Structures and Functions of Qβ Replicase: Translation Factors beyond Protein Synthesis
Abstract
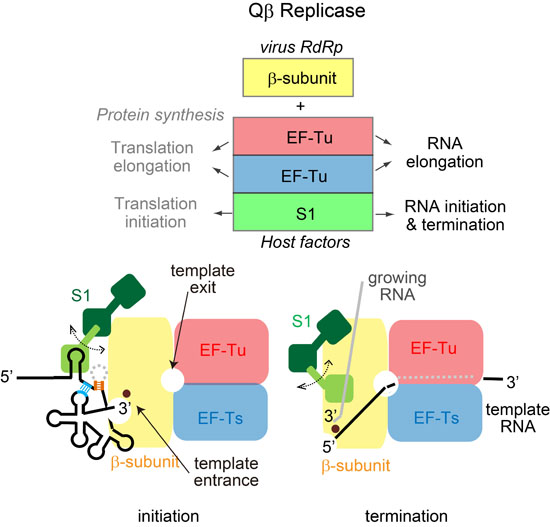
:1. Introduction
2. Overall Structure of the Core Qβ Replicase
2.1. Interactions between Subunits
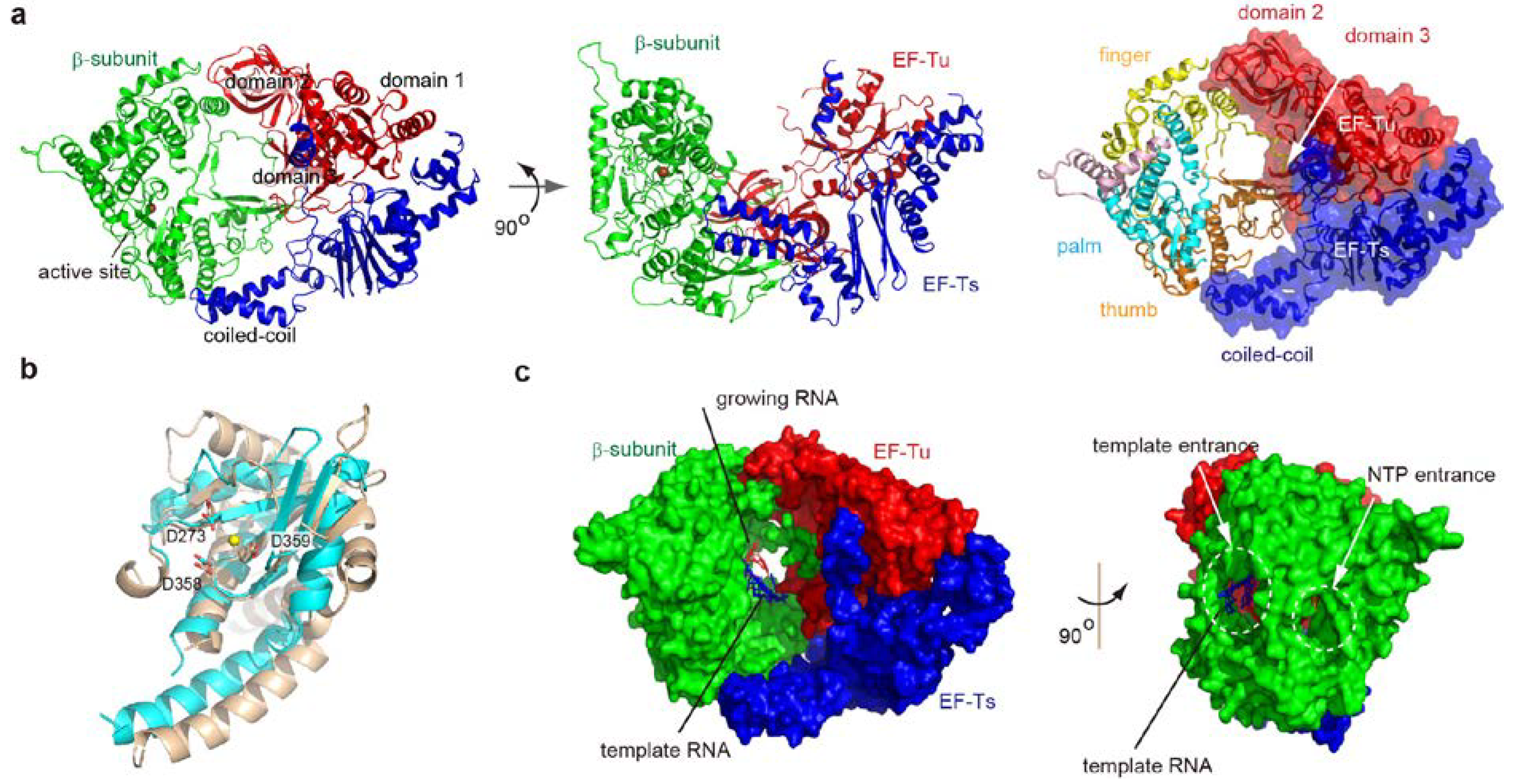
2.2. Structure of the β-Subunit
3. Initiation Stage of RNA Polymerization
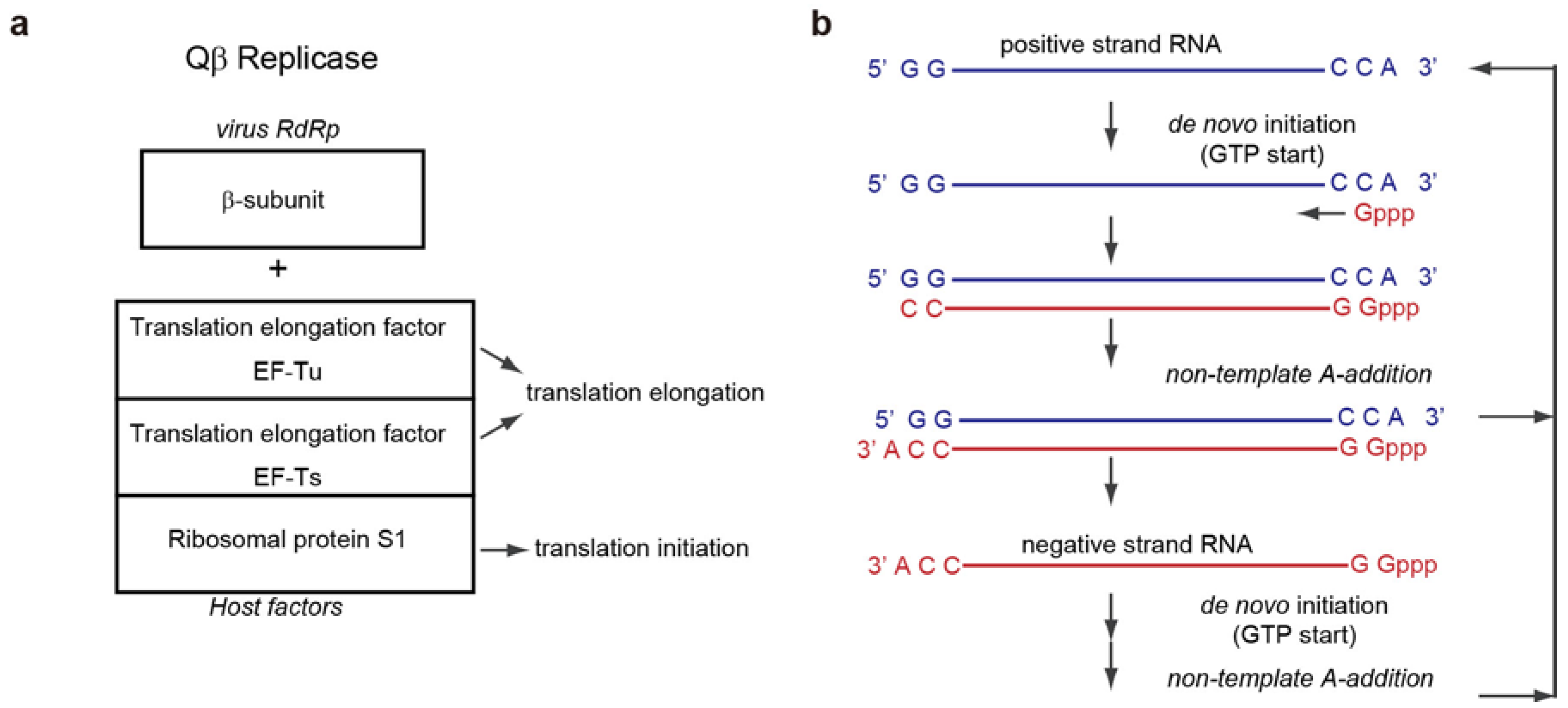
3.1. De Novo Initiation of RNA Synthesis
3.2. Structure of the Initiation Stage
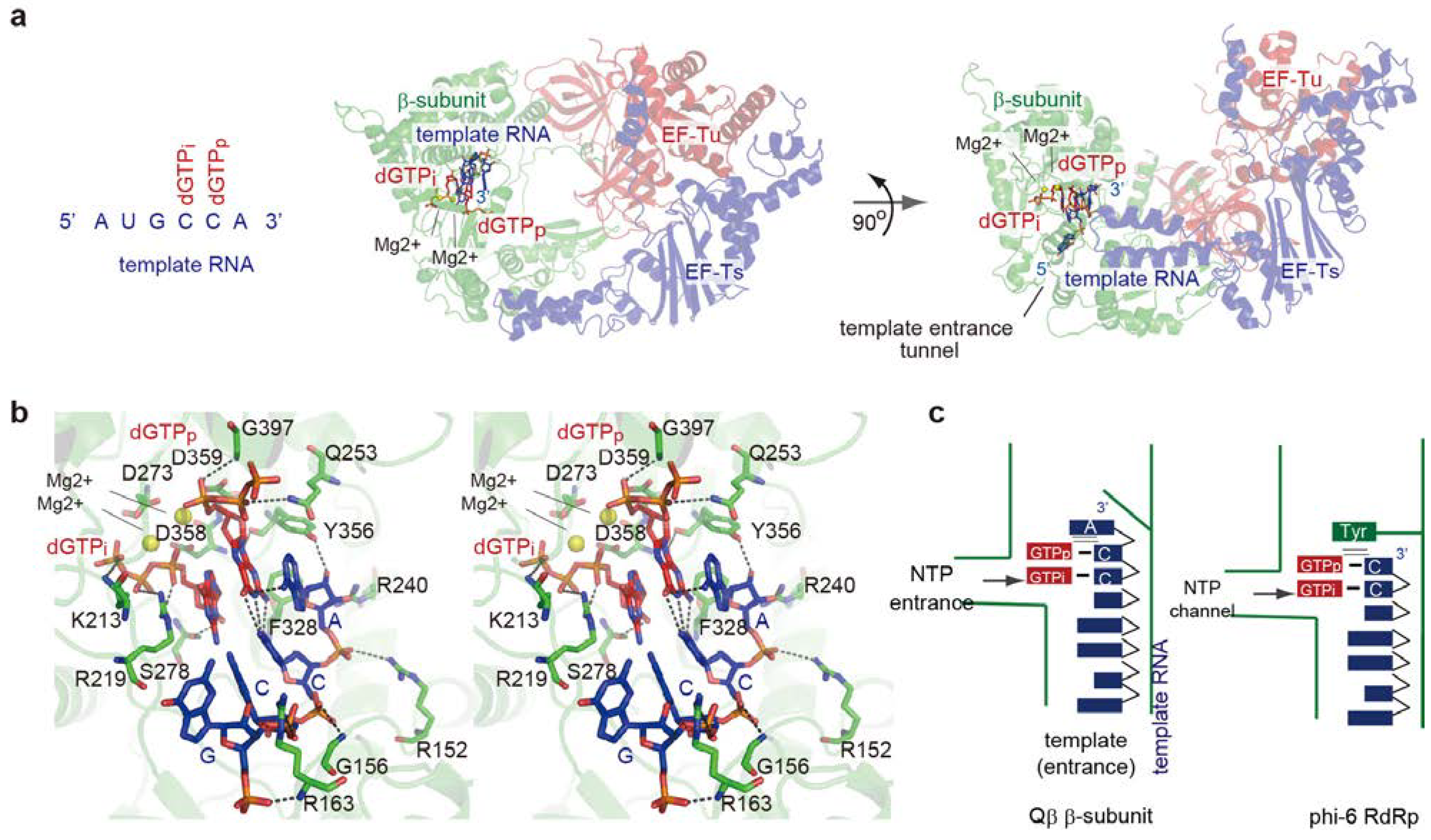
4. Elongation Stages of RNA Polymerization
4.1. Elongation of RNA Synthesis
4.2. Structure of Elongation Stages
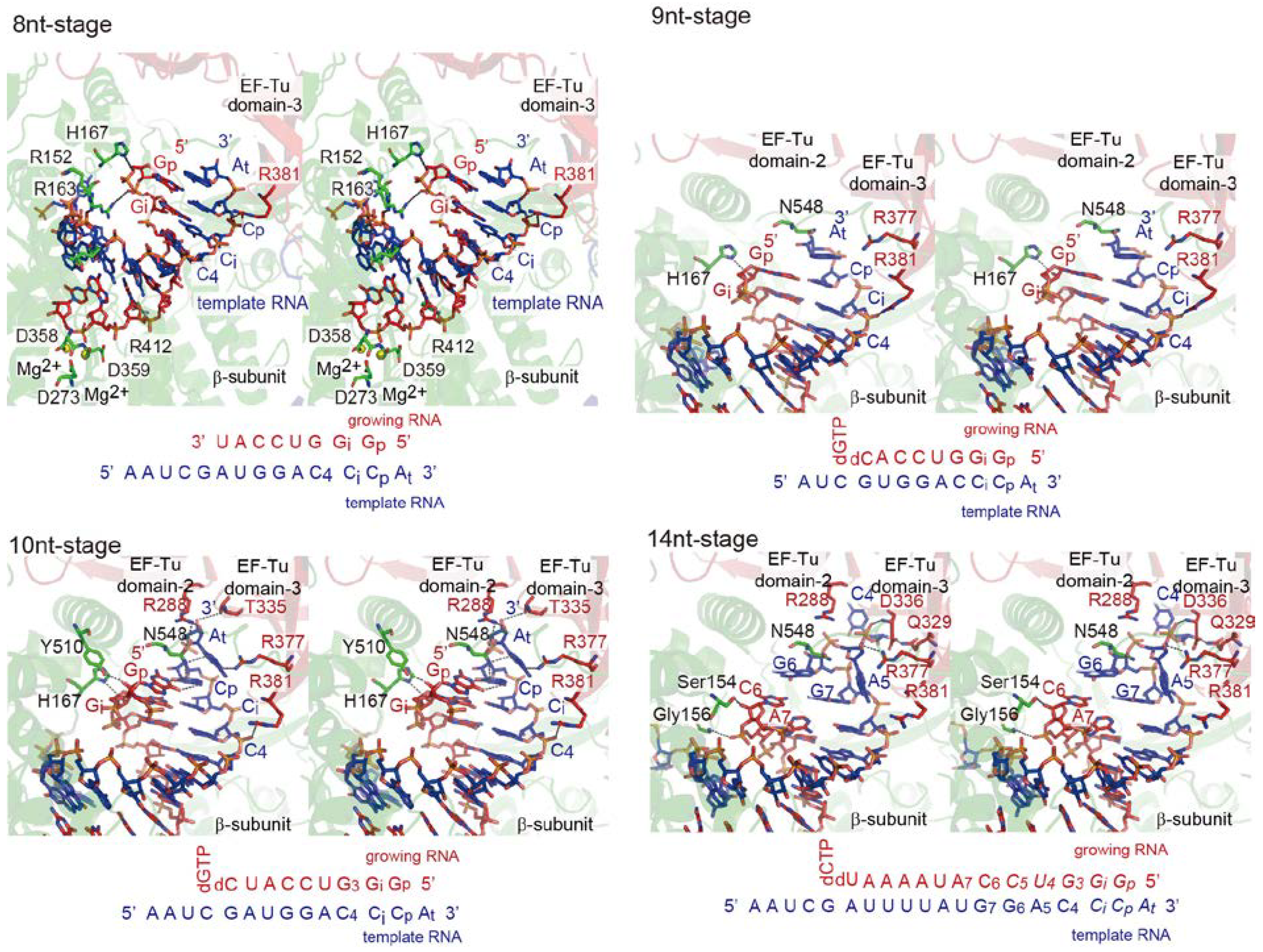
4.3. RNA Passages in the Core Qβ Replicase
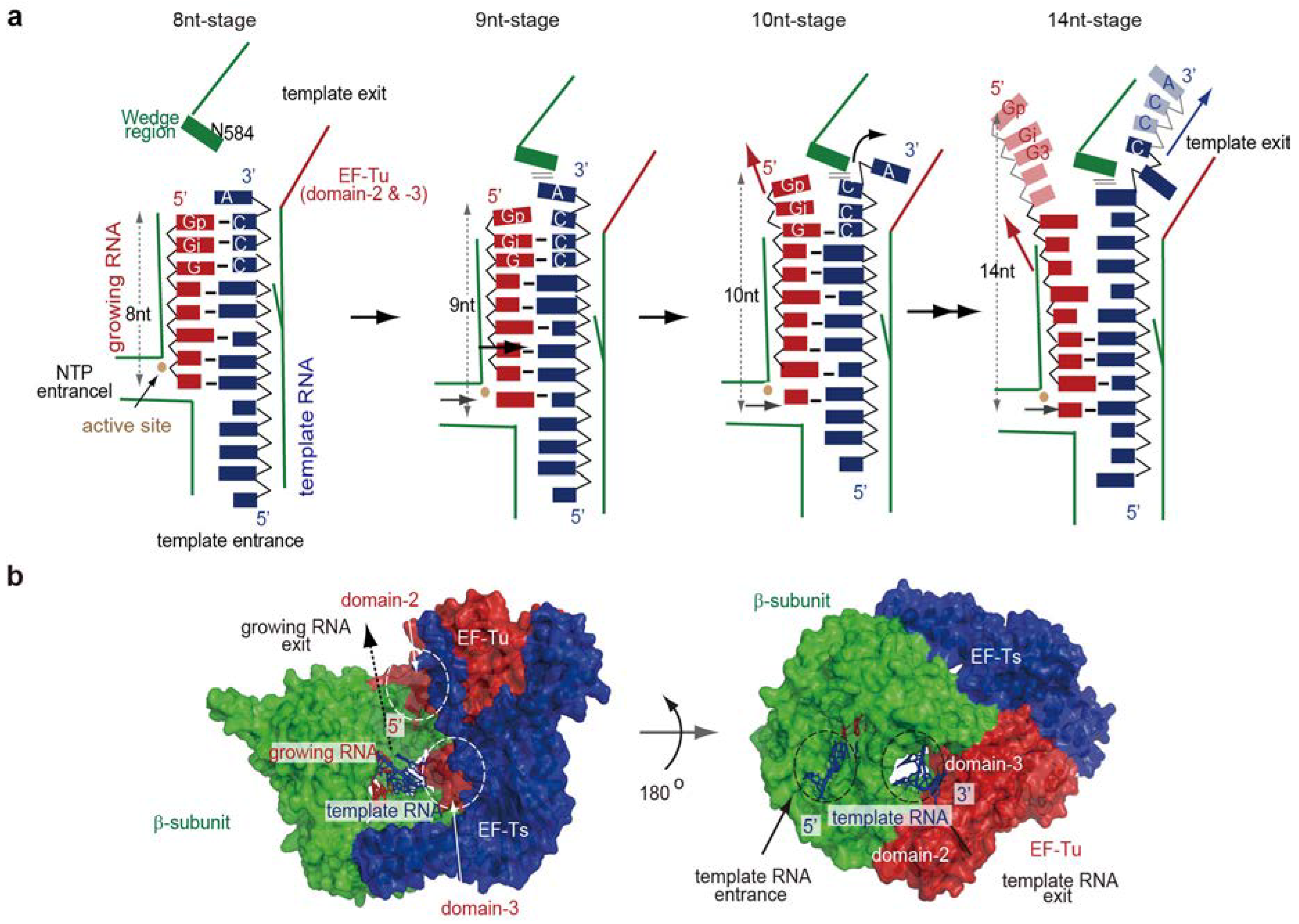
5. Termination Stage of RNA Polymerization
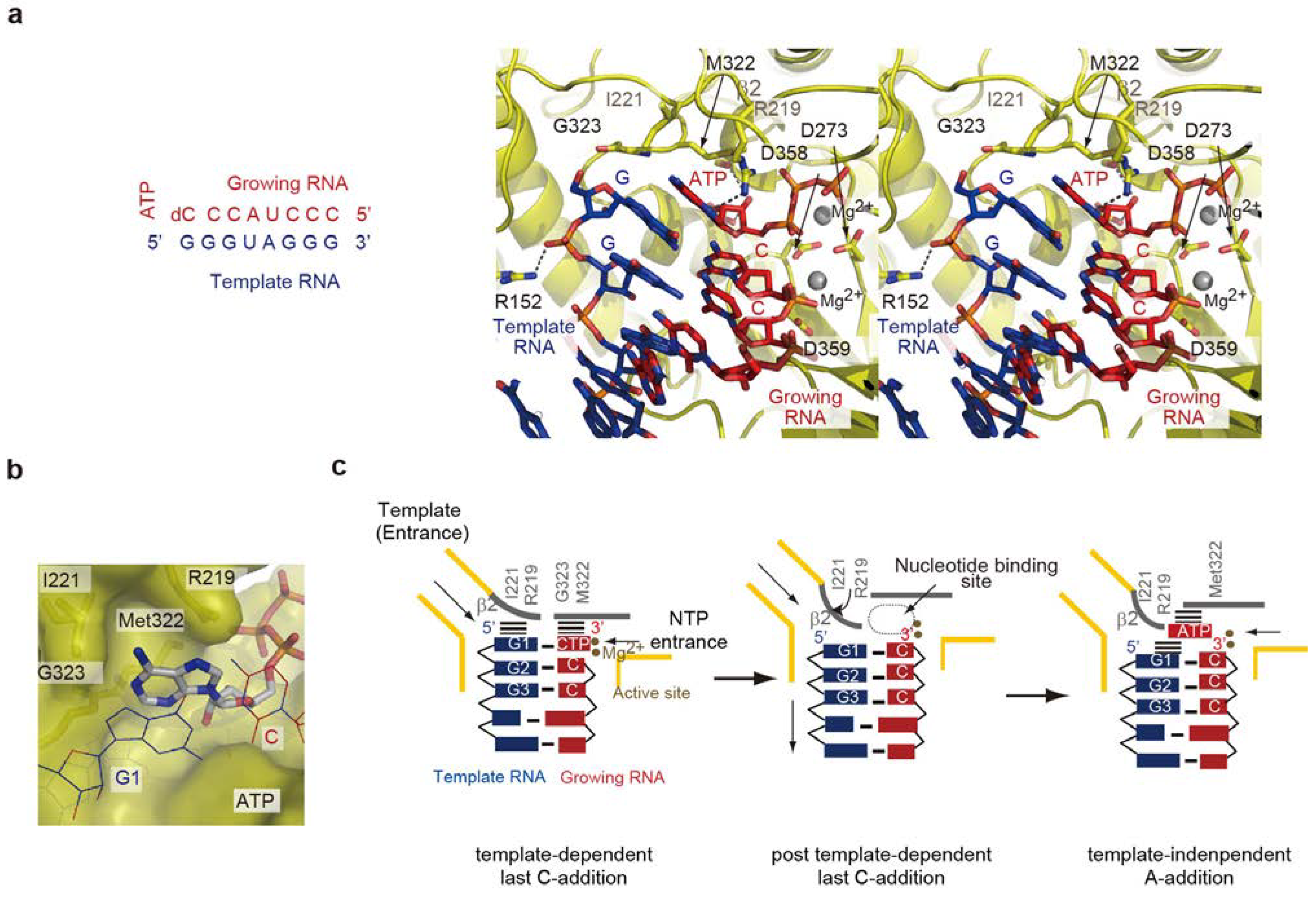
5.1. Non-Template 3'-A Addition
5.2. Structure of the Termination Stage
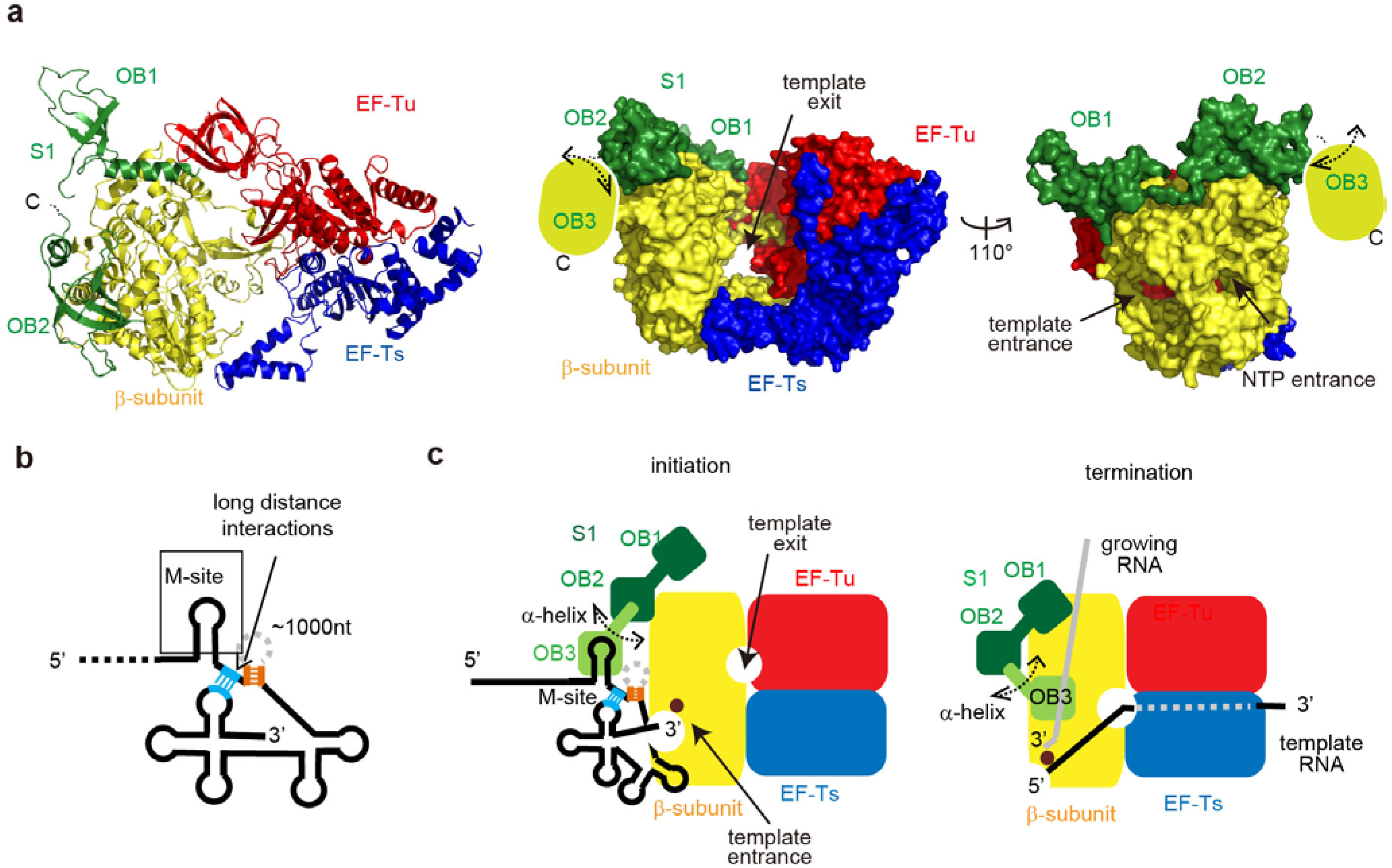
6. Qβ RNA Replication Initiation and Termination
6.1. Qβ RNA Replication
6.2. Structure of the Active Qβ Replicase Containing the S1 Fragment
7. Conclusions
Acknowledgments
Conflicts of Interest
References
- Blumenthal, T.; Carmichael, G.G. RNA replication: Function and structure of Qbeta-replicase. Annu. Rev. Biochem. 1979, 48, 525–548. [Google Scholar]
- Haruna, I.; Speigelman, S. Specific template requirements of RNA replicases. Proc. Natl. Acad. Sci. USA 1965, 54, 579–587. [Google Scholar] [CrossRef]
- Chetverin, A.B. Raradoxes of replication of RNA of a bacterial virus. Mol. Biol. 2011, 45, 139–149. [Google Scholar]
- Kondo, M.; Gallerani, R.; Weissmann, C. Subunit structure of Qβ replicase. Nature 1970, 228, 525–527. [Google Scholar]
- Kamen, R. Characterization of the subunits of Qβ replicase. Nature 1970, 228, 527–553. [Google Scholar]
- Blumenthal, T.; Landers, T.A.; Weber, K. Bacteriophage Q replicase contains the protein biosynthesis elongation factors EF-Tu and EF-Ts. Proc. Natl. Acad. Sci. USA 1972, 69, 1313–1317. [Google Scholar] [CrossRef]
