ClRTL1 Encodes a Chinese Fir RNase III–Like Protein Involved in Regulating Shoot Branching
Abstract
:1. Introduction


2. Results
2.1. Isolation and Identification of Shoot Branching Genes Using the cDNA-AFLP Technique
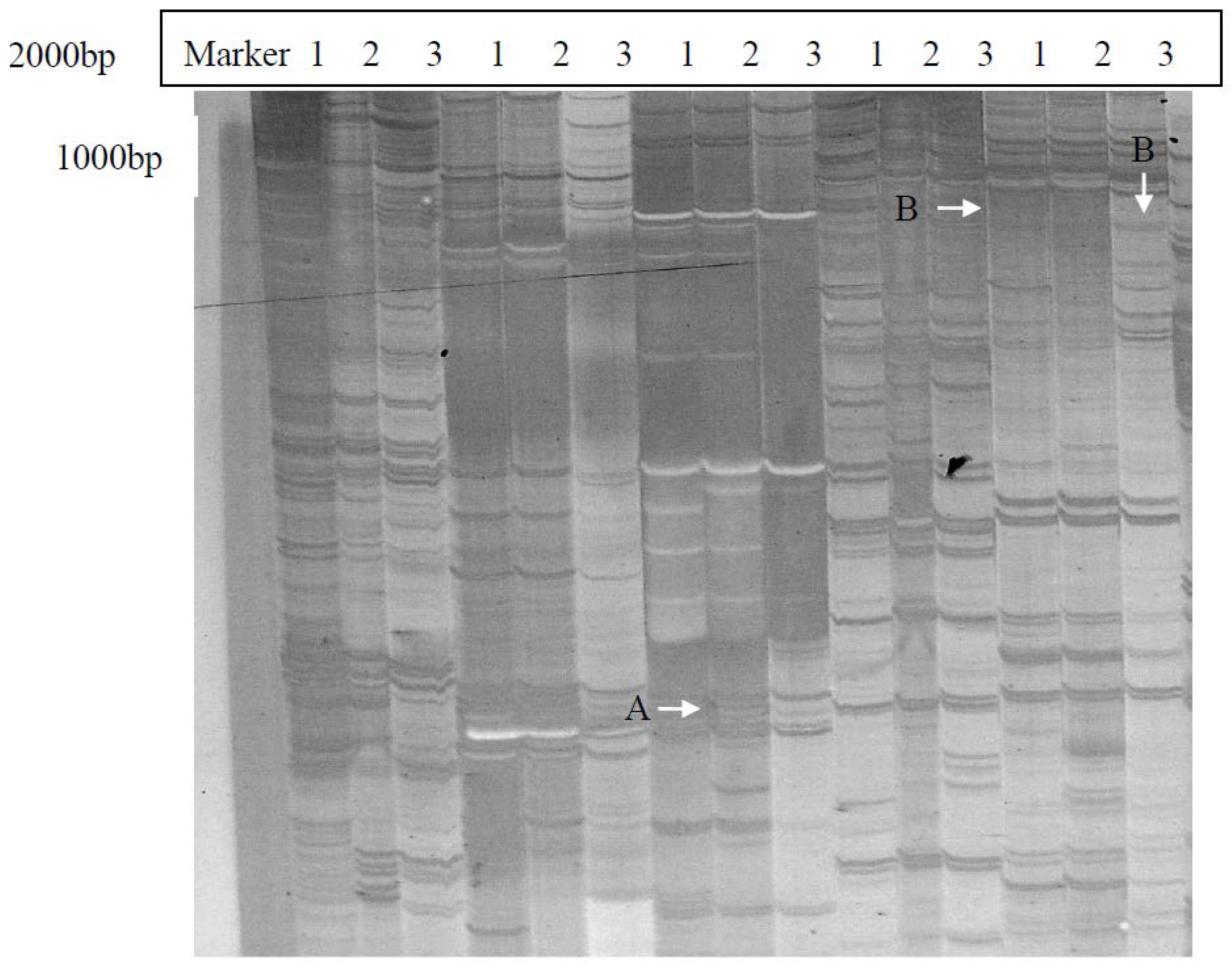

2.2. Cloning and Sequence Analysis of the Full-Length ClRTL1 cDNA Sequence
2.3. The Deduced Secondary Structure of ClRTL1

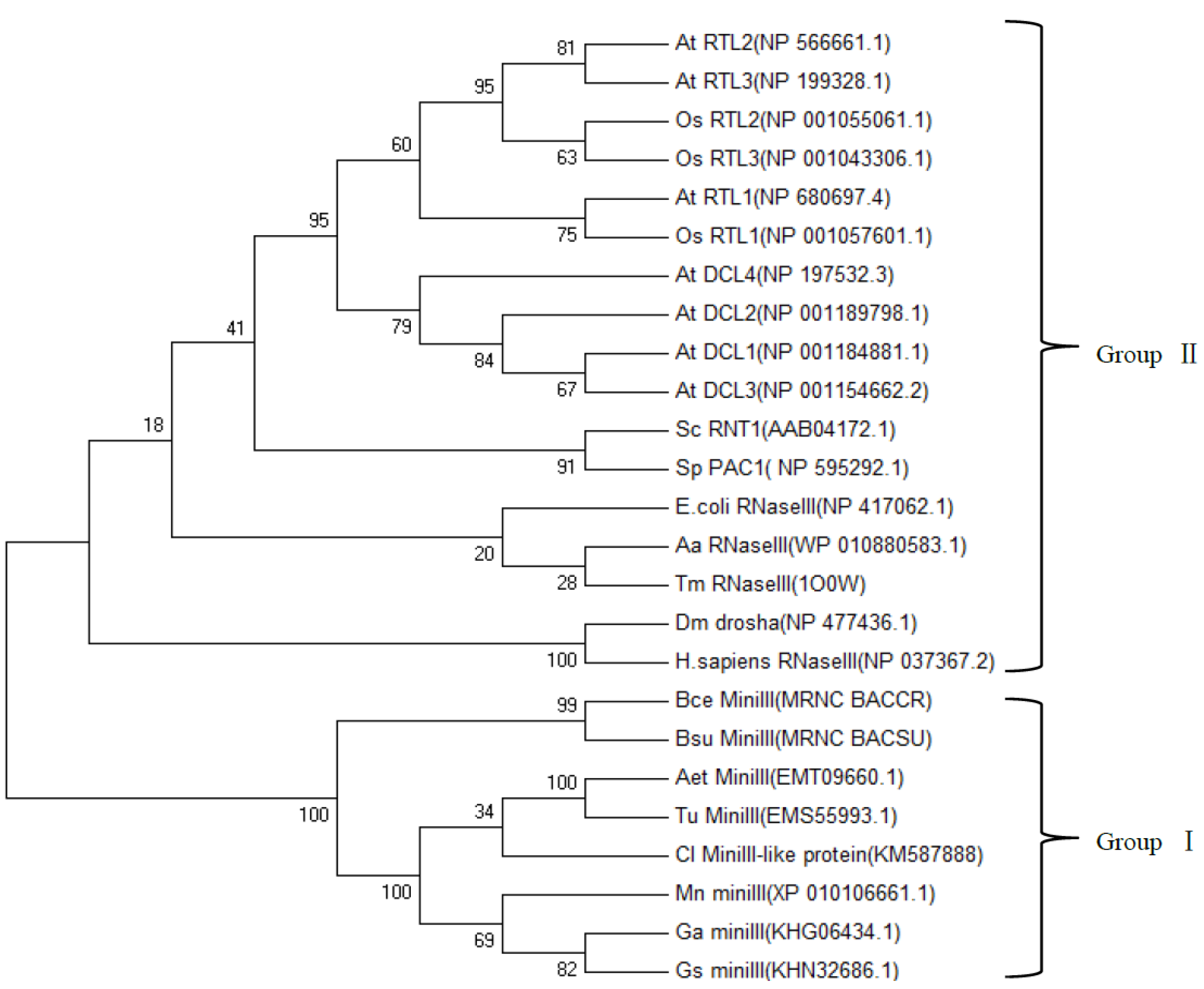
2.4. Phylogenetic Analysis of ClRTL1 and Its Homologous Proteins
2.5. Relative Expression Analysis of ClRTL1
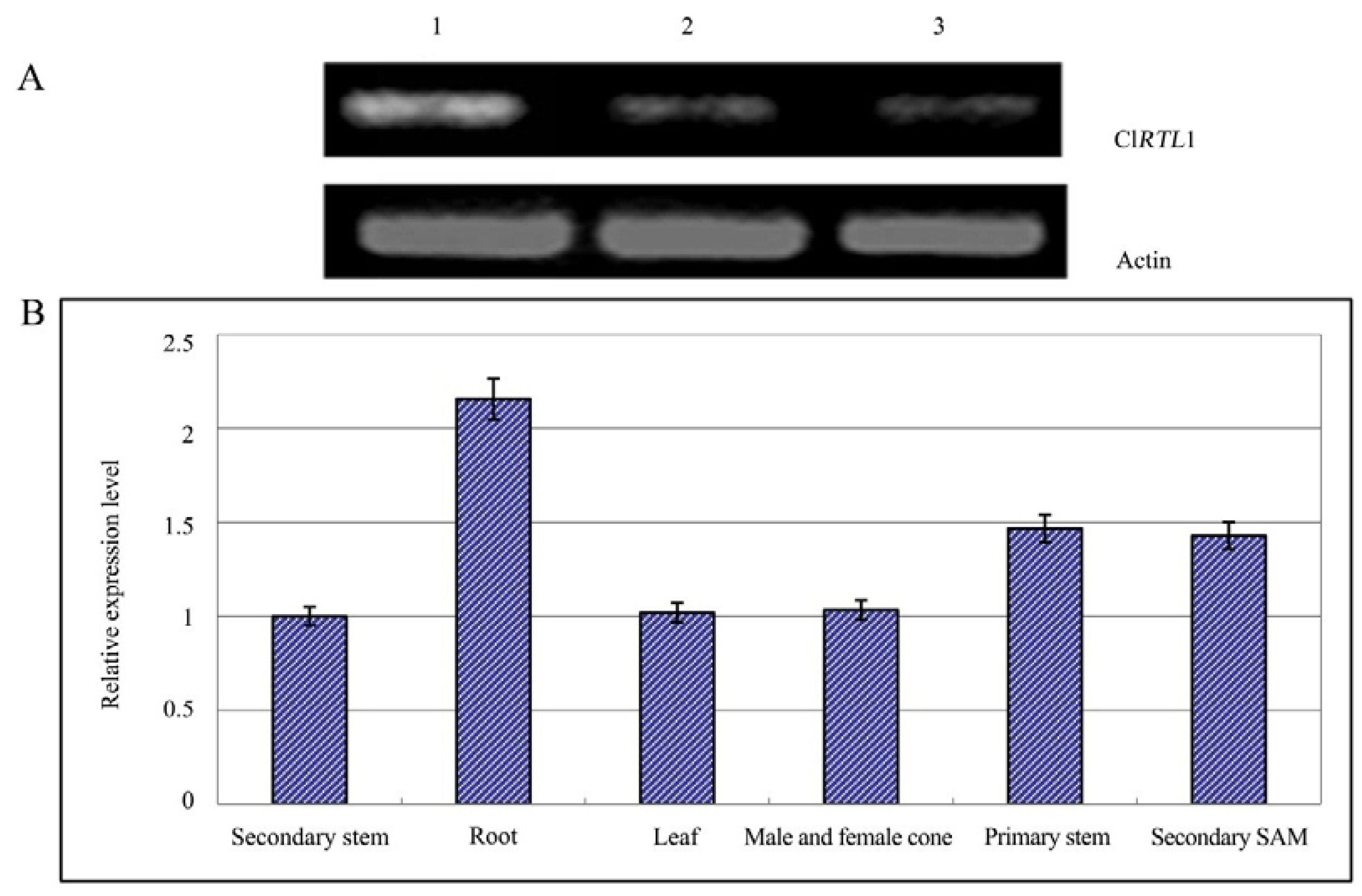
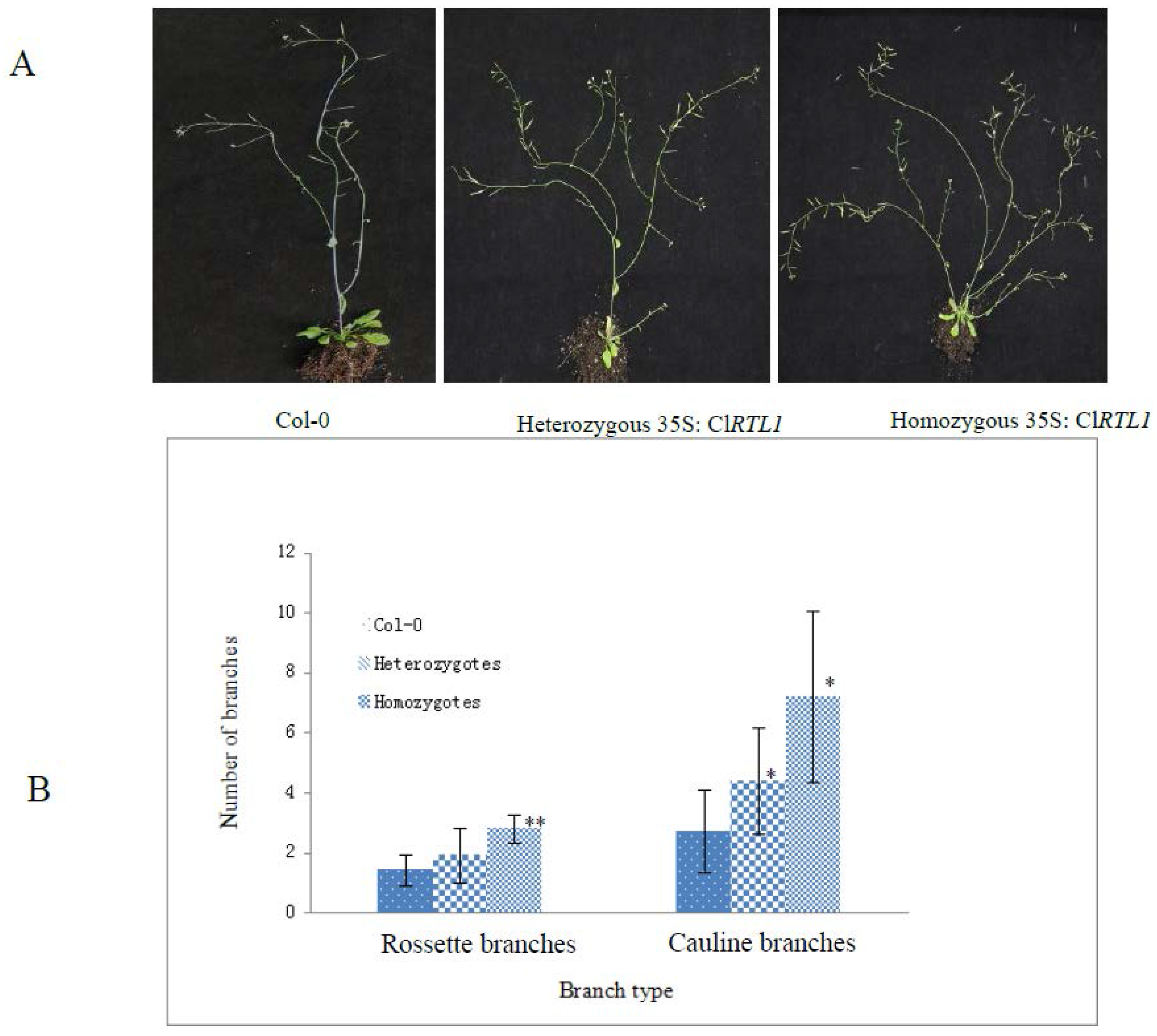
2.6. Phenotypes of CaMV 35S: ClRTL1 A. thaliana
3. Discussion
3.1. Effective Screening Strategy for ClRTL1 Identification
3.2. ClRTL1 Regulates the Development of Chinese Fir by Promoting Shoot Branching
3.3. ClRTL1 Encodes a mini-RNase III Protein with Conserved RNase III Enzyme Activity But with a Different Mechanism of RNA Recognition from the Typical RNase III
4. Experimental Section
4.1. Plant Materials
4.2. Screening for Branching-Related Genes with the cDNA-AFLP System
| Primer/Adapter | Nucleotide Sequences |
|---|---|
| EcoR I Adapters | 5′-CTCGTAGACTGCGTACC-3′ |
| 3′-CTGACGCATGGTTAA-5′ | |
| Mse I Adapters | 5′-GACGATGAGTCCTGAG-3′ |
| 3′-TACTCAGGACTCAT-5′ | |
| Pre-amplification primer E 00 | 5′-GACTGCGTACCAATTC-3′ |
| Pre-amplification primer M 00 | 5′-GATGAGTCCTGAGTAA-3′ |
| Selective-amplification primers E + 2 | 5′-GACTGCGTACCAATTCNN-3′ |
| Selective-amplification primers M + 2 | 5′-GATGA GTCCTGAGTAANN-3′ |
| GSP1 | 5′-TGATGCCCTGTTATACACTGCTGCTCC-3′ |
| GSP2 | 5′-TGGAGCAGCAGTGTATAACAGGGCATCATCT-3′ |
| RNase III F | 5′-GCGAACGAGGATGGCTGCTAC-3′ |
| RNase III R | 5′-ATCCCATCCTAACACCACTTG-3′ |
| RNase III forward primer R1 | 5′-GAACAGTAAGGCGTGCTGGAG-3′ |
| RNase III reverse primer R2 | 5′-CAAAACCCGAGGCTTCTCATT-3′ |
| Actin forward primer A1 | 5′-CAGCAACTGGGATGATATGG-3′ |
| Actin reverse primer A2 | 5′-ATTTCGCTTTCAGCAGTGGT-3′ |
| RNase III forward primer F4 | 5′-CTCCCATGACCAACCTGTTTCTG-3′ |
| RNase III forward primer R4 | 5′-ATTAAGGCATCCAAGCGATTTCC-3′ |
