Conservation Genetics of an Endangered Lady’s Slipper Orchid: Cypripedium japonicum in China
Abstract
:1. Introduction
2. Results
2.1. Genetic Diversity and Genetic Differentiation
| Populations | Na | Ne | H | I | PPL |
|---|---|---|---|---|---|
| DBS | 1.1284 (0.3361) | 1.0670 (0.2128) | 0.0393 (0.1194) | 0.0593 (0.1738) | 12.84% |
| SNJ | 1.1101 (0.3144) | 1.0724 (0.2282) | 0.0413 (0.1252) | 0.0609 (0.1810) | 11.01% |
| TMS | 1.0734 (0.2620) | 1.0510 (0.1871) | 0.0297 (0.1071) | 0.0435 (0.1563) | 7.34% |
| DMS | 1.1560 (0.3645) | 1.1041 (0.2713) | 0.0587 (0.1471) | 0.0863 (0.2114) | 15.60% |
| BTM | 1.0917 (0.2900) | 1.0697 (0.2274) | 0.0389 (0.1248) | 0.0563 (0.1794) | 9.17% |
| Average | 1.1119 (0.3134) | 1.0728 (0.2253) | 0.0416 (0.1247) | 0.0613 (0.1804) | 11.19% |
| Species Level | 1.3853 (0.4889) | 1.2132 (0.3283) | 0.1273 (0.1824) | 0.1928 (0.2656) | 38.53% |
| Ht | Hs | Gst | Nm | ||
| Mean | 0.1264 (0.0329) | 0.0416 (0.066) | 0.6712 | 0.2450 |
| SV | d.f. | SSD | MSD | VC | TVP | Φpt | p Value |
|---|---|---|---|---|---|---|---|
| Among populations | 4 | 579.734 | 144.934 | 5.624 | 70% | 0.698 | 0.001 |
| Within populations | 123 | 299.211 | 2.433 | 2.433 | 30% | ||
| Total | 127 | 878.945 | 8.056 | 100% |
2.2. Genetic Relationships and Population Structure
| Populations | DBS | SNJ | TMS | DMS | BTM |
|---|---|---|---|---|---|
| DBS | **** | 0.9055 | 0.8508 | 0.9128 | 0.8731 |
| SNJ | 0.0993 | **** | 0.9098 | 0.8887 | 0.9205 |
| TMS | 0.1616 | 0.0945 | **** | 0.8557 | 0.9012 |
| DMS | 0.0912 | 0.1180 | 0.1558 | **** | 0.8835 |
| BTM | 0.1357 | 0.0828 | 0.1040 | 0.1239 | **** |

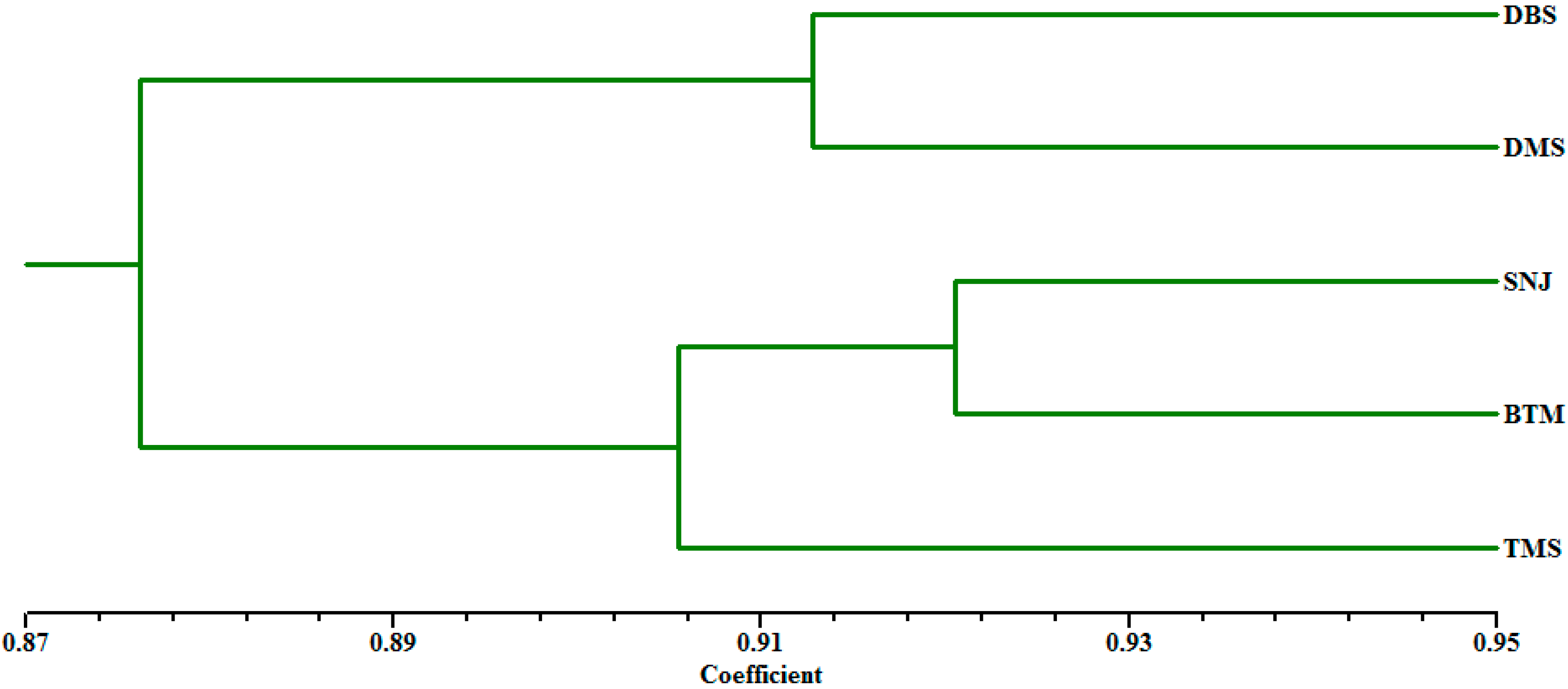
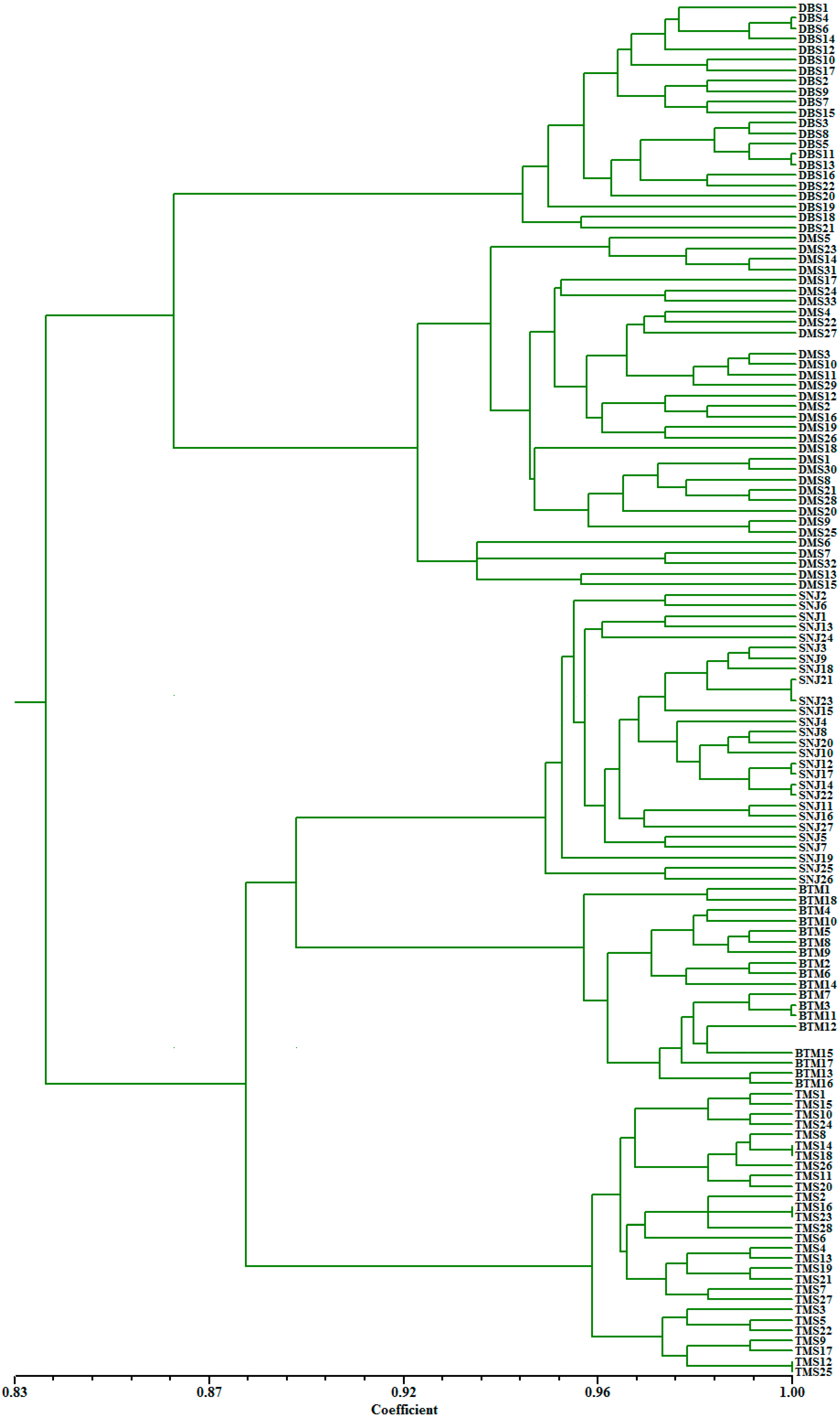
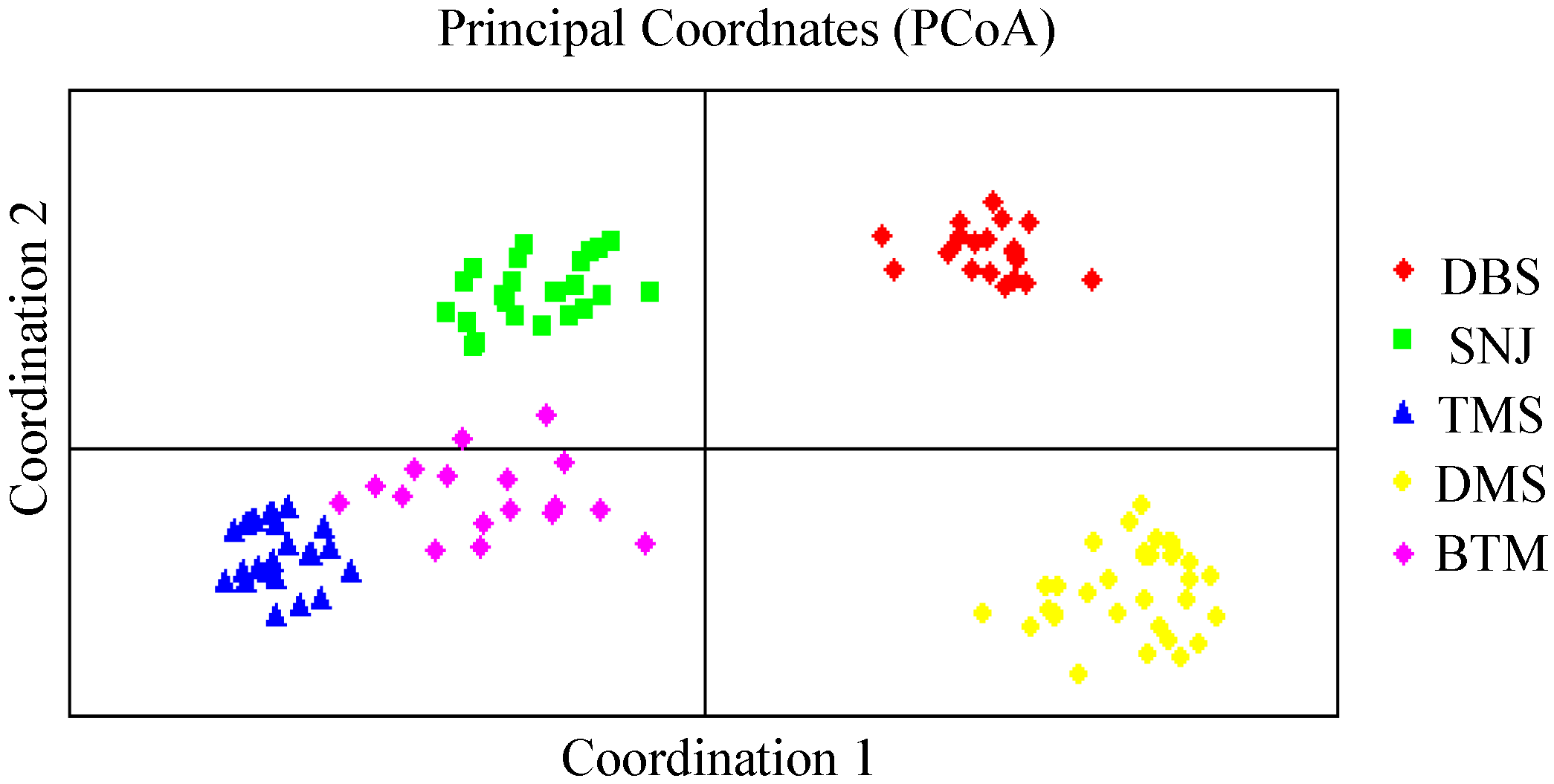
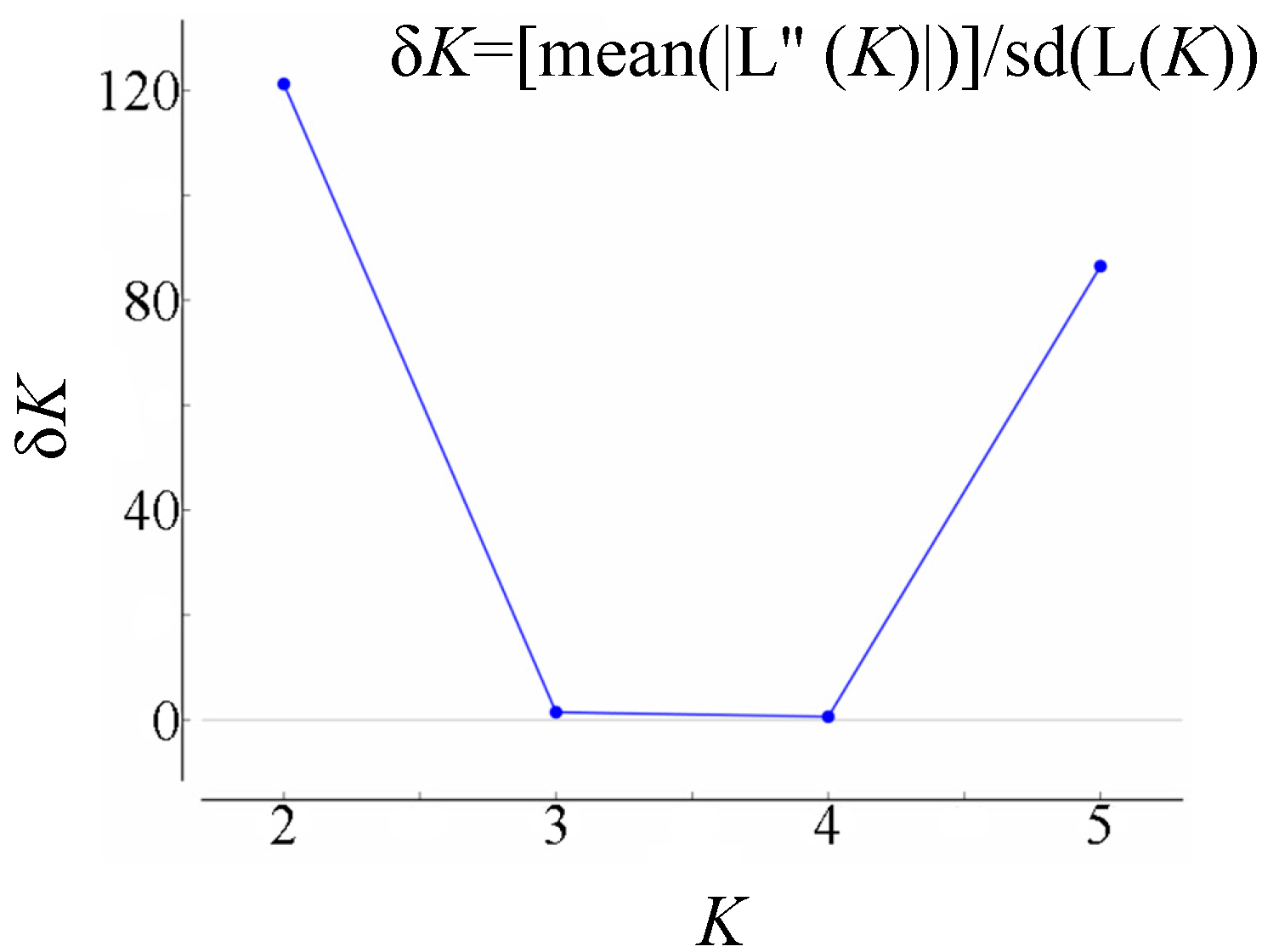
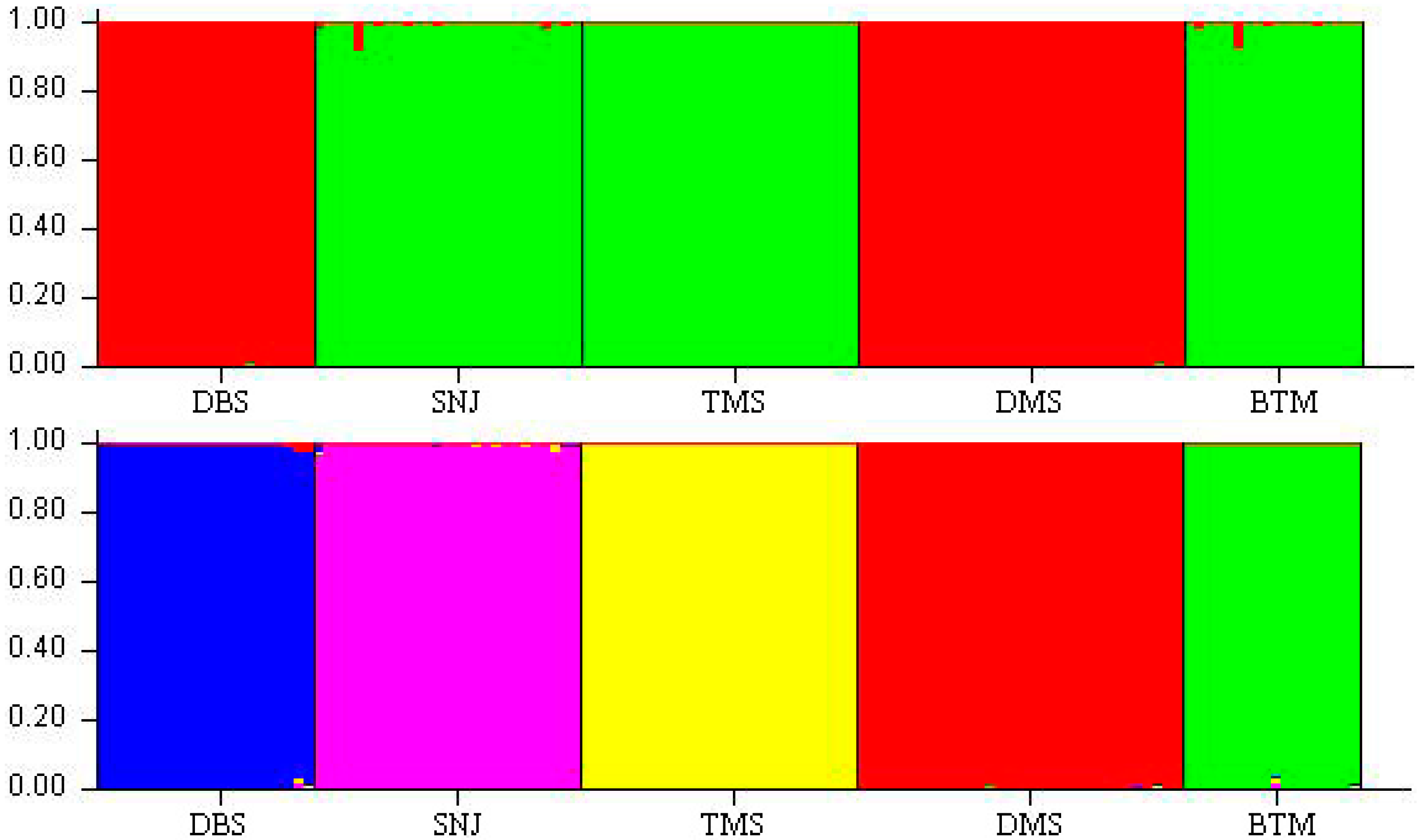
3. Discussion
3.1. Genetic Diversity and Differentiation
| Orchid Species | Hp | Ip | PPLp (%) | Hs | Is | PPLs (%) | Gst (Fst) | SR |
|---|---|---|---|---|---|---|---|---|
| Amitostigma hemipilioides | 0.603 | 0.2949 | 50.9 | 0.686 | 0.3873 | 64.7 | 0.367 | [26] |
| Brassavola tuberculata | - | - | - | 0.216 | 0.314 | 52.41 | - | [37] |
| Calanthe tsoongiana | 0.183 | 0.271 | 50.0 | 0.398 | 0.576 | 96.80 | 0.55 | [27] |
| Cattleya bicolor | - | - | - | 0.219 | 0.323 | 56.63 | - | [37] |