- Wahba, A.J.; Miller, M.J.; Niveleau, A.; Landers, T.A.; Carmichael, G.G.; Weber, K.; Hawley, D.A.; Slobin, L.I. Subunit I of Qβ replicase and 30 S ribosomal protein S1 of Escherichia coli, Evidence for the identity of the two proteins. J. Biol. Chem. 1974, 249, 3314–3316. [Google Scholar]
- Kamen, R.; Kondo, M.; Römer, W.; Weissmann, C. Reconstitution of Qβ replicase lacking subunit α with protein-synthesis-interference factor i. Eur. J. Biochem. 1972, 31, 44–51. [Google Scholar]
- Hori, K.; Harada, K.; Kuwano, M. Function of bacteriophage Qβ replicase containing an altered subunit IV. J. Mol. Biol. 1974, 86, 699–708. [Google Scholar] [CrossRef]
- Landers, T.A.; Blumenthal, T.; Weber, K. Function and structure in ribonucleic acid phage Qβ ribonucleic acid replicase. The roles of the different subunits in transcription of synthetic templates. J. Biol. Chem. 1974, 249, 5801–5808. [Google Scholar]
- Agirrezabala, X.; Frank, J. Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu. Q. Rev. Biophys. 2009, 42, 159–200. [Google Scholar] [CrossRef]
- Schuette, J.C.; Murphy, F.V., 4th; Kelley, A.C.; Weir, J.R.; Giesebrecht, J.; Connell, S.R.; Loerke, J.; Mielke, T.; Zhang, W.; Penczek, P.A.; et al. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J. 2009, 28, 755–765. [Google Scholar] [CrossRef]
- Schmeing, T.M.; Voorhees, R.M.; Kelley, A.C.; Gao, Y.G.; Murphy, F.V., 4th; Weir, J.R.; Ramakrishnan, V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 2009, 326, 688–694. [Google Scholar] [CrossRef]
- Subramanian, A.R. Structure and functions of ribosomal protein S1. Prog. Nucleic Acid Res. Mol. Biol. 1983, 28, 102–138. [Google Scholar]
- Farwell, M.A.; Robert, M.W.; Rabinowitz, J.C. The effect of ribosomal protein S1 from Escherichia coli and Micrococcus luteus on protein synthesis in vitro by E. coli and Bacillus subtilis. Mol. Microbiol. 1992, 6, 3375–3383. [Google Scholar] [CrossRef]
- Boni, I.V.; Isaeva, D.M.; Musychenko, M.L.; Tzareva, N.V. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 1991, 19, 155–162. [Google Scholar] [CrossRef]
