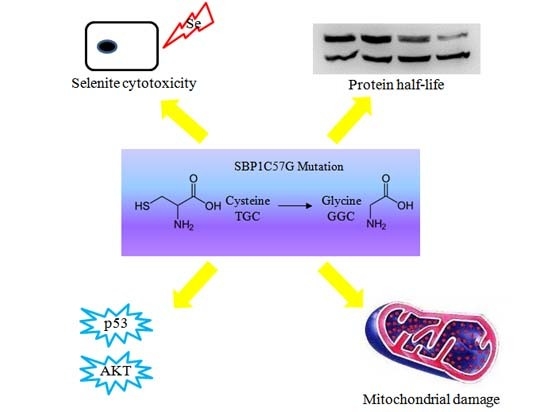
A Critical Role for Cysteine 57 in the Biological Functions of Selenium Binding Protein-1
Abstract
:1. Introduction
2. Results
2.1. SBP1GLY Was Impaired in Its Ability to Protect HCT116 Cells from Selenite-Induced Toxicity
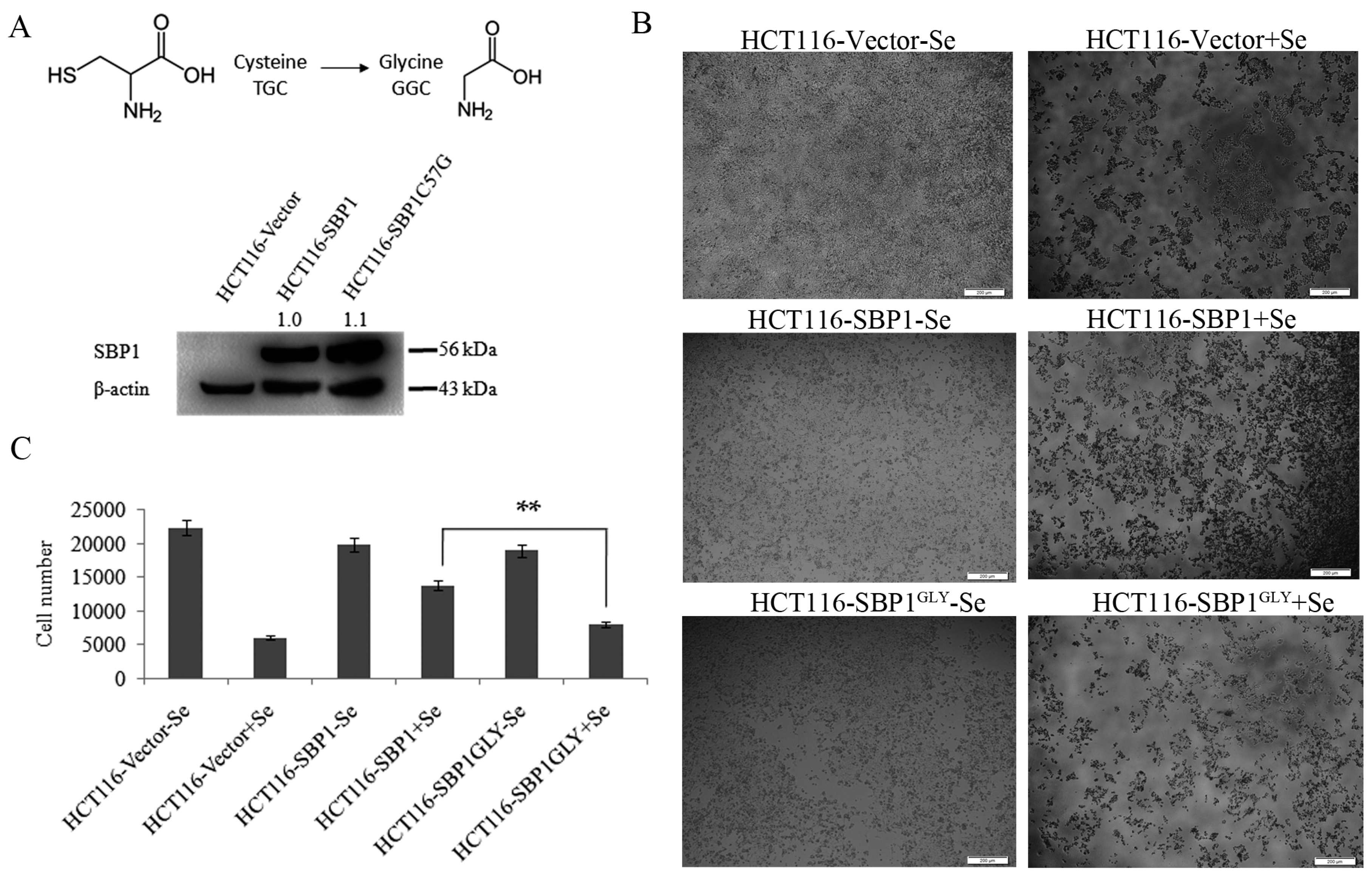
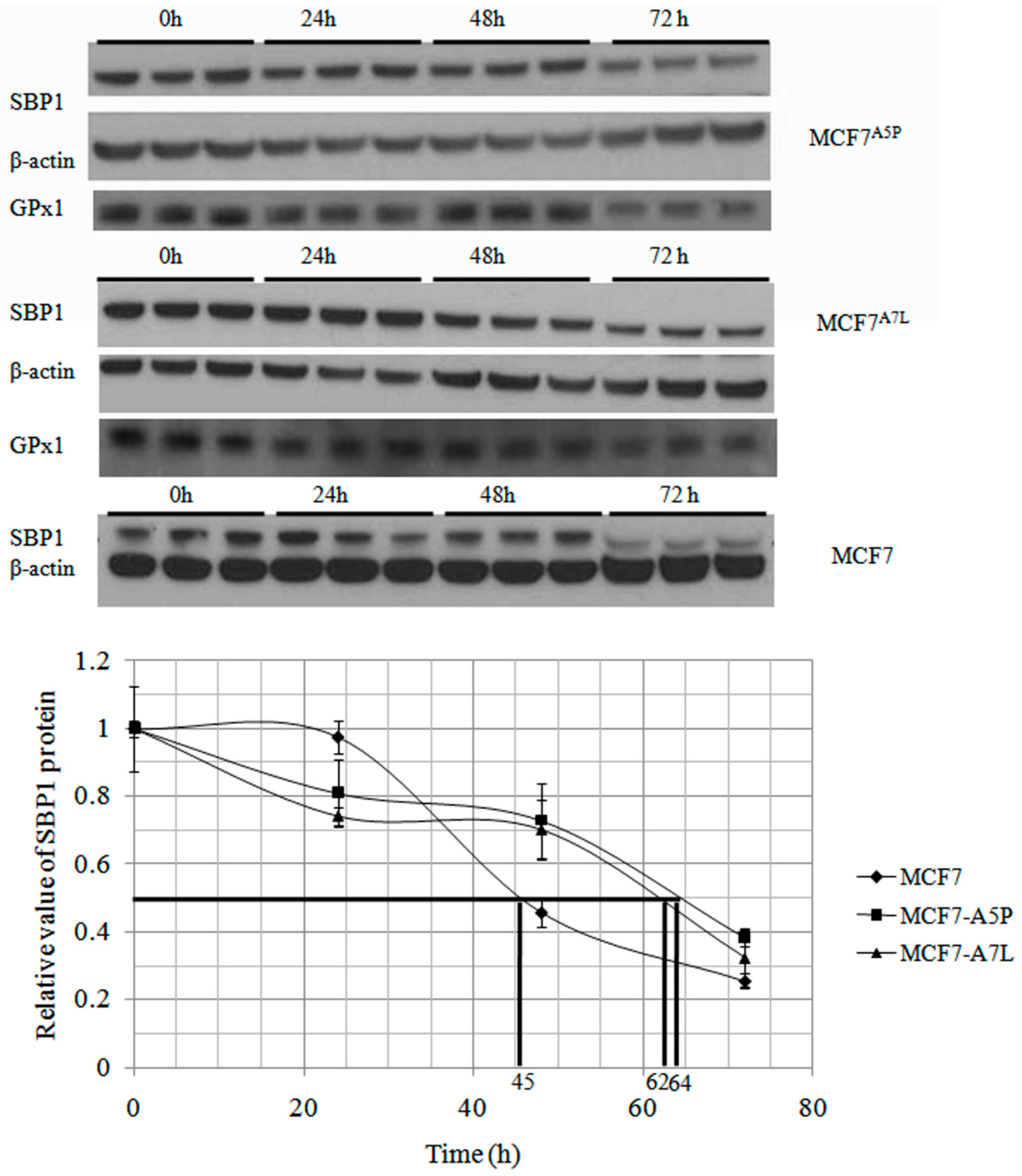
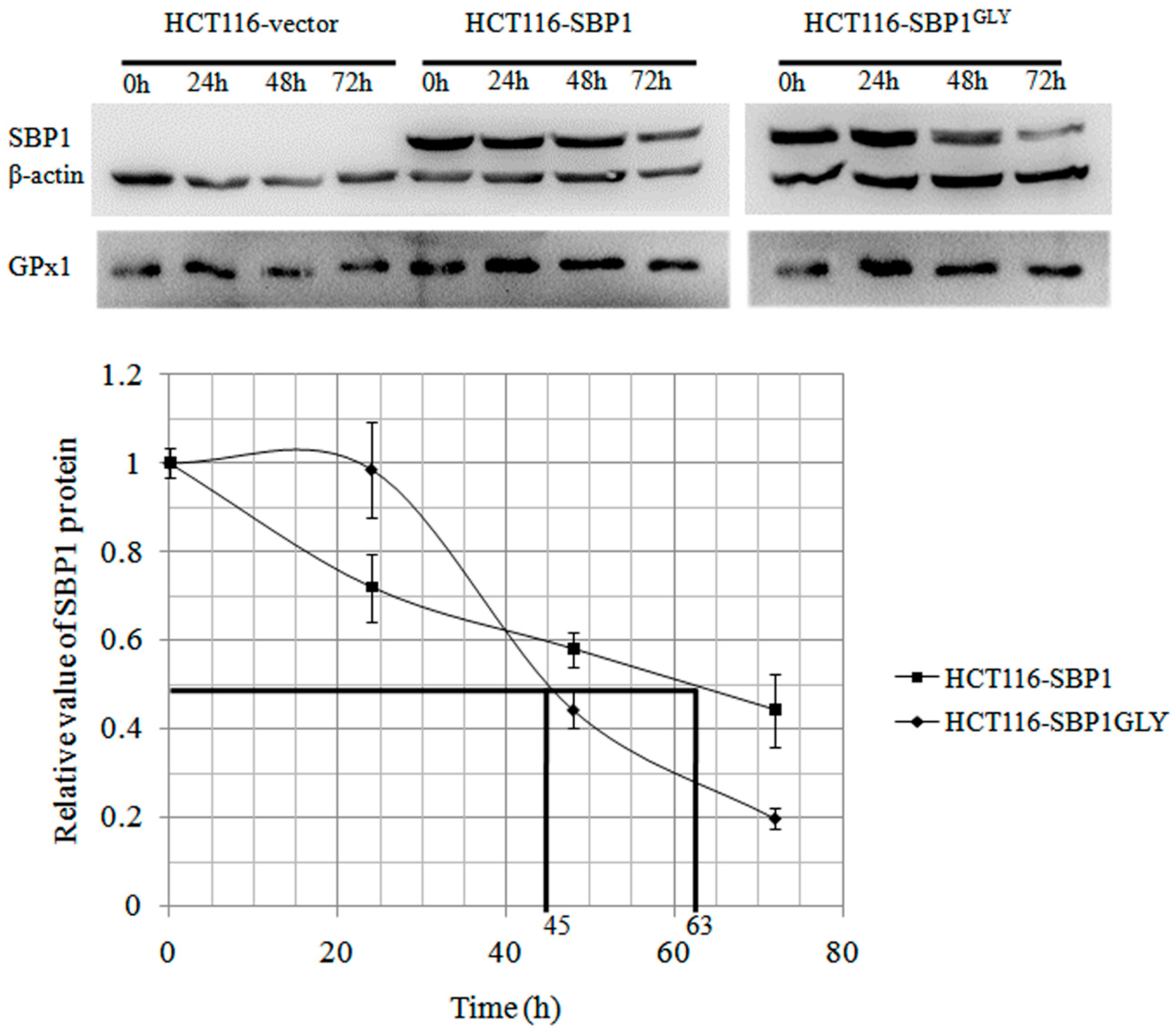
2.2. The Cytosolic Half-Life of SBP1 Is Affected by the Amino Acid at Position 57
2.3. Differential Affects of Wild Type SBP1 and SBP1GLY on Signaling Pathways Relevant to Carcinogenesis

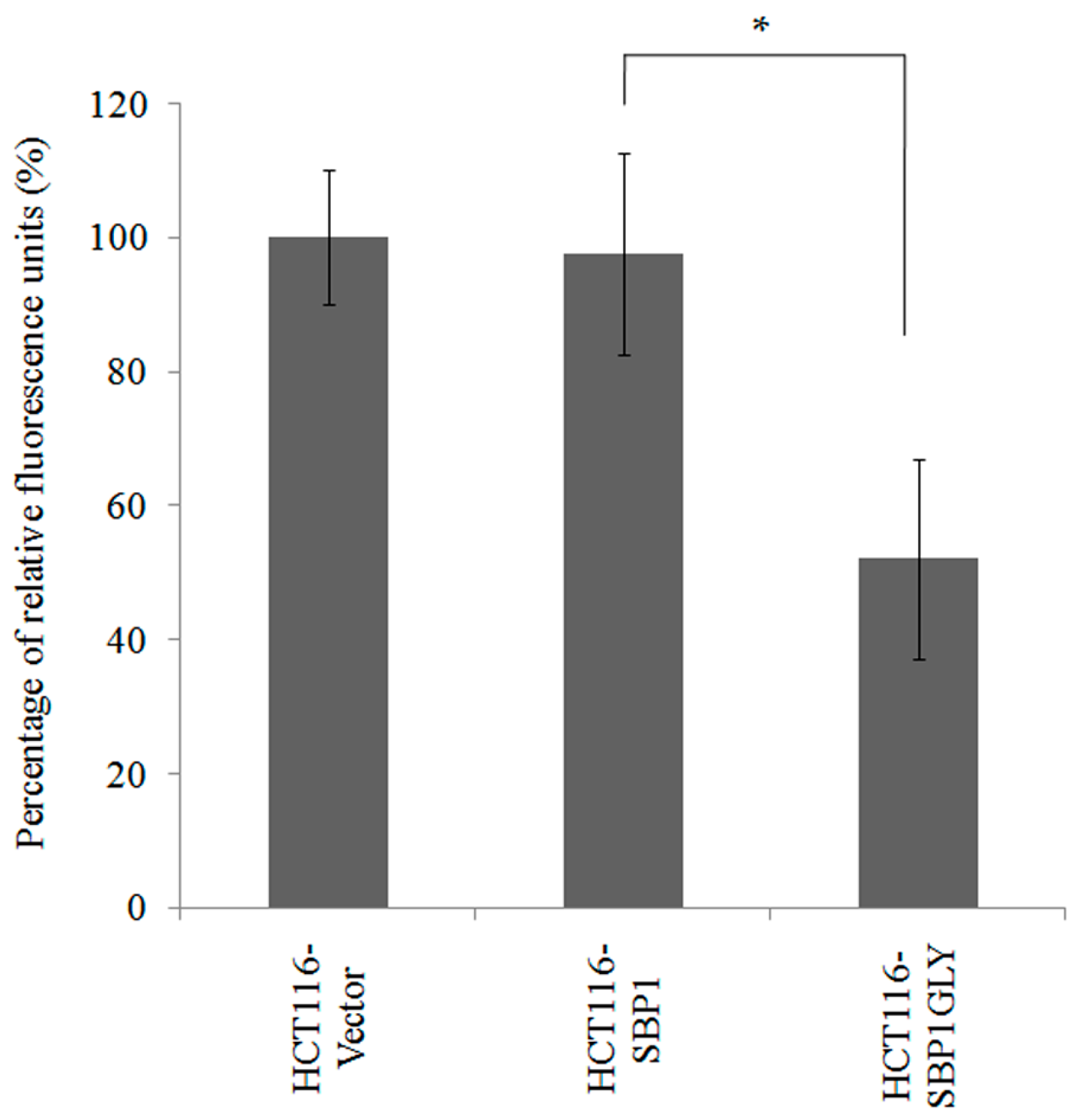
2.4. Expression of SBP1GLY Induced Mitochondrial Damage in HCT116 Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Reagents
4.2. Plasmid Construction
4.3. Immunoblot Analysis
4.4. Measurement of Selenite-Induced Cytotoxicity
4.5. Determination of the SBP1 Half-Life
4.6. Mitochondrial Damage Assay
4.7. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chang, P.W.; Tsui, S.K.; Liew, C.; Lee, C.C.; Waye, M.M.; Fung, K.P. Isolation, characterization, and chromosomal mapping of a novel cDNA clone encoding human selenium binding protein. J. Cell. Biochem. 1997, 64, 217–224. [Google Scholar] [CrossRef]