| Cattleya elongata | - | - | 56.8 | - | - | - | 0.18 | [28] |
| Cattleya granulosa | - | - | - | 0.163 | 0.237 | 30.72 | - | [37] |
| Cattleya labiata | - | - | - | 0.132 | 0.193 | 30.12 | - | [37] |
| Cattleya schofieldiana | - | - | - | 0.213 | 0.314 | 56.02 | - | [37] |
| Cymbidium goeringii | 0.1945 | 0.2958 | 63.06 | 0.2628 | 0.4037 | 88.19 | 0.244 | [29] |
| Dendrobium fimbriatum | 0.0871 | 0.1290 | 23.93 | 0.3227 | 0.4779 | 89.74 | 0.7443 | [38] |
| Gastrodia elata | 0.176 | 0.270 | 59.09 | 0.236 | 0.367 | 81.82 | 0.2725 | [39] |
| Octomeria crassifolia | 0.2648 | 0.4005 | 91.57 | 0.352 | 0.530 | - | 0.76 | [40] |
| Octomeria grandiflora | 0.2578 | 0.382 | 82.4 | 0.338 | 0.508 | - | 0.12 | [40] |
| Paphiopedilum micranthum | 0.2847 | 0.4236 | 80.28 | 0.3839 | 0.5646 | 91.66 | 0.2577 | [41] |
| Piperia yadonii (2006) | 0.062 | - | - | - | - | - | 0.424 | [30] |
| Piperia yadonii (2007) | 0.059 | - | - | - | - | - | 0.394 | [30] |
| Platanthera aquilonis | 0.084 | - | 22.61 | 0.184 | - | 61.7 | 0.70 | [42] |
| Platanthera dilatata | 0.1312 | - | 35.35 | 0.182 | - | 57.5 | 0.49 | [42] |
| Platanthera huronensis | 0.119 | - | 32.64 | 0.172 | - | 43.0 | 0.36 | [42] |
| Tipularia discolor | 0.0309 | - | 7.95 | - | - | - | 0.415 | [43] |
| Average | 0.1812 | 0.3084 | 50.51 | 0.2788 | 0.3997 | 64.36 | 0.4186 |
3.2. Conservation Implication
4. Materials and Methods
4.1. Study Organism
4.2. Population Sampling
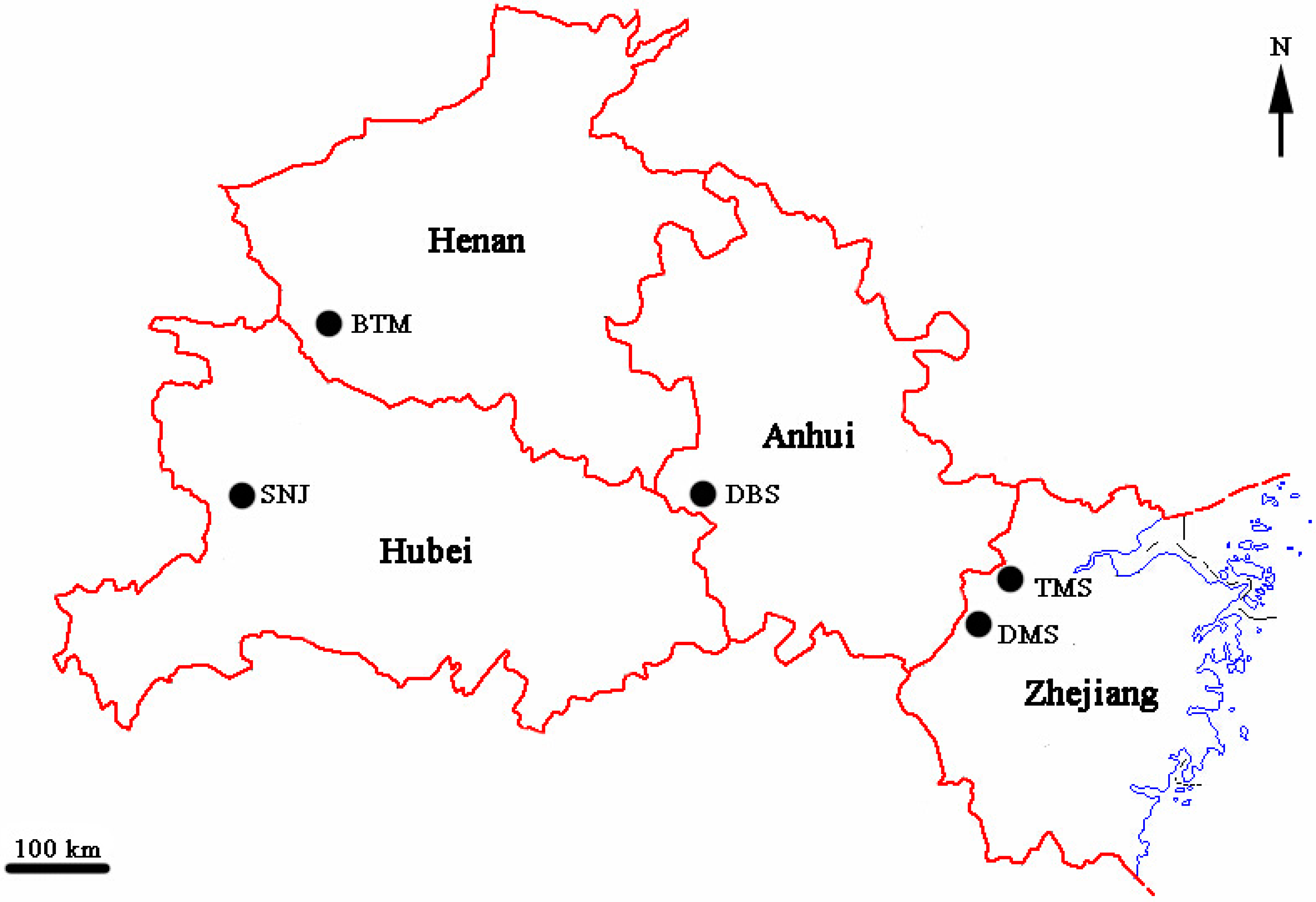
| Populations | Locality | Geographical Coordinate | Sample Size | Voucher Number |
|---|---|---|---|---|
| DBS | Dabieshan, Anhui province | 31°24'N, 116°36'E | 22 | Q526 |
| SNJ | Shennongjia, Hubei province | 31°21'N, 110°03'E | 27 | Q611 |
| TMS | Tianmushan, Zhejiang province | 30°21'N, 119°25'E | 28 | L605 |
| DMS | Damingshan, Zhejiang province | 30°03'N, 118°59'E | 33 | L522 |
| BTM | Baotianman, Henan province | 33°26'N, 111°38'E | 18 | Q518 |
4.3. Inter-Simple Sequence Repeat (ISSR) Amplification
| Primer Name | Sequence (5'→3') | Annealing Temperature (°C) | Primer Name | Sequence (5'→3') | Annealing Temperature (°C) |
|---|---|---|---|---|---|
| UBC807 | (AG)8T | 52 | UBC846 | (CA)8RT | 50 |
| UBC818 | (CA)8G | 52 | UBC849 | (GT)8YA | 52 |
| UBC827 | (AC)8G | 53 | UBC853 | (TC)8RA | 51 |