- Tedin, K.; Resch, A.; Blasi, U. Requirements for ribosomal protein S1 for translation initiation of mRNAs with and without a 5' leader sequence. Mol. Microbiol. 1997, 25, 189–199. [Google Scholar]
- Sorensen, M.A.; Fricke, J.; Pedersen, S. Ribosomal protein S1 is required for translation of most, if not all, natural mRNAs in Escherichia coli in vivo. J. Mol. Biol. 1998, 280, 561–569. [Google Scholar] [CrossRef]
- Komarova, A.V.; Tchufistova, L.S.; Supina, E.V.; Boni, I.V. Protein S1 counteracts the inhibitory effect of the extended Shine–Dalgarno sequence on translation. RNA 2002, 8, 1137–1147. [Google Scholar]
- Duval, M.; Korepanov, A.; Fuchsbauer, O.; Fechter, P.; Haller, A.; Fabbretti, A.; Choulier, L.; Micura, R.; Klaholz, B.P.; Romby, P.; et al. Escherichia coli ribosomal protein S1 unfolds structured mRNAs onto the ribosome for active translation initiation. PLoS Biol. 2013, 11, e1001731. [Google Scholar] [CrossRef]
- Dahlberg, J.E. Terminal sequences of bacteriophage RNAs. Nature 1968, 220, 548–552. [Google Scholar]
- Weissmann, C.; Billeter, M.A.; Goodman, H.M.; Hindley, J.; Weber, H. Structure and function of phage RNA. Annu. Rev. Biochem. 1973, 42, 303–328. [Google Scholar]
- Silverman, P.H. Replication of RNA viruses: Specific binding of the Q RNA polymerase to Q RNA. Arch. Biochem. Biophys. 1973, 157, 222–233. [Google Scholar]
- Blumenthal, T. Q beta replicase template specificity: Different templates require different GTP concentrations for initiation. Proc. Natl. Acad. Sci. USA 1980, 77, 2601–2605. [Google Scholar]
- Weber, H.; Weissmann, C. The 3'-termini of bacteriophage Qβ plus and minus strands. J. Mol. Biol. 1970, 51, 215–224. [Google Scholar]
- Bausch, J.N.; Kramer, F.R.; Miele, E.A.; Dobkin, C.; Mills, D.R. Terminal adenylation in the synthesis of RNA by Qβ replicase. J. Biol. Chem. 1983, 258, 1978–1984. [Google Scholar]