- Chen, G.; Wang, H.; Miller, C.T.; Thomas, D.G.; Gharib, T.G.; Misek, D.E.; Giordano, T.J.; Orringer, M.B.; Hanash, S.M.; Beer, D.G. Reduced selenium-binding protein 1 expression is associated with poor outcome in lung adenocarcinomas. J. Pathol. 2004, 202, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Ding, G.; Gu, C.; Zhou, J.; Kuang, M.; Ji, Y.; He, Y.; Kondo, T.; Fan, J. Decreased selenium-binding protein 1 enhances glutathione peroxidase 1 activity and downregulates HIF-1α to promote hepatocellular carcinoma invasiveness. Clin. Cancer Res. 2012, 18, 3042–3053. [Google Scholar] [CrossRef] [PubMed]
- Raucci, R.; Colonna, G.; Guerriero, E.; Capone, F.; Accardo, M.; Castello, G.; Costantini, S. Structural and functional studies of the human selenium binding protein-1 and its involvement in hepatocellular carcinoma. Biochim. Biophys. Acta 2011, 1814, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Goldberg, M.L.; Pohl, N.M.; Bi, X.; Tong, C.; Xiong, B.; Koh, T.J.; Diamond, A.M.; Yang, W. Functional and physical interaction between the selenium-binding protein 1 (SBP1) and the glutathione peroxidase 1 selenoprotein. Carcinogenesis 2010, 31, 1360–1366. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.C.; Park, D.C.; Ng, S.K.; Lee, J.Y.; Ni, X.; Ng, W.C.; Bandera, C.A.; Welch, W.R.; Berkowitz, R.S.; Mok, S.C.; et al. Selenium binding protein 1 in ovarian cancer. Int. J. Cancer 2006, 118, 2433–2440. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kang, H.J.; You, K.T.; Kim, S.H.; Lee, K.Y.; Kim, T.I.; Kim, C.; Song, S.Y.; Kim, H.J.; Lee, C.; et al. Suppression of human selenium-binding protein 1 is a late event in colorectal carcinogenesis and is associated with poor survival. Proteomics 2006, 6, 3466–3476. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Yang, W.; Li, M.; Byun, D.S.; Tong, C.; Nasser, S.; Zhuang, M.; Arango, D.; Mariadason, J.M.; Augenlicht, L.H. Expression of selenium-binding protein 1 characterizes intestinal cell maturation and predicts survival for patients with colorectal cancer. Mol. Nutr. Food Res. 2008, 52, 1289–1299. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.J.; Ma, Y.Y.; He, X.J.; Wang, H.J.; Ye, Z.Y.; Tao, H.Q. Suppression of selenium-binding protein 1 in gastric cancer is associated with poor survival. Hum. Pathol. 2011, 42, 1620–1628. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Dong, W.G.; Lin, J. Reduced selenium-binding protein 1 is associated with poor survival rate in gastric carcinoma. Med. Oncol. 2011, 28, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.Y.; Wang, Y.; Sytkowski, A.J. Human selenium binding protein-1 (hSP56) interacts with VDU1 in a selenium-dependent manner. Biochem. Biophys. Res. Commun. 