| UBC836 | (AG)8YA | 51 | UBC856 | (AC)8YA | 50 |
| UBC842 | (AG)8G | 53 | UBC859 | (CG)8RC | 51 |
| UBC844 | (CT)8RC | 52 | UBC865 | (CCG)6 | 58 |
| UBC845 | (CT)8RG | 52 | UBC868 | (GAA)6 | 47 |
4.4. Statistical Analyses
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Li, A.; Ge, S. Genetic variation and conservation of Changnienia amoena, an endangered orchid endemic to China. Plant Syst. Evol. 2006, 258, 251–260. [Google Scholar] [CrossRef]
- Luo, Y.B.; Jia, J.S.; Wang, C.L. A general review of the conservation status of chinese orchids. Biodivers. Sci. 2003, 11, 70–77. [Google Scholar]
- Chen, S.C.; Tsi, Z.H. The Orchid of China; Chinese Forestry Publisher: Beijing, China, 1998. [Google Scholar]
- Trapnell, D.W.; Hamrick, J.L. Mating patterns and gene flow in the neotropical epiphytic orchid, Laelia rubescens. Mol. Ecol. 2005, 14, 75–84. [Google Scholar]
- Sharma, I.K.; Jones, D.L.; French, C.J. Unusually high genetic variability revealed through allozymic polymorphism of an endemic and endangered australian orchid, Pterostylis aff. picta (Orchidaceae). Biochem. Syst. Ecol. 2003, 31, 513–526. [Google Scholar] [CrossRef]
- Brzosko, E.; Wróblewska, A.; Tałałaj, I.; Wasilewska, E. Genetic diversity of Cypripedium Calceolus in Poland. Plant Syst. Evol. 2011, 295, 83–96. [Google Scholar] [CrossRef]
- Wallace, L.E. Examining the effects of fragmentation on genetic variation in Platanthera leucophaea (Orchidaceae): Inferences from allozyme and random amplified polymorphic DNA markers. Plant Species Biol. 2002, 17, 37–49. [Google Scholar]
- Brzosko, E.; Wróblewska, A. Genetic diversity of nectar—Rewarding Platanthera chlorantha and nectarless Cephalanthera rubra. Bot. J. Linn. Soc. 2013, 171, 751–763. [Google Scholar] [CrossRef]
- Swarts, N.D.; Dixon, K.W. Terrestrial orchid conservation in the age of extinction. Ann. Bot. 2009, 104, 543–556. [Google Scholar] [CrossRef]
- Chen, S.C.; Tsi, Z.H.; Luo, Y.B. Native Orchids of China in Colour; Science Press: Beijing, China, 1999; p. 137. [Google Scholar]
- Sun, H.Q.; Cheng, J.; Zhang, F.M.; Luo, Y.B.; Ge, S. Reproductive success of non-rewarding Cypripedium japonicum benefits from low spatial dispersion pattern and asynchronous flowering. Ann. Bot. 2009, 103, 1227–1237. [Google Scholar] [CrossRef]