4.3. Cloning and Sequence Analysis of the Full-Length ClRTL1 cDNA Sequence
4.4. Secondary Structural Analysis of the Deduced Protein ClRTL1
4.5. Relative Expression Analysis of ClRTL1
4.6. Construction of Expression Vectors and Plant Transformation into A. thaliana
4.7. Isolation of 35S: Cl RNase III Transgenic Arabidopsis Lines
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- McSteen, P.; Leyser, O. Shoot branching. Annu. Rev. Plant Biol. 2005, 56, 353–374. [Google Scholar] [CrossRef] [PubMed]
- Mason, M.G.; Ross, J.J.; Babst, B.A.; Wienclaw, B.N.; Beveridge, C.A. Sugar demand, not auxin, is the initial regulator of apical dominance. Proc. Natl. Acad. Sci. USA 2014, 11, 6092–6097. [Google Scholar] [CrossRef] [PubMed]
- Yaish, M.W.F.; Guevara, D.R.; El-Kereamy, A.; Rothstein, S.J. Axillary Shoot Branching in Plants. In Plant Developmental Biology-Biotechnological Perspectives; Pua, E.C., Davey, M.R., Eds.; Springer: Berlin, Germany, 2010; pp. 37–52. [Google Scholar]
- Schumacher, K.; Schmitt, T.; Rossberg, M.; Schmitz, G.; Theres, K. The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. USA 1999, 96, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Greb, T.; Clarenz, O.; Schafer, E.; Muller, D.; Herrero, R.; Schmitz, G.; Theres, K. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 2003, 17, 1175–1187. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Qian, Q.; Fu, Z.; Wang, Y.; Xiong, G.; Zeng, D.; Wang, X.; Liu, X.; Teng, S.; Hiroshi, F.; et al. Control of tillering in rice. Nature 2003, 422, 618–621. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, G.; Tillmann, E.; Carriero, F.; Fiore, C.; Cellini, F.; Theres, K. The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proc. Natl. Acad. Sci. USA 2002, 99, 1064–1069. [Google Scholar] [CrossRef] [PubMed]
