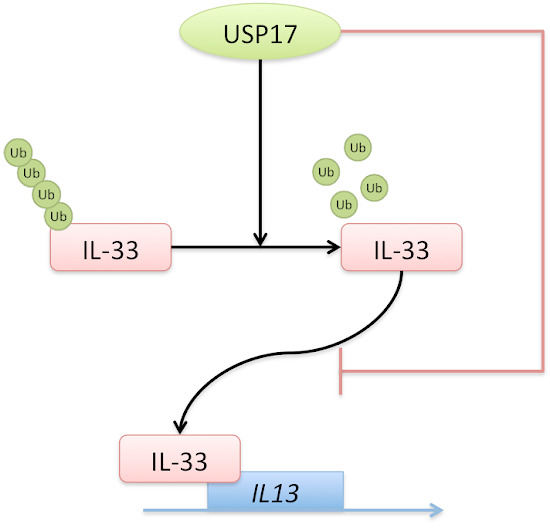
The Deubiquitinase USP17 Regulates the Stability and Nuclear Function of IL-33
Abstract
:1. Introduction
2. Results
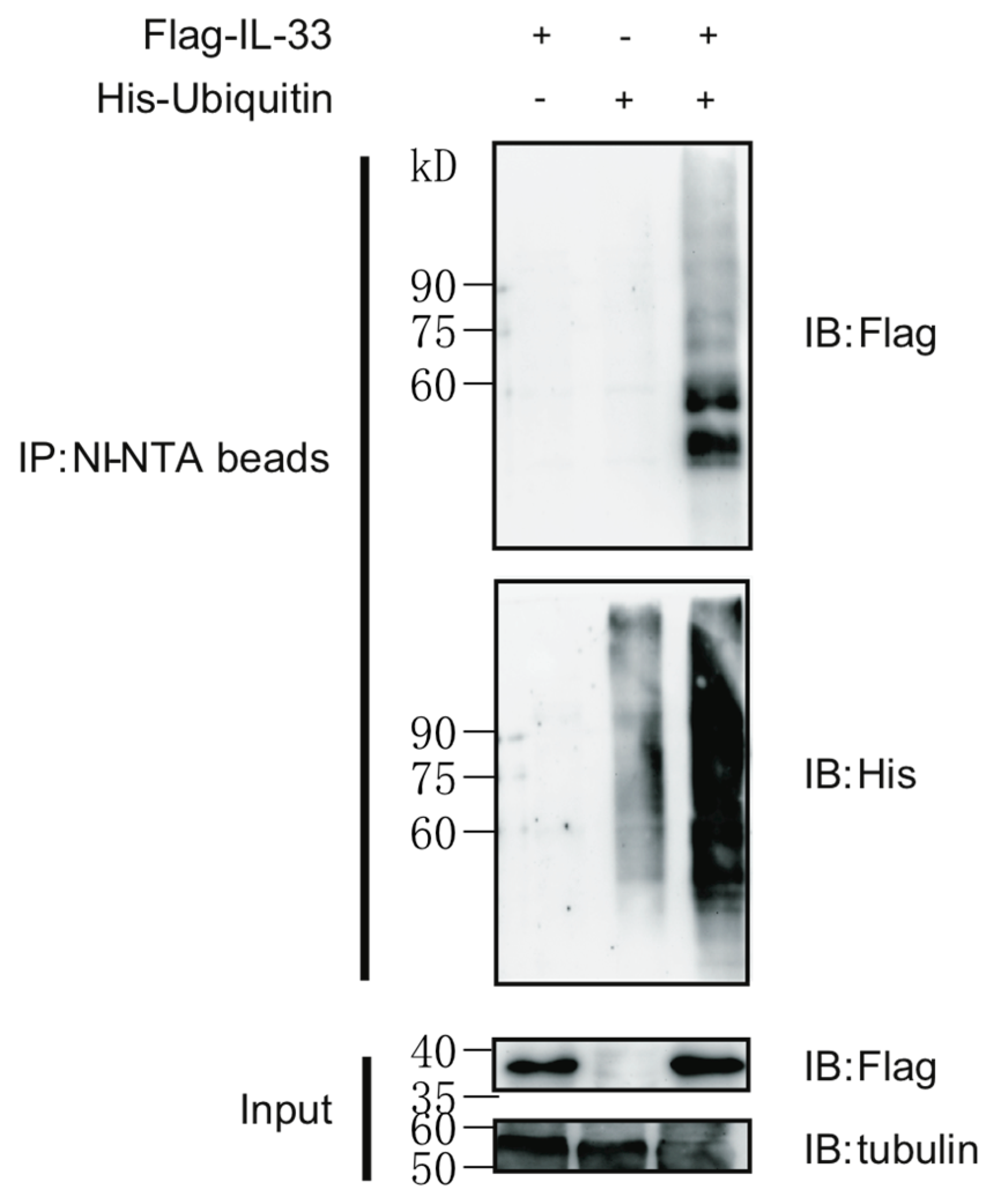
2.1. IL-33 Can Be Modified by Protein Polyubiquitination
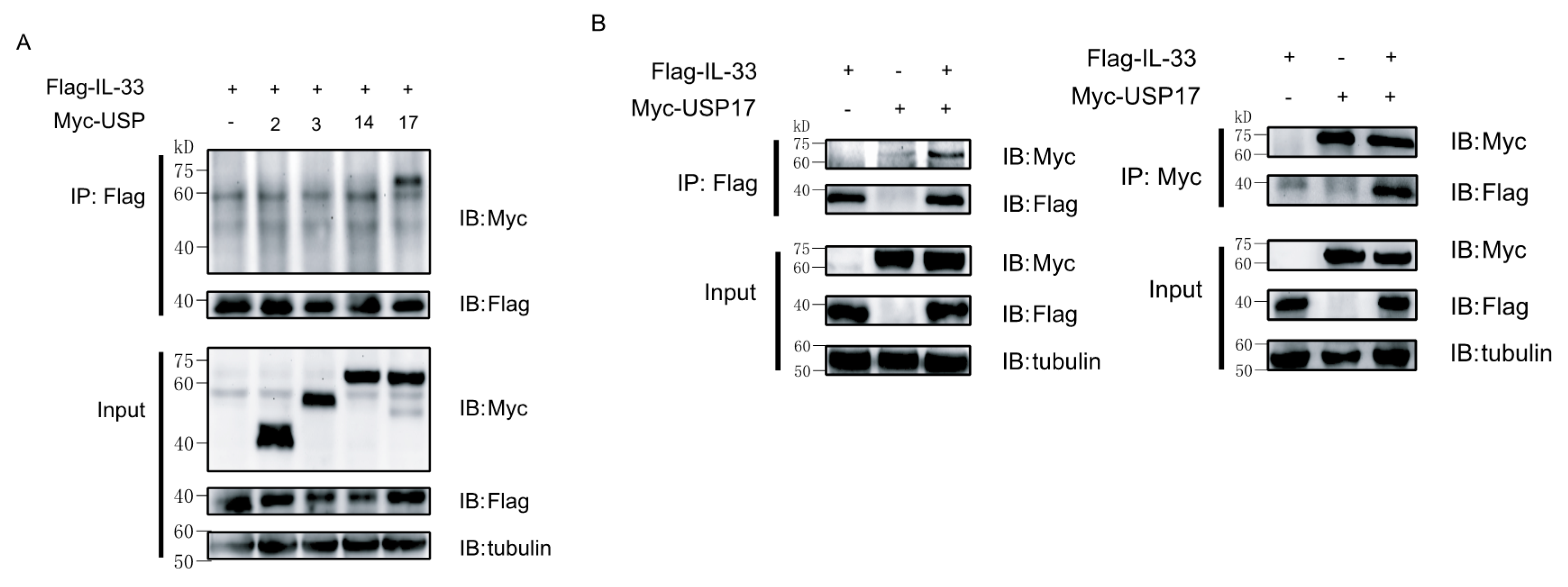