- Subject Database of China Plant. Available online: http://www.plant.csdb.cn/protectlist (accessed on 16 April 2014).
- Suetsugu, K.; Fukushima, S. Pollination biology of the endangered orchid Cypripedium japonicum in a fragmented forest of Japan. Plant Spec. Biol. [Online early access]. Available online: http://onlinelibrary.wiley.com/doi/10.1111/1442-1984.12016/abstract (accessed on 6 May 2013). [CrossRef]
- Chung, J.M.; Park, K.W.; Park, C.S.; Lee, S.H.; Chung, M.G.; Chung, M.Y. Contrasting levels of genetic diversity between the historically rare orchid Cypripedium japonicum and the historically common orchid Cypripedium macranthos in South Korea. Bot. J. Linn. Soc. 2009, 160, 119–129. [Google Scholar]
- Qian, X.; Li, Q.J.; Lian, J.J.; Wang, C.X.; Tian, M. Genetic diversity of endangered wild Cypripedium japonicum populations: An AFLP analysis. Chin. J. Ecol. 2013, 32, 1445–1450. [Google Scholar]
- Rodrigues, L.; van den Berg, C.; Póvoa, O.; Monteiro, A. Low genetic diversity and significant structuring in the endangered Mentha cervina populations and its implications for conservation. Biochem. Syst. Ecol. 2013, 50, 51–61. [Google Scholar] [CrossRef]
- Zietkiewicz, E.; Rafalski, A.; Labuda, D. Genome fingerprinting by simple sequence repeat (SSR)-floated polymerase chain reaction amplification. Genomics 1994, 20, 176–183. [Google Scholar]
- Wu, C.J.; Cheng, Z.Q.; Huang, X.Q.; Yin, S.-H.; Cao, K.M.; Sun, C.-R. Genetic diversity among and within populations of Oryza granulata from yunnan of China revealed by RAPD and ISSR markers: Implications for conservation of the endangered species. Plant Sci. 2004, 167, 35–42. [Google Scholar] [CrossRef]
- Yang, W.; de Oliveira, A.C.; Godwin, I.; Schertz, K.; Bennetzen, J.L. Comparison of DNA marker technologies in characterizing plant genome diversity: Variability in Chinese sorghums. Crop Sci. 1996, 36, 1669–1676. [Google Scholar]
- Bornet, B.; Branchard, M. Nonfloated inter simple sequence repeat (ISSR) markers: Reproducible and specific tools for genome fingerprinting. Plant Mol. Biol. Rep. 2001, 19, 209–215. [Google Scholar] [CrossRef]
- Farsani, T.M.; Etemadi, N.; Sayed-Tabatabaei, B.E.; Talebi, M. Assessment of genetic diversity of Bermudagrass (Cynodon dactylon) using ISSR markers. Int. J. Mol. Sci. 2012, 13, 383–392. [Google Scholar]
