Analysis
4.4. Validating of Rice Transcriptome Data for Pathway Tools Omics Viewer
4.5. Visualization of Rice Transcriptome Data Using Pathway Tools Omics Viewer
4.6. Animation the Comparative Transcriptome Analysis Using Pathway Tools Omics Viewer
4.7. Identification of the Differentially Regulated Individual Metabolic Pathways by the Pathway Tools Omics Viewer
4.8. Identification of Metabolic Pathways Potentially Related to SBPH Resistance by Pathway Tools Omics Viewer
4.9. Measurement of Free Amino Acids
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Choi, M.S.; Kim, Y.H.; Park, H.M.; Seo, B.Y.; Jung, J.K.; Kim, S.T.; Kim, M.C.; Shin, D.B.; Yun, H.T.; Choi, I.S.; et al. Expression of BrD1, a plant defensin from Brassica rapa, confers resistance against brown planthopper (Nilaparvata lugens) in transgenic rices. Mol. Cells 2009, 28, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Brar, D.S.; Virk, P.S.; Jena, K.K.; Khush, G.S. Breeding for resistance to planthoppers in rice. In Planthoppers: New Threats to the Sustainability of Intensive Rice Production Systems in Asia; Heong, K., Hardy, B., Eds.; International Rice Research Institute: Los Baños, Philippines, 2009; pp. 401–428. [Google Scholar]
- Yuan, H.; Chen, X.; Zhu, L.; He, G. Identification of genes responsive to brown planthopper Nilaparvata lugens Stål (Homoptera: Delphacidae) feeding in rice. Planta 2005, 221, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Du, B.; Zhang, W.; Liu, B.; Hu, J.; Wei, Z.; Shi, Z.; He, R.; Zhu, L.; Chen, R.; Han, B.; et al. Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc. Natl. Acad. Sci. USA 2009, 106, 22163–22168. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zhou, J.; Peng, X.; Xu, H.; Liu, C.; Du, B.; Yuan, H.; Zhu, L.; He, G. The Bphi008a gene interacts with the ethylene pathway and transcriptionally regulates MAPK genes in the response of rice to brown planthopper feeding. Plant Physiol. 2011, 156, 856–872. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Ju, H.; Zhou, G.; Zhu, C.; Erb, M.; Wang, X.; Wang, P.; Lou, Y. An EAR-motif-containing ERF transcription factor affects herbivore-induced signaling, defense and resistance in rice. Plant J. 2011, 68, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Ma, L.; Zhao, J.; Li, Z.; Sun, F.; Lu, X. Comparative transcriptome analysis of two rice varieties in response to rice stripe virus and small brown planthoppers during early interaction. PLoS ONE 2013, 8, e82126. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; Du, B.; Shangguan, X.; Zhao, Y.; Pan, Y.; Zhu, L.; He, Y.; He, G. BAC and RNA sequencing reveal the brown planthopper resistance gene BPH15 in a recombination cold spot that mediates a unique defense mechanism. BMC Genom. 2014, 15, 674. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhu, L.; He, G. Differential gene expression in response to brown planthopper feeding in rice. J. Plant Physiol. 2004, 161, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Hao, P.; Liu, C.; Wang, Y.; Chen, R.; Tang, M.; Du, B.; Zhu, L.; He, G. Herbivore-induced callose deposition on the sieve plates of rice: An important mechanism for host resistance. Plant Physiol. 2008, 146, 1810–1820. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, X.; Yuan, H.; Chen, R.; Zhu, L.; He, R.; He, G. Responses of two contrasting genotypes of rice to brown planthopper. Mol. Plant Microbe Interact. 2008, 21, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, H.; Si, Y.; Zhang, H.; Guo, H.; Miao, X. Microarray analysis of broad-spectrum resistance derived from an indica cultivar Rathu Heenati. Planta 2012, 235, 829–840. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Ratcliffe, O.J. A genomic perspective on plant transcription factors. Curr. Opin. Plant Biol. 2000, 3, 423–434. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, H.; Li, H.; Zhang, H.; Miao, X. Identification of transcription factors potential related to brown planthopper resistance in rice via microarray expression profiling. BMC Genom. 2012, 13, 687. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hao, J.; Chen, X.; Hao, Z.; Wang, X.; Lou, Y.; Peng, Y.; Guo, Z. Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol. Biol. 2007, 65, 799–815. [Google Scholar] [CrossRef] [PubMed]
- Dharmawardhana, P.; Ren, L.; Amarasinghe, V.; Monaco, M.; Thomason, J.; Ravenscroft, D.; McCouch, S.; Ware, D.; Jaiswal, P. A genome scale metabolic network for rice and accompanying analysis of tryptophan, auxin and serotonin biosynthesis regulation under biotic stress. Rice 2013, 6, 15. [Google Scholar] [CrossRef] [PubMed]
- Paley, S.M.; Karp, P.D. The Pathway Tools cellular overview diagram and Omics Viewer. Nucleic Acids Res. 2006, 34, 3771–3778. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Dreher, K.; Karthikeyan, A.; Chi, A.; Pujar, A.; Caspi, R.; Karp, P.; Kirkup, V.; Latendresse, M.; Lee, C.; et al. Creation of a genome-wide metabolic pathway database for Populus trichocarpa using a new approach for reconstruction and curation of metabolic pathways for plants. Plant Physiol. 2010, 153, 1479–1491. [Google Scholar] [CrossRef] [PubMed]
- Oliveros, J. VENNY. An interactive tool for comparing lists with Venn Diagrams. Available online: http://bioinfogp.cnb.csic.es/tools/venny_old/index.html (accessed on 13 November 2014).