- Kidmose, R.T.; Vasiliev, N.N.; Chetverin, A.B.; Andersen, G.R.; Knudsen, C.R. Structure of the Qβ replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins. Proc. Natl. Acad. Sci. USA 2010, 107, 10884–10889. [Google Scholar]
- Takeshita, D.; Tomita, K. Assembly of Qβ viral RNA polymerase with host translational elongation factors EF-Tu and -Ts. Proc. Natl. Acad. Sci. USA. 2010, 107, 15733–15738. [Google Scholar]
- Takeshita, D.; Tomita, K. Molecular basis for RNA polymerization by Qβ replicase. Nat. Struct. Mol. Biol. 2012, 19, 229–237. [Google Scholar]
- Takeshita, D.; Yamashita, S.; Tomita, K. Mechanism for template-independent terminal adenylation activity of Qβ replicase. Structure 2012, 20, 1661–1669. [Google Scholar]
- Takeshita, D.; Yamashita, S.; Tomita, K. Molecular insights into replication initiation by Qβ replicase using ribosomal protein S1. Nucleic Acids Res. 2014. [Google Scholar] [CrossRef]
- Ferrer-Orta, C.; Arias, A.; Escarmís, C.; Verdaguer, N. A comparison of viral RNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 2006, 16, 27–34. [Google Scholar] [CrossRef]
- O’Reilly, E.K.; Kao, C.C. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology 1998, 252, 287–303. [Google Scholar]
- Kawashima, T.; Berthet-Colominas, C.; Wulff, M.; Cusack, S.; Leberman, R. The structure of the Escherichia coli EF-Tu:EF-Ts complex at 2.5 Å resolution. Nature 1996, 379, 511–518. [Google Scholar] [CrossRef]
- Nissen, P.; Kjeldgaard, M.; Thirup, S.; Polekhina, G.; Reshetnikova, L.; Clark, B.F.; Nyborg, J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 1995, 270, 1464–1472. [Google Scholar]
- Nissen, P.; Kjeldgaard, M.; Thirup, S.; Clark, B.F.; Nyborg, J. The ternary complex of aminoacylated tRNA and EF-Tu-GTP. Recognition of a bond and a fold. Biochimie 1996, 78, 921–933. [Google Scholar] [CrossRef]
- Karring, H.; Mathu, S.G.; van Duin, J.; Clark, B.F.; Kraal, B.; Knudsen, C.R. Qβ-phage resistance by deletion of the coiled-coil motif in elongation factor Ts. J. Biol. Chem. 2004, 279, 1878–1884. [Google Scholar]