- Keller, T.; Abbott, J.; Moritz, T.; Doner, P. Arabidopsis REGULATOR OF AXILLARYMERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 2006, 18, 598–611. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, K.; Maekawa, M.; Ujiie, S.; Satake, Y.; Furutani, I.; Okamoto, H.; Shimamoto, K.; Kyozuka, J. LAX and SPA: Major regulators of shoot branching in rice. Proc. Natl. Acad. Sci. USA 2003, 100, 11765–11770. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, L.; McSteen, P.; Doebley, J.; Hake, S. Expression patterns and mutant phenotype of teosinte brancbedl correlate with growth suppression in maize and teosinte. Genetics 2002, 162, 1927–1935. [Google Scholar] [PubMed]
- Takeda, T.; Suwa, Y.; Suzukj, M.; Kitano, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Matsuoka, M. The OsTB1 gene negatively regulates lateral branching in rice. Plant 2003, 33, 513–520. [Google Scholar] [CrossRef]
- Braun, N.; de Saint Germain, A.; Pillot, J.-P.; Boutet-Mercey, S.; Dalmais, M.; Antoniadi, I.; Li, X.; Maia-Grondard, A.; le Signor, C.; Bouteiller, N.; et al. The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol. 2012, 158, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Reintanz, B.; Lehnen, M.; Reichelt, M.; Gershenzon, J.; Kowalczyk, M.; Sandberg, G.; Godde, M.; Uhl, R.; Palme, K. bus, a Bushy Arabidopsis knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. Plant Cell 2001, 13, 351–367. [Google Scholar] [CrossRef] [PubMed]
- Tantikanjana, T.; Young, J.W.H.; Letham, D.S.; Griffith, M.; Hussain, M.; Ljung, K.; Sandberg, G.; Sundaresan, V. Control of axillary bud initiation and shoot architecture in Arabidopsis through the SUPERSHOOT gene. Genes Dev. 2001, 15, 1577–1588. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, H.; Jiang, C.J.; Sakakibara, H.; Kojima, M.; Honda, I.; Ajisaka, H.; Nishijima, T.; Koshioka, M.; Homma, T.; Mander, L.N.; et al. Overexpression of a petunia zinc-finger gene alters cytokinin metabolism and plant forms. Plant J. 2005, 41, 512–523. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Yang, Y.J.; Zhou, J. Genes involved in the synthesis and signaling pathway of strigolactone, a shoot branching inhibitor. Biol. Plant. 2012, 56, 210–214. [Google Scholar] [CrossRef]
- Chen, Y.W.; Shi, J.S. Some fundamental strategies on Chinese fir genetic improvement (I). J. Nanjing For. Univ. 1983, 4, 4–18. [Google Scholar]