2009, 379, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Schild, F.; Kieffer-Jaquinod, S.; Palencia, A.; Cobessi, D.; Sarret, G.; Zubieta, C.; Jourdain, A.; Dumas, R.; Forge, V.; Testemale, D.; et al. Biochemical and biophysical characterization of the selenium-binding and reducing site in Arabidopsis thaliana homologue to mammals selenium-binding protein 1. J. Biol. Chem. 2014, 289, 31765–31776. [Google Scholar] [CrossRef] [PubMed]
- Porat, A.; Sagiv, Y.; Elazar, Z. A 56-kDa selenium-binding protein participates in intra-Golgi protein transport. J. Biol. Chem. 2000, 275, 14457–14465. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fang, W.; Huang, Y.; Hu, F.; Ying, Q.; Yang, W.; Xiong, B. Reduction of selenium-binding protein 1 sensitizes cancer cells to selenite via elevating extracellular glutathione: A novel mechanism of cancer-specific cytotoxicity of selenite. Free Radic. Biol. Med. 2015, 79, 186–196. [Google Scholar] [CrossRef] [PubMed]
- Ansong, E.; Yang, W.; Diamond, A.M. Molecular cross-talk between members of distinct families of selenium containing proteins. Mol. Nutr. Food Res. 2014, 58, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Bera, S.; Weinberg, F.; Ekoue, D.N.; Ansenberger-Fricano, K.; Mao, M.; Bonini, M.G.; Diamond, A.M. Natural allelic variations in glutathione peroxidase-1 affect its subcellular localization and function. Cancer Res. 2014, 74, 5118–5126. [Google Scholar] [CrossRef] [PubMed]