2.2. Ubiquitin-Specific Protease 17 (USP17) Associates with IL-33
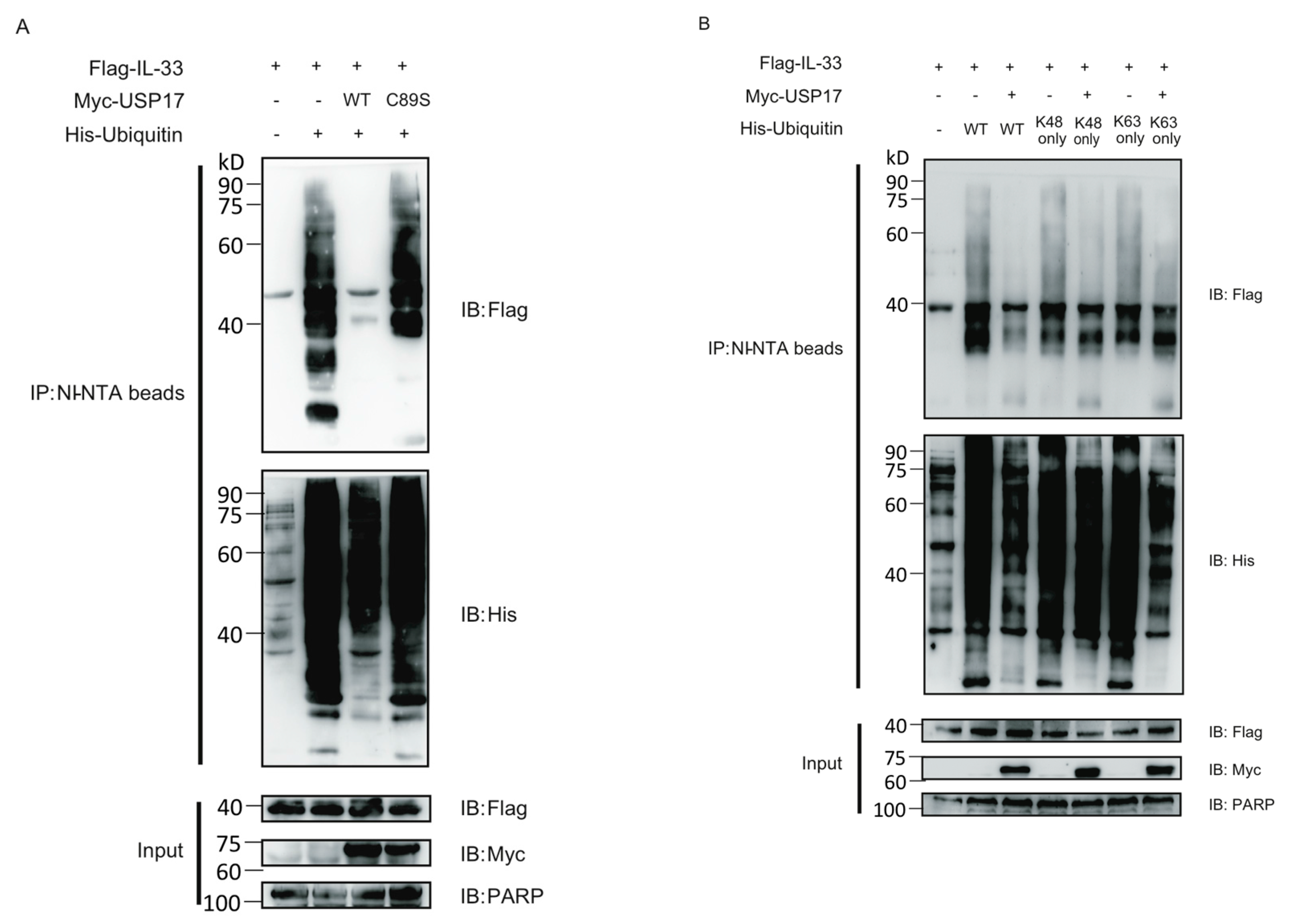
2.3. USP17 Deubiquitinates IL-33
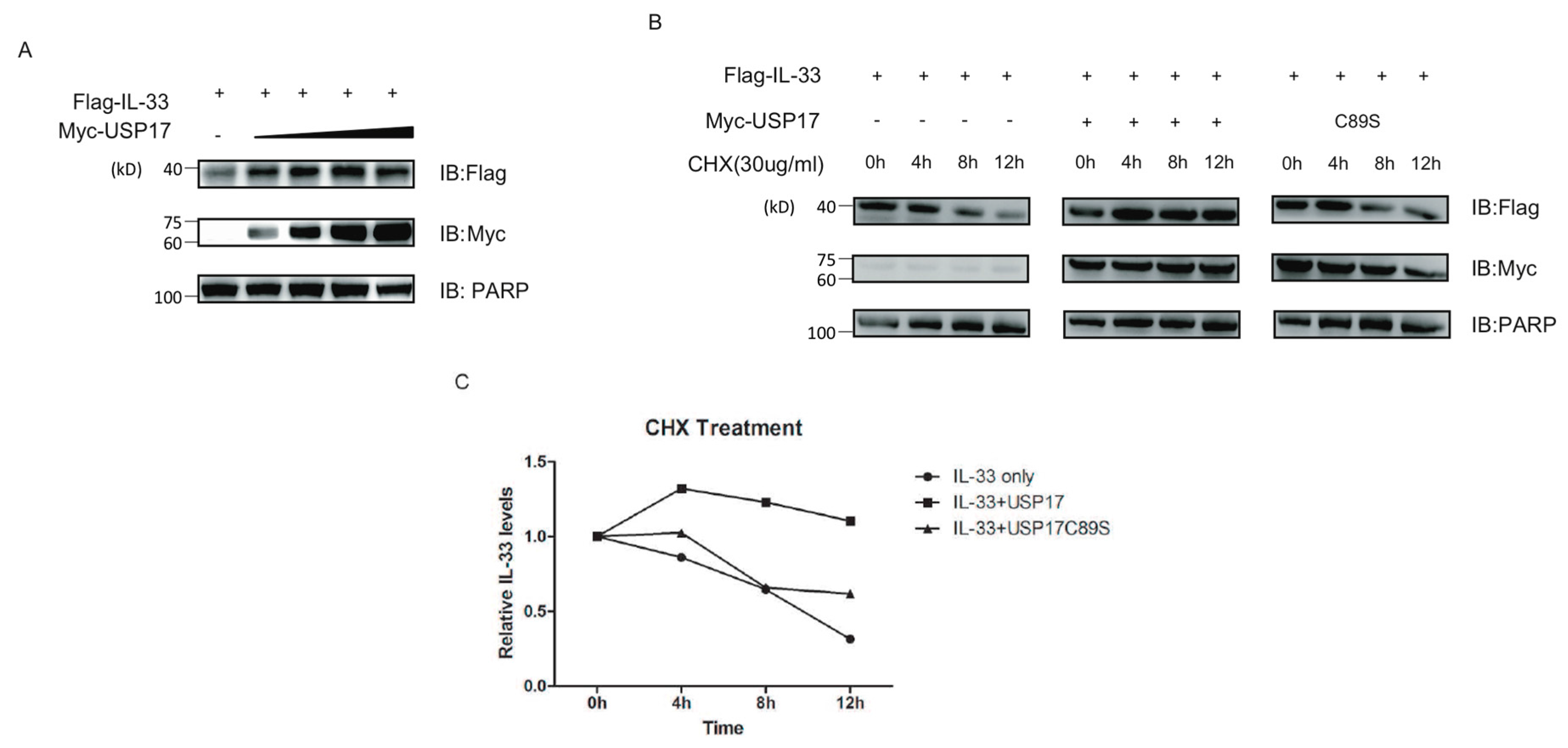
2.4. USP17 Stabilizes IL-33
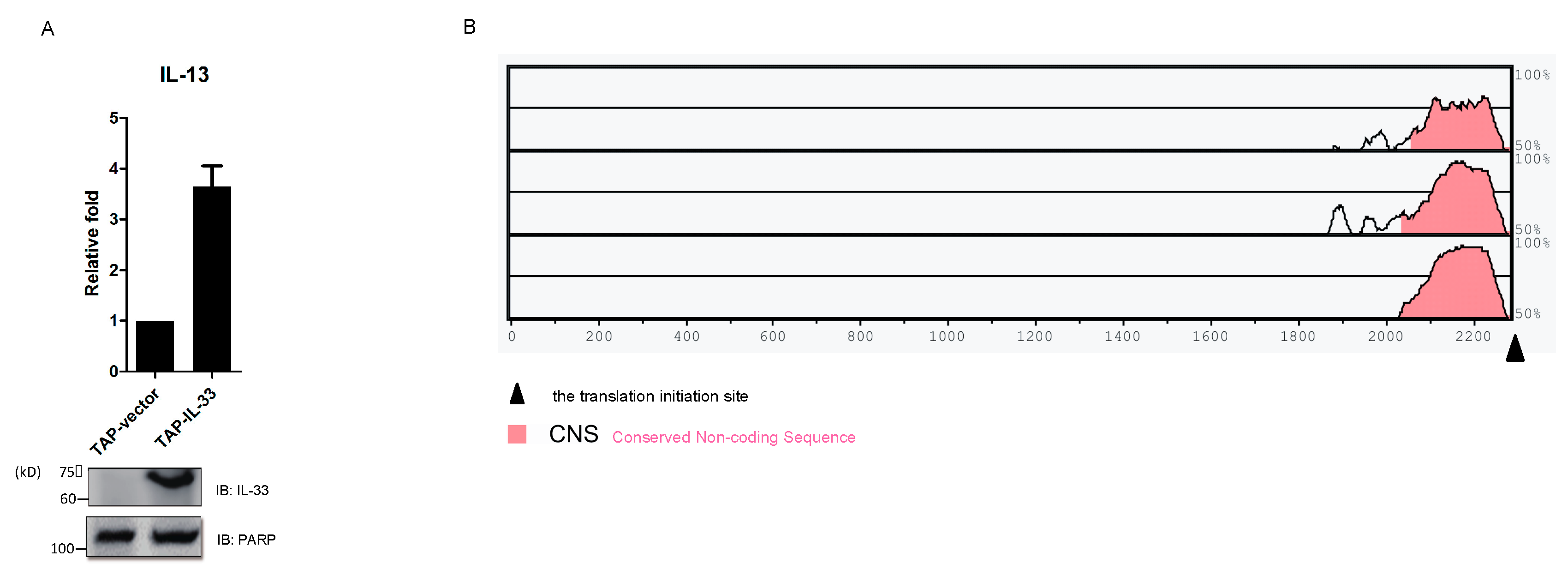
2.5. IL-33 Upregulates IL-13 by Chromatin Binding to the IL13 Gene Locus