- Filippov, V.; Solovyev, V.; Filippva, M.; Gill, S.S. A novel type of RNase III family proteins in eukaryotes. Gene 2000, 245, 213–221. [Google Scholar] [CrossRef]
- Robertson, H.D.; Webster, R.E.; Zinder, N.D. Purification and properties of ribonuclease III from Escherichia coli. J. Biol. Chem. 1968, 243, 82–91. [Google Scholar] [PubMed]
- Elela, S.A.; Igel, H.; Ares, M., Jr. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell 1996, 85, 115–124. [Google Scholar] [CrossRef]
- Jacobsen, S.E.; Running, M.P.; Meyerowitz, E.M. Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 1999, 126, 5231–5243. [Google Scholar] [PubMed]
- Wu, H.; Xu, H.; Miraglia, L.J.; Crooke, S.T. Human RNase III is a 160 kDa protein involved in preribosomal RNA processing. J. Biol. Chem. 2000, 275, 36957–36965. [Google Scholar] [CrossRef] [PubMed]
- Gan, J.; Tropea, J.E.; Austin, B.P.; Court, D.L.; Waugh, D.S.; Ji, X. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell 2006, 124, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Redko, Y.; Bechhofer, D.H.; Condon, C. Mini-III, an unusual member of the RNase III family of enzymes, catalyses 23S ribosomal RNA maturation in B. subtilis. Mol. Microbiol. 2008, 68, 1096–1106. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shi, J.S. Development and Optimization of cDNA-AFLP Reaction System for Young Stem of Cunninghamia lanceolata (Lamb.) Hook. In Proceedings of the International Conference on Plant Vascular Biology and Agriculture, Chongqiong, China, 21–21 June 2009.
- Ray, A.; Lang, J.D.; Golden, T.; Ray, S. SHORT INTEGUMENT (SIN1), a gene required for ovule development in Arabidopsis, also controls flowering time. Development 1996, 122, 2631–2638. [Google Scholar] [PubMed]
- Blaszczyk, J.; Tropea, J.E.; Bubunenko, M.; Routzahn, K.M.; Waugh, D.S.; Court, D.L.; Ji, X. Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure 2001, 9, 1225–1236. [Google Scholar] [CrossRef]