- Yang, H.Q.; An, M.Y.; Gu, Z.J.; Tian, B. Genetic diversity and differentiation of Dendrocalamus membranaceus (poaceae: Bambusoideae), a declining bamboo species in yunnan, China, as based on inter-simple sequence repeat (ISSR) analysis. Int. J. Mol. Sci. 2012, 13, 4446–4457. [Google Scholar] [CrossRef]
- Golkar, P.; Arzani, A.; Rezaei, A.M. Genetic variation in safflower (Carthamus tinctorious L.) for seed quality-related traits and inter-simple sequence repeat (ISSR) markers. Int. J. Mol. Sci. 2011, 12, 2664–2677. [Google Scholar] [CrossRef]
- Liu, J.F.; Shi, S.Q.; Chang, E.M.; Yang, W.J.; Jiang, Z.P. Genetic diversity of the critically endangered Thuja sutchuenensis revealed by ISSR markers and the implications for conservation. Int. J. Mol. Sci. 2013, 14, 14860–14871. [Google Scholar]
- Wei, L.; Wu, X.J. Genetic variation and population differentiation in a medical herb Houttuynia cordata in China revealed by inter-simple sequence repeats (ISSRs). Int. J. Mol. Sci. 2012, 13, 8159–8170. [Google Scholar] [CrossRef]
- Yang, Q.; Fu, Y.; Wang, Y.Q.; Wang, Y.; Zhang, W.H.; Li, X.Y.; Zhang, J. Genetic diversity and differentiation in the critically endangered orchid (Amitostigma hemipilioides): Implications for conservation. Plant Syst. Evol. 2014, 300, 871–879. [Google Scholar] [CrossRef]
- Qian, X.; Wang, C.; Tian, M. Genetic diversity and population differentiation of Calanthe tsoongiana, a rare and endemic orchid in China. Int. J. Mol. Sci. 2013, 14, 20399–20413. [Google Scholar] [CrossRef]
- Da Cruz, D.T.; Selbach-Schnadelbach, A.; Lambert, S.M.; Ribeiro, P.L.; Borba, E.L. Genetic and morphological variability in Cattleya elongata Barb. Rodr.(Orchidaceae), endemic to the campo rupestre vegetation in northeastern Brazil. Plant Syst. Evol. 2011, 294, 87–98. [Google Scholar] [CrossRef]
- Yao, X.H.; Gao, L.; Yang, B. Genetic diversity of wild Cymbidium goeringii (Orchidaceae) populations from Hubei based on inter-simple sequence repeats analysis. Front. Biol. China 2007, 2, 419–424. [Google Scholar]
- George, S.; Sharma, J.; Yadon, V.L. Genetic diversity of the endangered and narrow endemic Piperia yadonii (Orchidaceae) assessed with ISSR polymorphisms. Am. J. Bot. 2009, 96, 2022–2030. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Godt, M.J.W. Effects of life history traits on genetic diversity in plant species. Phil. Trans. Roy. Soc. B 1996, 351, 1291–1298. [Google Scholar] [CrossRef]