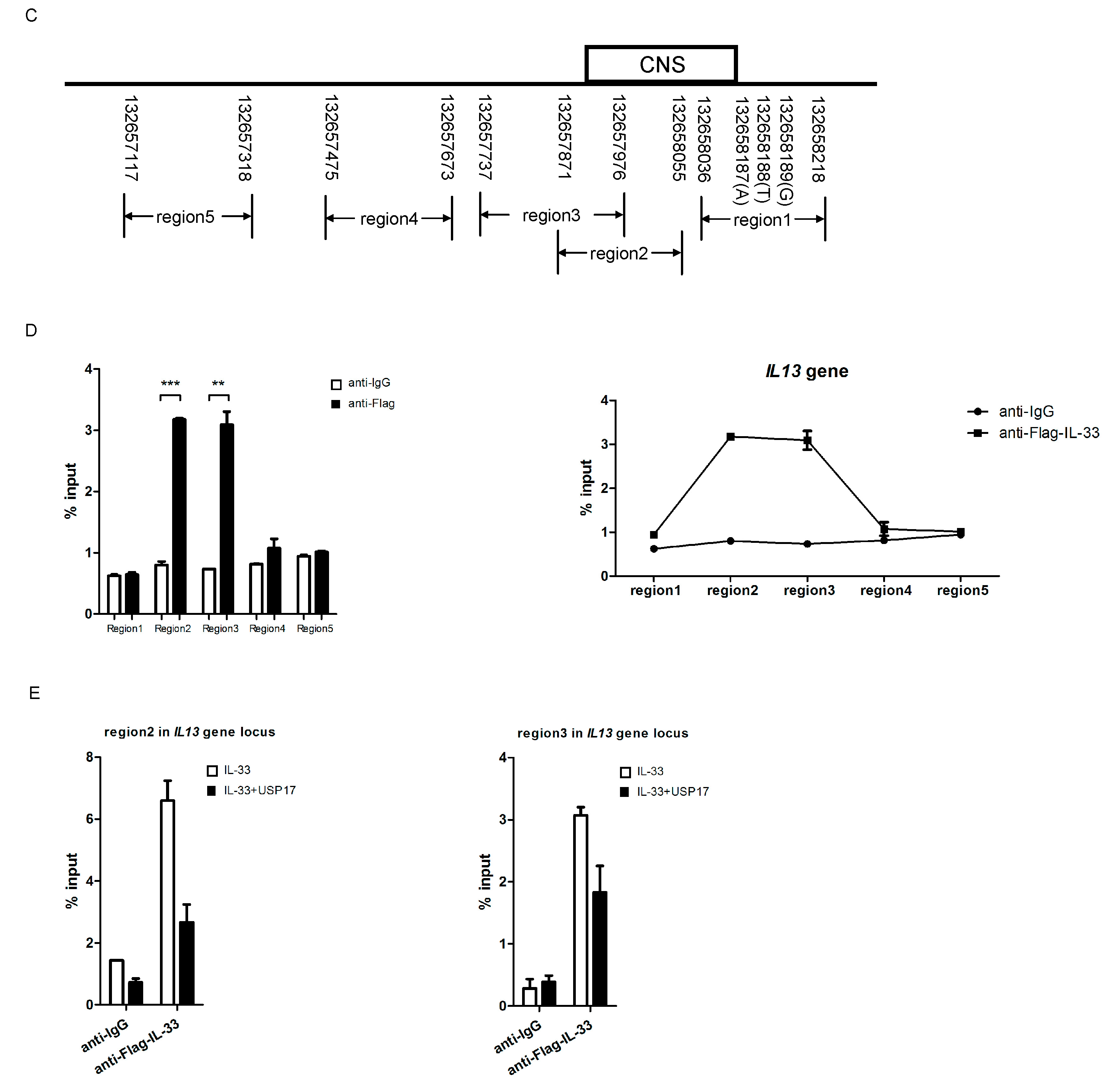
2.6. USP17 Downregulates the Chromatin Binding of IL-33 to the IL13 Gene Locus
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Transfection
4.2. Antibodies and Reagents
4.3. Plasmid Constructs and Stable Cell Line Construction