- Ying, Q.; Ansong, E.; Diamond, A.M.; Lu, Z.; Yang, W.; Bie, X. Quantitative proteomic analysis reveals that anti-cancer effects of selenium-binding protein 1 in vivo are associated with metabolic pathways. PLoS ONE 2015, 10, e0126285. [Google Scholar] [CrossRef] [PubMed]
- Ansong, E.; Ying, Q.; Ekoue, D.N.; Deaton, R.; Hall, A.R.; Kajdacsy-Balla, A.; Yang, W.; Gann, P.H.; Diamond, A.M. Evidence that selenium binding protein 1 is a tumor suppressor in prostate cancer. PLoS ONE 2015, 10, e0127295. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, P.; Diamond, A.M. Molecular mechanisms by which selenoproteins affect cancer risk and progression. Biochim. Biophys. Acta 2009, 1790, 1546–1554. [Google Scholar] [CrossRef] [PubMed]
- Rayman, M.P. Selenoproteins and human health: Insights from epidemiological data. Biochim. Biophys. Acta 2009, 1790, 1533–1540. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, P.; Goldberg, M.; Herman, L.; Lee, B.S.; Wang, H.; Brown, R.L.; Foster, C.B.; Peters, U.; Diamond, A.M. Molecular consequences of genetic variations in the glutathione peroxidase 1 selenoenzyme. Cancer Res. 2009, 69, 8183–8190. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.J.; Diamond, A.M. Role of glutathione peroxidase 1 in breast cancer: Loss of heterozygosity and allelic differences in the response to selenium. Cancer Res. 2003, 63, 3347–3351. [Google Scholar] [PubMed]