- Arimura, G.; Tashiro, K.; Kuhara, S.; Nishioka, T.; Ozawa, R.; Takabayashi, J. Gene responses in bean leaves induced by herbivory and by herbivore-induced volatiles. Biochem. Biophys. Res. Commun. 2000, 277, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, A.A. Induced responses to herbivory and increased plant performance. Science 1998, 279, 1201–1202. [Google Scholar] [CrossRef] [PubMed]
- Dicke, M. Induced responses to herbivory by R. Karban and I.T. Baldwin. Trends Ecol. Evol. 1998, 13, 83. [Google Scholar] [CrossRef]
- Chapman, R. The role of the leaf surface in food selection by acridids and other insects. Colloq. Int. Centre Natl. Rech. Sci. 1977, 265, 133–149. [Google Scholar]
- Woodhead, S.; Padgham, D.E. The effect of plant surface characteristics on resistance of rice to the brown planthopper, Nilaparvata lugens. Entomol. Exp. Appl. 1988, 47, 15–22. [Google Scholar] [CrossRef]
- Samuels, L.; Kunst, L.; Jetter, R. Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 2008, 59, 683–707. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, T.; Wynne, G.D. The effects of stress on plant cuticular waxes. New Phytol. 2006, 171, 469–499. [Google Scholar] [CrossRef] [PubMed]
- Raffaele, S.; Leger, A.; Roby, D. Very long chain fatty acid and lipid signaling in the response of plants to pathogens. Plant Signal. Behav. 2009, 4, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Qi, J.; Ren, N.; Cheng, J.; Erb, M.; Mao, B.; Lou, Y. Silencing OsHI-LOX makes rice more susceptible to chewing herbivores, but enhances resistance to a phloem feeder. Plant J. 2009, 60, 638–648. [Google Scholar] [CrossRef] [PubMed]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D. Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Pangesti, N.; Pineda, A.; Pieterse, C.M.; Dicke, M.; van Loon, J.J. Two-way plant mediated interactions between root-associated microbes and insects: From ecology to mechanisms. Front. Plant Sci. 2013, 4, 414. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xu, J.; Leng, Y.; Xiong, G.; Hu, J.; Zhang, G.; Huang, L.; Wang, L.; Guo, L.; Li, J.; et al. Quantitative trait loci identification, fine mapping and gene expression profiling for ovicidal response to whitebacked planthopper (Sogatella furcifera Horváth) in rice (Oryza sativa L.). BMC Plant Biol. 2014, 14, 145. [Google Scholar] [CrossRef] [PubMed]
- Erb, M.; Meldau, S.; Howe, G.A. Role of phytohormones in insect-specific plant reactions. Trends Plant Sci. 2012, 17, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Sano, H.; Seo, S.; Koizumi, N.; Niki, T.; Iwamura, H.; Ohashi, Y. Regulation by cytokinins of endogenous levels of jasmonic and salicylic acids in mechanically wounded tobacco plants. Plant Cell Physiol. 1996, 37, 762–769. [Google Scholar] [CrossRef]
- Strack, D.; Vogt, T.; Schliemann, W. Recent advances in betalain research. Phytochemistry 2003, 62, 247–269. [Google Scholar] [CrossRef]
- Casique-Arroyo, G.; Martinez-Gallardo, N.; de La Gonzalez, V.L.; Delano-Frier, J.P. Betacyanin biosynthetic genes and enzymes are differentially induced by (a)biotic stress in Amaranthus hypochondriacus. PLoS ONE 2014, 9, e99012. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, I.; Kissen, R.; Bones, A.M. Phytoalexins in defense against pathogens. Trends Plant Sci. 2012, 17, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Kabuto, C.; Sasaki, N.; Tsunagawa, M.; Aizawa, H.; Fujita, K.; Kato, Y.; Kitahara, Y. Momilactones, growth inhibitors from rice, oryza sativa L. Tetrahedron Lett. 1973, 14, 3861–3864. [Google Scholar] [CrossRef]