- Caldas, T.D.; Yaagoubi, A.E.; Richarme, G. Chaperone properties of bacterial elongation factor EF-Tu. J. Biol. Chem. 1998, 273, 11478–11482. [Google Scholar]
- Kudlicki, W.; Coffman, A.; Kramer, G.; Hardesty, B. Renaturation of rhodanese by translational elongation factor (EF) Tu. J. Biol. Chem. 1997, 272, 32206–32210. [Google Scholar]
- Suzuki, H.; Ueda, T.; Taguchi, H.; Takeuchi, N. Chaperone properties of mammalian mitochondrial translation elongation factor Tu. J. Biol. Chem. 2007, 282, 4076–4084. [Google Scholar]
- Koonin, E.V. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 1991, 72, 2197–2206. [Google Scholar] [CrossRef]
- Brautigam, C.A.; Steitz, T.A. Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr. Opin. Struct. Biol. 1998, 8, 54–63. [Google Scholar] [CrossRef]
- Steitz, T.A.; Steitz, J.A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. USA 1993, 90, 6498–6502. [Google Scholar] [CrossRef]
- Rensing, U.; August, J.T. The 3'-Terminus and the replication of phage RNA. Nature 1969, 224, 853–856. [Google Scholar]
- Schaffner, W.; Ruegg, K.J.; Weissmann, C. Nanovariant RNAs: Nucleotide sequence and interaction with bacteriophage Qβ replicase. J. Mol. Biol. 1997, 117, 877–907. [Google Scholar]
- Makeyev, E.V.; Bamford, D.H. Replicase activity of purified recombinant P2 of double-stranded RNA bacteriophage 6. EMBO J. 2000, 19, 124–133. [Google Scholar]
- Makeyev, E.V.; Bamford, D.H. The polymerase subunit of a dsRNA virus plays a central role in the regulation of viral RNA metabolism. EMBO J. 2000, 19, 6275–6284. [Google Scholar]
- Butcher, S.J.; Grimes, J.M.; Makeyev, E.V.; Bamford, D.H.; Stuart, D.I. A mechanism for initiating RNA-dependent RNA polymerization. Nature 2011, 410, 235–240. [Google Scholar]
- Salgado, P.S.; Makeyev, E.V.; Butcher, S.J.; Bamford, D.H.; Stuart, D.I.; Grimes, J.M. The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase. Structure 2004, 12, 307–316. [Google Scholar]