4.4. Immunoprecipitation and Immune Blot
4.5. Ubiquitin Pull-down Assay
4.6. Quantitative PCR
- IL-33 forward: 5′-GAATCAGGTGACGGTGTTGA-3′;
- IL-33 reverse: 5′-TCCAGGATCAGTCTTGCATTC3′;
- β-actin forward: 5′-GGACTTCGAGCAAGAGATGG-3′;
- β-actin reverse: 5′-AGCACTGTGTTGGCGTACAG-3′.
4.7. Rapid Micro-Chromatin Immunoprecipitation Assay on Quantitative PCR
- Region1-F: 5′-CGACACTGGATTTTCCACAA-3′;
- Region1-R: 5′-GCCAACAGGAGAGGATTGAG-3′;
- Region2-F: 5′-GGGTTGTCACAGGCAAAACT-3′;
- Region2-R: 5′-TTGTGGAAAATCCAGTGTCG-3′;
- Region3-F: 5′-ACTCCCCAAATTCCCATAGC-3′;
- Region3-R: 5′-CCAAGATCTGGGTGGGTTTA-3′;
- Region4-F: 5′-GTGAGACCCCATCTCCAAAA-3′;
- Region4-R: 5′-GGCTTGCCTAATTCCTCCTT-3′;
- Region5-F: 5′-GAGGGAAGAGCAGGAAAAGG-3′;
- Region5-R: 5′-GCAGCCTTAGTCCAGGTCAG-3′;
- Region6-F: 5′-GCTTCGAGTGTGGACAGAGA-3′;
- Region6-R: 5′-TTCTCCCCTACCCACTTCCT-3′;
- Region7-F: 5′-GATAAGGGGCGTTGACTCAC-3′;
- Region7-R: 5′-CGTGTGACCCCTCTACGG-3′.