- Yamane, H. Biosynthesis of phytoalexins and regulatory mechanisms of it in rice. Biosci. Biotechnol. Biochem. 2013, 77, 1141–1148. [Google Scholar] [CrossRef] [PubMed]
- Kanno, H.; Hasegawa, M.; Kodama, O. Accumulation of salicylic acid, jasmonic acid and phytoalexins in rice, Oryza sativa, infested by the white-backed planthopper, Sogatella furcifera (Hemiptera: Delphacidae). Appl. Entomol. Zool. 2012, 47, 27–34. [Google Scholar] [CrossRef]
- Verhoeyen, M.E.; Bovy, A.; Collins, G.; Muir, S.; Robinson, S.; de Vos, C.H.; Colliver, S. Increasing antioxidant levels in tomatoes through modification of the flavonoid biosynthetic pathway. J. Exp. Bot. 2002, 53, 2099–2106. [Google Scholar] [CrossRef] [PubMed]
- Adjei-Afriyie, F.; Kim, C.S.; Takemura, M.; Ishikawa, M.; Horiike, M. Isolation and identification of the probing stimulants in the rice plant for the white-back planthopper, Sogatella furcifera (Homoptera: Delphacidae). Biosci. Biotechnol. Biochem. 2000, 64, 443–446. [Google Scholar] [CrossRef] [PubMed]
- Adjei-Afriyie, F.; Kim, C.S.; Takemura, M.; Ishikawa, M.; Tebayashi, S.; Horiike, M. Probing stimulants from the rice plant towards the smaller brown planthopper, Laodelphax striatellus (Fallén) (Homoptera: Delphacidae). Z. Naturforschung C 2000, 55, 1038–1043. [Google Scholar] [CrossRef]
- Steinbrenner, A.D.; Gomez, S.; Osorio, S.; Fernie, A.R.; Orians, C.M. Herbivore-induced changes in tomato (Solanum lycopersicum) primary metabolism: A whole plant perspective. J. Chem. Ecol. 2011, 37, 1294–1303. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Polyamines and abiotic stress tolerance in plants. Plant Signal. Behav. 2010, 5, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Kusano, T.; Yamaguchi, K.; Berberich, T.; Takahashi, Y. Advances in polyamine research in 2007. J. Plant Res. 2007, 120, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Jander, G.; de Vos, M. Non-protein amino acids in plant defense against insect herbivores: Representative cases and opportunities for further functional analysis. Phytochemistry 2011, 72, 1531–1537. [Google Scholar] [CrossRef] [PubMed]
- Malik, N.S.; Perez, J.L.; Kunta, M.; Patt, J.M.; Mangan, R.L. Changes in free amino acids and polyamine levels in Satsuma leaves in response to Asian citrus psyllid infestation and water stress. Insect Sci. 2014, 21, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Cascales-Minana, B.; Munoz-Bertomeu, J.; Flores-Tornero, M.; Anoman, A.D.; Pertusa, J.; Alaiz, M.; Osorio, S.; Fernie, A.R.; Segura, J.; Ros, R. The phosphorylated pathway of serine biosynthesis is essential both for male gametophyte and embryo development and for root growth in Arabidopsis. Plant Cell 2013, 25, 2084–2101. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Thornburg, R. Wound-inducible genes in plants. In Inducible Gene Expression; Reynolds, P.H.S., Ed.; Book News: Portland, OR, USA, 1999; pp. 127–158. [Google Scholar]
- Sardans, J.; Gargallo-Garriga, A.; Perez-Trujillo, M.; Parella, T.J.; Seco, R.; Filella, I.; Penuelas, J. Metabolic responses of Quercus ilex seedlings to wounding analysed with nuclear magnetic resonance profiling. Plant Biol. 2014, 16, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.J.; Chen, K.W.; Ruaan, M.K. Mixed acid fermentation of glucose as a mechanism of emphysematous urinary tract infection. J. Urol. 1991, 146, 148–151. [Google Scholar] [PubMed]