- Lin, L.; Hu, Z.Y.; Ni, S.; Li, J.Y.; Qiu, Y.X. Genetic diversity of Camellia japonica (Theaceae), a species endangered to East Asia, detected by inter-simple sequence repeat (ISSR). Biochem. Syst. Ecol. 2013, 50, 199–206. [Google Scholar] [CrossRef]
- Guo, W.; Jeong, J.; Kim, Z.; Wang, R.; Kim, E.; Kim, S. Genetic diversity of Lilium tsingtauense in China and Korea revealed by ISSR markers and morphological characters. Biochem. Syst. Ecol. 2011, 39, 352–360. [Google Scholar] [CrossRef]
- Culley, T.M.; Sbita, S.J.; Wick, A. Population genetic effects of urban habitat fragmentation in the perennial herb Viola pubescens (Violaceae) using ISSR markers. Ann. Bot. 2007, 100, 91–100. [Google Scholar] [CrossRef]
- Liu, W.S.; Wei, W.; Dong, M. Clonal and genetic diversity of Carex moorcroftii on the Qinghai-Tibet plateau. Biochem. Syst. Ecol. 2009, 37, 370–377. [Google Scholar] [CrossRef]
- Li, A.; Ge, S. Genetic variation and clonal diversity of Psammochloa villosa (Poaceae) detected by ISSR markers. Ann. Bot. 2001, 87, 585–590. [Google Scholar] [CrossRef]
- Fajardo, C.G.; de Almeida Vieira, F.; Molina, W.F. Interspecific genetic analysis of orchids in Brazil using molecular markers. Plant Syst. Evol. 2014. [Google Scholar] [CrossRef]
- Ma, J.M.; Yin, S.H. Genetic diversity of Dendrobium fimbriatum (Orchidaceae), an endangered species, detected by inter-simple sequence repeat (ISSR). Acta Bot. Yunnanica 2009, 31, 35–41. [Google Scholar]
- Wu, H.F.; Li, Z.Z.; Huang, H.W. Genetic differentiation among natural populations of Gastrodia elata (Orchidaceae) in Hubei and germplasm assessment of the cultivated populations. Biodivers. Sci. 2006, 14, 315–326. [Google Scholar] [CrossRef]
- Barbosa, A.R.; Silva-Pereira, V.; Borba, E.L. High genetic variability in self-incompatible myophilous Octomeria (Orchidaceae, Pleurothallidinae) species. Braz. J. Bot. 2013, 36, 179–187. [Google Scholar]
- Huang, J.L.; Li, S.Y.; Hu, H. ISSR and SRAP markers reveal genetic diversity and population structure of an endangered slipper orchid, Paphiopedilum micranthum (Orchidaceae). Plant Divers. Resour. 2014, 36, 209–218. [Google Scholar]