- Li, H.L.; Chelladurai, B.S.; Zhang, K.; Nicholson, A.W. Ribonuclease III cleavage of a bacteriophage T7 processing signal. Divalent cation specificity, and specific anion effects. Nucleic Acids Res. 1993, 21, 1919–1925. [Google Scholar] [CrossRef] [PubMed]
- Rotondo, G.; Frendewey, D. Purification and characterization of the Pac1 ribonuclease of Schizosaccharomyces pombe. Nucleic Acids Res. 1996, 24, 2377–2386. [Google Scholar] [CrossRef] [PubMed]
- Provost, P.; Dishart, D.; Doucet, J.; Frendewey, D.; Samuelsson, B.; Rådmark, O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002, 21, 5864–5874. [Google Scholar] [CrossRef] [PubMed]
- Kreuze, J.F.; Savenkov, E.I.; Cuellar, W.; Li, X.; Valkonen, J.P. Viral class 1 RNase III involved in suppression of RNA silencing. J. Virol. 2005, 79, 7227–7238. [Google Scholar] [CrossRef] [PubMed]
- Dellagi, A.; Birch, P.R.J.; Heilbronn, J.; Lyon, G.D. cDNA-AFLP analysis of differential gene-expression in the prokaryotic plant pathogen Erwinia-Carotovora. Microbiology 2000, 146, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Durrant, W.E.; Rowland, O.; Piedras, P.; Hammondkosack, K.E.; Jones, J.D.G. cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene-expression profiles. Plant Cell 2000, 12, 963–977. [Google Scholar] [CrossRef] [PubMed]
- Van der Biezen, E.A.; Juwana, H.; Parker, J.E.; Jones, J.D.G. cDNA-AFLP display for the isolation of Peronospora parasitica genes expressed during infection in Arabidopsis thaliana. Mol. Plant Microbe Interact. 2000, 13, 895–898. [Google Scholar] [CrossRef] [PubMed]
- Golden, T.A.; Schauer, S.E.; Lang, J.D.; Pien, S.; Mushegian, A.R. Short integuments1/suspensor1/carpel factory, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol. 2002, 130, 808–822. [Google Scholar] [CrossRef] [PubMed]
- Schauer, S.E.; Jacobsen, S.E.; Meinke, D.W.; Ray, A. DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 2002, 7, 487–491. [Google Scholar] [CrossRef]
- Xie, Z.; Allen, E.; Wilken, A.; Carrington, J.C. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2005, 102, 12984–12989. [Google Scholar] [CrossRef] [PubMed]