- Trchounian, A.; Gary, S.R. Novel insights into the bioenergetics of mixed-acid fermentation: Can hydrogen and proton cycles combine to help maintain a proton motive force? IUBMB Life 2014, 66, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hugler, M.; Wirsen, C.O.; Fuchs, G.; Taylor, C.D.; Sievert, S.M. Evidence for autotrophic CO2 fixation via the reductive tricarboxylic acid cycle by members of the epsilon subdivision of proteobacteria. J. Bacteriol. 2005, 187, 3020–3027. [Google Scholar] [CrossRef] [PubMed]
- Stuttmann, J.; Hubberten, H.M.; Rietz, S.; Kaur, J.; Muskett, P.; Guerois, R.; Bednarek, P.; Hoefgen, R.; Parker, J.E. Perturbation of Arabidopsis amino acid metabolism causes incompatibility with the adapted biotrophic pathogen Hyaloperonospora arabidopsidis. Plant Cell 2011, 23, 2788–2803. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, R.; Oritani, T.; Nishi, A. Studies on nitrogen metabolism in crop plants. VIII. Occurrence of kinetic-like factor in root exudates of rice plant. Proc. Crop Sci. Soc. Jpn. 1970, 39, 363–369. [Google Scholar] [CrossRef]
- Schubert, K.R. Enzymes of purine biosynthesis and catabolism in glycine max: I. Comparison of activities with N2 fixation and composition of xylem exudates during nodule development. Plant Physiol. 1981, 68, 1115–1122. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Dong, Y.; Yang, L.; Ma, B.; Ma, R.; Huang, F.; Wang, C.; Hu, H.; Li, C.; Yan, C.; et al. Small brown planthopper resistance loci in wild rice (Oryza officinalis). Mol. Genet. Genom. 2014, 289, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.W.; Gao, D.M.; Chen, S.X. Studies on techniques of rapid detecting rice stripe virus in Laodelphax striatellus. Zhejiang J. Agric. Sci. 1994, 6, 226–229. [Google Scholar]
- Tusher, V.G.; Tibshirani, R.; Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 2001, 98, 5116–5121. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Ouyang, S.; Wang, A.; Zhu, W.; Maiti, R.; Lin, H.; Hamilton, J.; Haas, B.; Sultana, R.; Cheung, F.; et al. The institute for genomic research Osa1 rice genome annotation database. Plant Physiol. 2005, 138, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Korpelainen, H.; Li, C. Physiological and biochemical responses to high Mn concentrations in two contrasting Populus cathayana populations. Chemosphere 2007, 68, 686–694. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Yang, L.; Li, M.; Ma, B.; Yan, C.; Chen, J. Omics-Based Comparative Transcriptional Profiling of Two Contrasting Rice Genotypes during Early Infestation by Small Brown Planthopper. Int. J. Mol. Sci. 2015, 16, 28746-28764. https://doi.org/10.3390/ijms161226128
Zhang W, Yang L, Li M, Ma B, Yan C, Chen J. Omics-Based Comparative Transcriptional Profiling of Two Contrasting Rice Genotypes during Early Infestation by Small Brown Planthopper. International Journal of Molecular Sciences. 2015; 16(12):28746-28764. https://doi.org/10.3390/ijms161226128
Chicago/Turabian StyleZhang, Weilin, Ling Yang, Mei Li, Bojun Ma, Chengqi Yan, and Jianping Chen. 2015. "Omics-Based Comparative Transcriptional Profiling of Two Contrasting Rice Genotypes during Early Infestation by Small Brown Planthopper" International Journal of Molecular Sciences 16, no. 12: 28746-28764. https://doi.org/10.3390/ijms161226128
APA StyleZhang, W., Yang, L., Li, M., Ma, B., Yan, C., & Chen, J. (2015). Omics-Based Comparative Transcriptional Profiling of Two Contrasting Rice Genotypes during Early Infestation by Small Brown Planthopper. International Journal of Molecular Sciences, 16(12), 28746-28764. https://doi.org/10.3390/ijms161226128