- Wallace, L.E. A comparison of genetic variation and structure in the allopolyploid Platanthera huronensis and its diploid progenitors, Platanthera aquilonis and Platanthera dilatata. Can. J. Bot. 2004, 82, 244–252. [Google Scholar] [CrossRef]
- Smith, J.L.; Hunter, K.L.; Hunter, R.B. Genetic variation in the terrestrial orchid Tipularia discolor. Southeast. Nat. 2002, 1, 17–26. [Google Scholar] [CrossRef]
- Shefferson, R.P.; Weiss, M.; Kull, T.; Taylor, D.L. High specificity generally characterizes mycorrhizal association in rare lady’s slipper orchids, genus Cypripedium. Mol. Ecol. 2005, 14, 613–626. [Google Scholar]
- Jacquemyn, H.; Vandepitte, K.; Brys, R.; Honnay, O.; Roldán-Ruiz, I. Fitness variation and genetic diversity in small, remnant populations of the food deceptive orchid Orchis purpurea. Biol. Conserv. 2007, 139, 203–210. [Google Scholar] [CrossRef]
- Izawa, T.; Kawahara, T.; Takahashi, H. Genetic diversity of an endangered plant, Cypripediummacranthos var. rebunense (Orchidaceae): Background genetic research for future conservation. Conserv. Genet. 2007, 8, 1369–1376. [Google Scholar]
- Brzosko, E.; Wróblewska, A.; Ratkiewicz, M.; Till-Bottraud, I.; Nicole, F.; Baranowska, U. Genetic diversity of Cypripedium calceolus at the edge and in the centre of its range in Europe. Ann. Bot. Fenn. 2009, 46, 201–214. [Google Scholar] [CrossRef]
- Chung, M.Y.; Park, C.W.; Chung, M.G. Extremely low levels of allozyme variation in southern Korean populations of the two rare and endangered lithophytic or epiphytic Bulbophyllum drymoglossum and Sarcanthus scolopendrifolius (Orchidaceae): Implications for conservation. Biodivers. Conserv. 2007, 16, 775–786. [Google Scholar] [CrossRef]
- Qiu, Y.X.; Fu, C.X.; Comes, H.P. Plant molecular phylogeography in China and adjacent regions: Tracing the genetic imprints of quaternary climate and environmental change in the world’s most diverse temperate flora. Mol. Phylogenet. Evol. 2011, 59, 225–244. [Google Scholar] [CrossRef]
- Wallace, L.E.; Case, M.A. Contrasting allozyme diversity between northern and southern populations of Cypripedium parviflorum (Orchidaceae): Implications for Pleistocene refugia and taxonomic boundaries. Syst. Bot. 2000, 25, 281–296. [Google Scholar] [CrossRef]