- Court, D. RNA processing and degradation by RNase III. In Control of Messenger RNA Stability; Belasco, J., Brawerman, G., Eds.; Academic: New York, NY, USA, 1993. [Google Scholar]
- Nicholson, A.W. The ribonuclease III family: forms and functions in RNA maturation, decay, and gene silencing. In RNAi: A Guide to Gene Silencing; Hannon, G.J., Ed.; Cold Spring Harbor: New York, NY, USA, 2003; pp. 149–174. [Google Scholar]
- Lee, Y.; Ahn, C.; Han, J.J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initialtes micro RNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Lichner, Z.; Silhavy, D.; Burgya’n, J. Double-stranded RNA binding proteins could suppress RNA interference-mediated antiviral defences. J. Gen. Virol. 2003, 84, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Carthew, R.W. Gene silencing by double-stranded RNA. Curr. Opin. Cell Biol. 2001, 13, 244–248. [Google Scholar] [CrossRef]
- Carmell, M.A.; Hannon, G.J. RNase III enzymes and the initiation of gene silencing. Nat. Struct. Mol. Biol. 2004, 11, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Kiyota, E.; Okada, R.; Kondo, N.; Hiraguri, A.; Moriyama, H.; Fukuhara, T. An Arabidopsis RNase III-like protein, AtRTL2, cleaves double-stranded RNA in vitro. J. Plant Res. 2011, 124, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single processing center models for human Dicer and bacterial RNase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Dunoyer, P.; Himber, C.; Voinnet, O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 2005, 37, 1356–1360. [Google Scholar] [CrossRef] [PubMed]
- Mlotshwa, S.; Pruss, G.J.; Peragine, A.; Endres, M.W.; Li, J.; Chen, X.; Scott Poethig, R.; Bowman, L.H.; Vance, V. DICER-LIKE2 plays a primary role in transitive silencing of transgenes in Arabidopsis. PLoS ONE 2008, 3, e1755. [Google Scholar] [CrossRef] [PubMed]
- Dunn, J.J. Ribonuclease III. In The Enzymes; Boyer, P.D., Ed.; Academic Press: New York, NY, USA, 1982; pp. 485–499. [Google Scholar]
- Robertson, H.D. Escherichia coli ribonuclease III cleavage sites. Cell 1982, 30, 669–672. [Google Scholar] [CrossRef]