- Nissen, P.; Thirup, S.; Kjeldgaard, M.; Nyborg, J. The crystal structure of Cys–tRNACys–EF-Tu–GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure 1999, 7, 143–156. [Google Scholar]
- Biebricher, C.K.; Luce, R. Sequence analysis of RNA species synthesized by Q.beta. replicase without template. Biochemistry 1993, 32, 4848–4854. [Google Scholar]
- Tomita, K.; Fukai, S.; Ishitani, R.; Ueda, T.; Takeuchi, N.; Vassylyev, D.G.; Nureki, O. Structural basis for template-independent RNA polymerization. Nature 2004, 430, 700–704. [Google Scholar]
- Xiong, Y.; Steitz, T.A. Mechanism of transfer RNA maturation by CCA-adding enzyme without using an oligonucleotide template. Nature 2004, 430, 640–645. [Google Scholar]
- Tomita, K.; Ishitani, R.; Fukai, S.; Nureki, O. Complete crystallographic analysis of the dynamics of CCA sequence addition. Nature 2006, 443, 956–960. [Google Scholar]
- Toh, Y.; Numata, T.; Watanabe, K.; Takeshita, D.; Nureki, O.; Tomita, K. Molecular basis for maintenance of fidelity during the CCA-adding reaction by a CCA-adding enzyme. EMBO J. 2008, 27, 1944–1952. [Google Scholar]
- Toh, Y.; Takeshita, D.; Numata, T.; Fukai, S.; Nureki, O.; Tomita, K. Mechanism for the definition of elongation and termination by the class II CCA-adding enzyme. EMBO J. 2009, 28, 3353–3365. [Google Scholar]
- Pan, B.; Xiong, Y.; Steitz, T.A. How the CCA-adding enzyme selects adenine over cytosine at position 76 of tRNA. Science 2010, 330, 937–940. [Google Scholar]
- Toh, Y.; Takeshita, D.; Nagaike, T.; Numata, T.; Tomita, K. Mechanism for the alteration of the substrate specificities of template-independent RNA polymerases. Structure 2011, 19, 232–243. [Google Scholar]
- Yamashita, S.; Takeshita, D.; Tomita, K. Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase. Structure 2014, 22, 315–325. [Google Scholar]
- Tomita, K.; Yamashita, S. Molecular mechanisms of template-independent RNA polymerization by tRNA nucleotidyltransferases. Front. Genet. 2014, 5. [Google Scholar] [CrossRef]