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
References
- Baekkevold, E.S.; Roussigné, M.; Yamanaka, T.; Johansen, F.E.; Jahnsen, F.L.; Amalric, F.; Brandtzaeg, P.; Erard, M.; Haraldsen, G.; Girard, J.P. Molecular characterization of NF-HEV, a nuclear factor preferentially expressed in human high endothelial venules. Am. J. Pathol. 2003, 163, 69–79. [Google Scholar] [CrossRef]
- Schmitz, J.; Owyang, A.; Oldham, E.; Song, Y.; Murphy, E.; McClanahan, T.K.; Zurawski, G.; Moshrefi, M.; Qin, J.; Li, X.; et al. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity 2005, 23, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Nakae, S.; Morita, H.; Ohno, T.; Arae, K.; Matsumoto, K.; Saito, H. Role of interleukin-33 in innate-type immune cells in allergy. Allergol. Int. 2013, 62, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Lefrançais, E.; Cayrol, C. Mechanisms of IL-33 processing and secretion: Differences and similarities between IL-1 family members. Eur. Cytokine Netw. 2012, 23, 120–127. [Google Scholar] [PubMed]
- Liew, F.Y.; Pitman, N.I.; McInnes, I.B. Disease-associated functions of IL-33: The new kid in the IL-1 family. Nat. Rev. Immunol. 2010, 10, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Carriere, V.; Roussel, L.; Ortega, N.; Lacorre, D.A.; Americh, L.; Aguilar, L.; Bouche, G.; Girard, J.P. IL-33, the IL-1-like cytokine ligand for ST2 receptor, is a chromatin-associated nuclear factor in vivo. Proc. Natl. Acad. Sci. USA 2007, 104, 282–287. [Google Scholar] [CrossRef] [PubMed]
- Roussel, L.; Erard, M.; Cayrol, C.; Girard, J.P. Molecular mimicry between IL-33 and KSHV for attachment to chromatin through the H2A-H2B acidic pocket. EMBO Rep. 2008, 9, 1006–1012. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Mohs, A.; Thomas, M.; Klare, J.; Ross, R.; Schmitz, M.L.; Martin, M.U. The dual function cytokine IL-33 interacts with the transcription factor NF-κB to dampen NF-κB-stimulated gene transcription. J. Immunol. 2011, 87, 1609–1616. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.S.; Park, J.A.; Kim, J.; Rho, S.S.; Park, H.; Kim, Y.M.; Kwon, Y.G. Nuclear IL-33 is a transcriptional regulator of NF-κB p65 and induces endothelial cell activation. Biochem. Biophys. Res. Commun. 2012, 421, 305–311. [Google Scholar] [CrossRef]
- Tao, L.; Chen, C.; Song, H.; Piccioni, M.; Shi, G.; Li, B. Deubiquitination and stabilization of IL-33 by USP21. Int. J. Clin. Exp. Pathol. 2014, 7, 4930–4937. [Google Scholar] [PubMed]
- Hammond-Martel, I.; Yu, H.; Affarel, B. Roles of ubiquitin signaling in transcription regulation. Cell Signal. 2012, 24, 410–421. [Google Scholar] [CrossRef]
- Ikeda, F.; Dikic, I. Atypical ubiquitin chains: New molecular signals. Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep. 2008, 9, 536–542. [Google Scholar] [CrossRef] [PubMed]
- Muratani, M.; Tansey, W.P. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 2003, 4, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Thrower, J.S.; Hoffman, L.; Rechsteiner, M.; Pickart, C.M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000, 19, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Hochrainer, K.; Lipp, J. Ubiquitylation within signaling pathways in- and outside of inflammation. Thromb. Haemost. 2007, 97, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Pickart, C.M. Back to the future with ubiquitin. Cell 2004, 116, 181–190. [Google Scholar] [CrossRef]
- Komander, D.; Clague, M.J.; Urbé, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Sippl, W.; Collura, V.; Colland, F. Ubiquitin-specific proteases as cancer drug targets. Future Oncol. 2011, 7, 619–632. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.H.; Ramakrishna, S.; Baek, K.H. Molecular mechanisms and functions of cytokine-inducible deubiquitinating enzymes. Cytokine Growth Factor Rev. 2013, 24, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Burrows, J.F.; McGrattan, M.J.; Johnston, J.A. The DUB/USP17 deubiquitinating enzymes, a multigene family within a tandemly repeated sequence. Genomics 2005, 85, 524–529. [Google Scholar] [CrossRef] [PubMed]
- Burrows, J.F.; Kelvin, A.A.; McFarlane, C.; Burden, R.E.; McGrattan, M.J.; de la Vega, M.; Govender, U.; Quinn, D.J.; Dib, K.; Gadina, M.; et al. USP17 regulates Ras activation and cell proliferation by blocking RCE1 activity. J. Biol. Chem. 2009, 284, 9587–9595. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, S.; Suresh, B.; Lee, E.J.; Lee, H.J.; Ahn, W.S.; Baek, K.H. Lys-63-specific deubiquitination of SDS3 by USP17 regulates HDAC activity. J. Biol. Chem. 2011, 286, 10505–10514. [Google Scholar] [CrossRef] [PubMed]
- Pereg, Y.; Liu, B.Y.; O’Rourke, K.M.; Sagolla, M.; Dey, A.; Komuves, L.; French, D.M.; Dixit, V.M. Ubiquitin hydrolase Dub3 promotes oncogenic transformation by stabilizing Cdc25A. Nat. Cell Biol. 2010, 12, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Vista Tools. Available online: http://genome.lbl.gov/vista/index.shtml (accessed on 16 November 2015).