- Robertson, H.D.; Dunn, J.J. Ribonnucleic acid processing activity of Escherichia coli ribonuclease III. J. Biol. Chem. 1975, 250, 3050–3056. [Google Scholar] [PubMed]
- Calin-Jageman, I.; Nicholson, A.W. RNA structure dependent uncoupling of substrate recognition and cleavage by Escherichia coli ribonuclease III. Nucleic Acids Res. 2003, 31, 2381–2392. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, S.; Fernandez, L.; Kameyama, L.; Inada, T.; Nakamura, Y.; Pappas, A.; Court, D.L. Genetic uncoupling of the dsRNA-binding and RNA cleavage activity of the Escherichia coli endoribonuclease RNase III-the effect of dsRNA binding on gene expression. Mol. Microbiol. 1998, 28, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Guarneros, G. Retroregulation of bacteriophage lambda int gene expression. Curr. Top. Microbiol. Immunol. 1988, 136, 1–19. [Google Scholar] [PubMed]
- Oppenheim, A.B.; Komitzer, D.; Altuvia, S.; Court, D.L. Posttranscriptional control of the lysogenic pathway in bacteriophage lambda. Prog. Nucleic Acid Res. Mol. Biol. 1993, 46, 37–49. [Google Scholar] [PubMed]
- Kharratt, A.; Macia, M.J.; Gibson, T.J.; Nilges, M.; Pastore, A. Structure of the dsRNA binding domain of E. coli RNase III. EMBO J. 1995, 14, 3572–3584. [Google Scholar]
- Sun, W.; Jun, E.; Nicholson, A.W. Intrinsic double-stranded-RNA processing activity of Escherichia coli ribonuclease III lacking the dsRNA-binding domain. Biochemistry 2001, 40, 14976–14984. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Jun, E.; Nicholson, A.W. Mutational analysis of the nuclease domain of Escherichia coli ribonuclease III. Identification of conserved acidic residues that are important for catalytic function in vitro. Biochemistry 2004, 43, 13054–13062. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.J.; Jeff, P.; John, C. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Bachem, C.W.B.; van der Hoeven, R.S.; de Bruijn, S.M.; Vreugdenhil, D.; Zabeau, M.; Visser, R.G.F. Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: Analysis of gene expression during potato tuber development. Plant J. 1996, 9, 745–753. [Google Scholar] [CrossRef] [PubMed]