- Bycroft, M.; Hubbard, T.J.; Proctor, M.; Freund, S.M; Murzin, A.G. The solution structure of the S1 RNA binding domain: A member of an ancient nucleic acid-binding fold. Cell 1997, 88, 235–242. [Google Scholar]
- Meyer, F.; Weber, H.; Weissmann, C. Interactions of Qβ replicase with Qβ RNA. J. Mol. Biol. 1981, 153, 631–660. [Google Scholar]
- Barrera, I.; Schuppli, D.; Sogo, J.M.; Weber, H. Different mechanisms of recognition of bacteriophage Qβ plus and minus strand RNAs by Qβ replicase. J. Mol. Biol. 1993, 232, 512–521. [Google Scholar] [CrossRef]
- Miranda, G.; Schuppli, D.; Barrera, I.; Hausherr, C.; Sogo, J.M.; Weber, H. Recognition of bacteriophage Qβ plus strand RNA as a template by Qβ replicase: Role of RNA interactions mediated by ribosomal proteins S1 and host factor. J. Mol. Biol. 1997, 267, 1089–1103. [Google Scholar]
- Schuppli, D.; Miranda, G.; Qiu, S.; Weber, H. A branched stem-loop structure in the M-site of bacteriophage Qβ RNA is important for template recognition by Qβ replicase holoenzyme. J. Mol. Biol. 1998, 283, 585–593. [Google Scholar]
- Klovins, J.; Berzins, V.; van Duin, J. A long-range interaction in Qβ RNA that bridges the thousand nucleotides between the M-site and the 3' end is required for replication. RNA 1998, 4, 948–957. [Google Scholar]
- Klovins, J.; van Duin, J. A long-range pseudoknot in Qβ RNA is essential for replication. J. Mol. Biol. 1999, 294, 875–884. [Google Scholar]
- Guerrier-Takada, C.; Subramanian, A.R.; Cole, P.E. The activity of discrete fragments of ribosomal protein S1 in Q beta replicase function. J. Biol. Chem. 1983, 258, 13649–13652. [Google Scholar]
- Vasilyev, N.N.; Kutlubaeva, Z.S.; Ugarov, V.I.; Chetverina, H.V.; Chetverin, A.B. Ribosomal protein S1 functions as a termination factor in RNA synthesis by Qβ phage replicase. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef]
- Lai, M.M. Cellular factors in the transcription and replication of viral RNA genomes: A parallel to DNA-dependent RNA transcription. Virology 1998, 244, 1–12. [Google Scholar]