- Schnell, J.D.; Hicke, L. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J. Biol. Chem. 2003, 278, 35857–35860. [Google Scholar] [CrossRef] [PubMed]
- Flick, K.; Ouni, I.; Wohlschlegel, J.A.; Capati, C.; McDonald, W.H.; Yates, J.R.; Kaiser, P. Proteolysis-independent regulation of the transcription factor Met4 by a single Lys 48-linked ubiquitin chain. Nat. Cell Biol. 2004, 6, 634–641. [Google Scholar] [CrossRef]
- Yuan, J.; Luo, K.; Zhang, L.; Cheville, J.C.; Lou, Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell 2010, 140, 384–396. [Google Scholar] [CrossRef]
- Vviñas-Castells, R.; Frías, Á.; Robles-Lanuza, E.; Zhang, K.; Longmore, G.D.; de García Herreros, A.; Díaz, V.M. Nuclear ubiquitination by FBXL5 modulates Snail1 DNA binding and stability. Nucleic Acids Res. 2014, 42, 1079–1094. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.; Park, S.Y.; Cho, J.H.; Park, J.W.; Sohn, J.H.; Kim, Y.J.; Oh, J.W.; Han, J.S. The TLR4-associated phospholipase D1 activation is crucial for Der f 2-induced IL-13 production. Allergy 2015. [Google Scholar] [CrossRef] [PubMed]
- Ro, E.J.; Cha, P.H.; Kim, H.Y.; Cho, Y.H.; Park, J.W.; Han, J.S.; Choi, K.Y. House dust mite allergen Der f 2 induces interleukin-13 expression by activating the PI3K/Akt pathway. Immunol. Res. 2013, 56, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, C.M. IL-33 family members and asthma—Bridging innate and adaptive immune responses. Curr. Opin. Immunol. 2010, 22, 800–806. [Google Scholar] [CrossRef] [PubMed]
- Dahl, J.A.; Collas, P. A rapid micro chromatin immunoprecipitation assay (microChIP). Nat. Protoc. 2008, 3, 1032–1045. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ni, Y.; Tao, L.; Chen, C.; Song, H.; Li, Z.; Gao, Y.; Nie, J.; Piccioni, M.; Shi, G.; Li, B. The Deubiquitinase USP17 Regulates the Stability and Nuclear Function of IL-33. Int. J. Mol. Sci. 2015, 16, 27956-27966. https://doi.org/10.3390/ijms161126063
Ni Y, Tao L, Chen C, Song H, Li Z, Gao Y, Nie J, Piccioni M, Shi G, Li B. The Deubiquitinase USP17 Regulates the Stability and Nuclear Function of IL-33. International Journal of Molecular Sciences. 2015; 16(11):27956-27966. https://doi.org/10.3390/ijms161126063
Chicago/Turabian StyleNi, Yingmeng, Lianqin Tao, Chen Chen, Huihui Song, Zhiyuan Li, Yayi Gao, Jia Nie, Miranda Piccioni, Guochao Shi, and Bin Li. 2015. "The Deubiquitinase USP17 Regulates the Stability and Nuclear Function of IL-33" International Journal of Molecular Sciences 16, no. 11: 27956-27966. https://doi.org/10.3390/ijms161126063
APA StyleNi, Y., Tao, L., Chen, C., Song, H., Li, Z., Gao, Y., Nie, J., Piccioni, M., Shi, G., & Li, B. (2015). The Deubiquitinase USP17 Regulates the Stability and Nuclear Function of IL-33. International Journal of Molecular Sciences, 16(11), 27956-27966. https://doi.org/10.3390/ijms161126063