- SMART. Available online: http://smart.embl.de/ (accessed on 27 May 2015).
- Combet, C.; Blanchet, C.; Geourjon, C.; Deleage, G. NPS@: Network protein sequence analysis. Trends Biochem. Sci. 2000, 25, 147–150. [Google Scholar] [CrossRef]
- ExPASy. Available online: http://expasy.org/spdbv/ (accessed on 27 May 2015).
- Hoekema, A.; Hirsch, P.R.; Hooykaas, P.J.J.; Schilperoort, R.A. A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 1983, 303, 179–180. [Google Scholar] [CrossRef]
- Bechtold, N.; Pelletier, G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 1998, 82, 259–266. [Google Scholar] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Sridevi, G.; Parameswari, C.; Rajamuni, P.; Veluthambi, K. Identification of hemizygous and homozygous transgenic rice plants in T1 generation by DNA blot analysis. Plant Biotechnol. 2006, 23, 531–534. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Su, Q.; Zheng, R.; Liu, G.; Lu, Y.; Bian, L.; Chen, J.; Shi, J. ClRTL1 Encodes a Chinese Fir RNase III–Like Protein Involved in Regulating Shoot Branching. Int. J. Mol. Sci. 2015, 16, 25691-25710. https://doi.org/10.3390/ijms161025691
Li X, Su Q, Zheng R, Liu G, Lu Y, Bian L, Chen J, Shi J. ClRTL1 Encodes a Chinese Fir RNase III–Like Protein Involved in Regulating Shoot Branching. International Journal of Molecular Sciences. 2015; 16(10):25691-25710. https://doi.org/10.3390/ijms161025691
Chicago/Turabian StyleLi, Xia, Qian Su, Renhua Zheng, Guangxin Liu, Ye Lu, Liming Bian, Jinhui Chen, and Jisen Shi. 2015. "ClRTL1 Encodes a Chinese Fir RNase III–Like Protein Involved in Regulating Shoot Branching" International Journal of Molecular Sciences 16, no. 10: 25691-25710. https://doi.org/10.3390/ijms161025691
APA StyleLi, X., Su, Q., Zheng, R., Liu, G., Lu, Y., Bian, L., Chen, J., & Shi, J. (2015). ClRTL1 Encodes a Chinese Fir RNase III–Like Protein Involved in Regulating Shoot Branching. International Journal of Molecular Sciences, 16(10), 25691-25710. https://doi.org/10.3390/ijms161025691