- Quadt, R.; Kao, C.C.; Browning, K.S.; Hershberger, R.P.; Ahlquist, P. Characterization of a host protein associated with brome mosaic virus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA 1993, 90, 1498–1502. [Google Scholar]
- Osaman, T.A.M.; Buck, K.W. The tobacco mosaic virus RNA polymerase complex contains a plant protein related to the RNA-binding subunit of yeast eIF-3. J. Virol. 1997, 71, 6075–6082. [Google Scholar]
- Das, T.; Mathur, M.; Gupta, A.K.; Janssen, G.M.C.; Banerjee, A.K. RNA polymerase of vesicular stomatitis virus specifically associates with translation elongation factor-1 αβγ for its activity. Proc. Natl. Acad. Sci. USA 1998, 95, 1460–1465. [Google Scholar]
- Harris, K.S; Xiang, W.; Alexander, L.; Lane, W.S.; Paul, A.V.; Wimmer, E. Interaction of poliovirus polypeptide 3CDpro with the 5' and 3' termini of the poliovirus genome: Identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem. 1994, 269, 27004–27014. [Google Scholar]
- Weiner, A.M.; Maizels, N. tRNA-like structures tag the ends of genomic RNA molecules for replication: Implications for the origin of protein synthesis. Proc. Natl. Acad. Sci. USA 1987, 84, 7383–7387. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Tomita, K. Structures and Functions of Qβ Replicase: Translation Factors beyond Protein Synthesis. Int. J. Mol. Sci. 2014, 15, 15552-15570. https://doi.org/10.3390/ijms150915552
Tomita K. Structures and Functions of Qβ Replicase: Translation Factors beyond Protein Synthesis. International Journal of Molecular Sciences. 2014; 15(9):15552-15570. https://doi.org/10.3390/ijms150915552
Chicago/Turabian StyleTomita, Kozo. 2014. "Structures and Functions of Qβ Replicase: Translation Factors beyond Protein Synthesis" International Journal of Molecular Sciences 15, no. 9: 15552-15570. https://doi.org/10.3390/ijms150915552
APA StyleTomita, K. (2014). Structures and Functions of Qβ Replicase: Translation Factors beyond Protein Synthesis. International Journal of Molecular Sciences, 15(9), 15552-15570. https://doi.org/10.3390/ijms150915552