- Pohl, N.M.; Tong, C.; Fang, W.; Bi, X.; Li, T.; Yang, W. Transcriptional regulation and biological functions of selenium-binding protein 1 in colorectal cancer in vitro and in nude mouse xenografts. PLoS ONE 2009, 4, e7774. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ying, Q.; Ansong, E.; Diamond, A.M.; Yang, W. A Critical Role for Cysteine 57 in the Biological Functions of Selenium Binding Protein-1. Int. J. Mol. Sci. 2015, 16, 27599-27608. https://doi.org/10.3390/ijms161126043
Ying Q, Ansong E, Diamond AM, Yang W. A Critical Role for Cysteine 57 in the Biological Functions of Selenium Binding Protein-1. International Journal of Molecular Sciences. 2015; 16(11):27599-27608. https://doi.org/10.3390/ijms161126043
Chicago/Turabian StyleYing, Qi, Emmanuel Ansong, Alan M. Diamond, and Wancai Yang. 2015. "A Critical Role for Cysteine 57 in the Biological Functions of Selenium Binding Protein-1" International Journal of Molecular Sciences 16, no. 11: 27599-27608. https://doi.org/10.3390/ijms161126043
APA StyleYing, Q., Ansong, E., Diamond, A. M., & Yang, W. (2015). A Critical Role for Cysteine 57 in the Biological Functions of Selenium Binding Protein-1. International Journal of Molecular Sciences, 16(11), 27599-27608. https://doi.org/10.3390/ijms161126043