- Kennedy, A.H.; Walker, G.L. The population genetic structure of the showy lady’s slipper orchid (Cypripedium reginae Walter) in its glaciated and unglaciated ranges. Castanea 2007, 72, 248–261. [Google Scholar] [CrossRef]
- Yeh, F.; Yang, R.; Boyle, T. POPGENE version 1.32. In Microsoft Window-Based Freeware for Population Genetic Analysis; Molecular Biology and Biotechnology Center, University of Alberta: Edmonton, AB, Canada, 1999. [Google Scholar]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef]
- Lewinton, R.C. The apportionment of human diversity. Evol. Biol. 1972, 6, 381–398. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987; pp. 187–192. [Google Scholar]
- Rohlf, F.J. NTSYSpc, Numerical Taxonomy and Multivariate Analysis SystemVersion 2.2 User Guide; Exeter Software: Setauket, NY, USA, 2005. [Google Scholar]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer. Res. 1967, 27, 209–220. [Google Scholar]
- Miller, M.P. Tools for Population Genetic Analyses (TFPGA) Version 1.3: A Windows Program for the Analysis of Allozyme and Molecular Population Genetic Data; Utah State University: Logan, UT, USA, 1997. [Google Scholar]
- Miller, M.P.; Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure from multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar]
- Earl, D.A.; von Holdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Qian, X.; Li, Q.-J.; Liu, F.; Gong, M.-J.; Wang, C.-X.; Tian, M. Conservation Genetics of an Endangered Lady’s Slipper Orchid: Cypripedium japonicum in China. Int. J. Mol. Sci. 2014, 15, 11578-11596. https://doi.org/10.3390/ijms150711578
Qian X, Li Q-J, Liu F, Gong M-J, Wang C-X, Tian M. Conservation Genetics of an Endangered Lady’s Slipper Orchid: Cypripedium japonicum in China. International Journal of Molecular Sciences. 2014; 15(7):11578-11596. https://doi.org/10.3390/ijms150711578
Chicago/Turabian StyleQian, Xin, Quan-Jian Li, Fen Liu, Mao-Jiang Gong, Cai-Xia Wang, and Min Tian. 2014. "Conservation Genetics of an Endangered Lady’s Slipper Orchid: Cypripedium japonicum in China" International Journal of Molecular Sciences 15, no. 7: 11578-11596. https://doi.org/10.3390/ijms150711578
APA StyleQian, X., Li, Q.-J., Liu, F., Gong, M.-J., Wang, C.-X., & Tian, M. (2014). Conservation Genetics of an Endangered Lady’s Slipper Orchid: Cypripedium japonicum in China. International Journal of Molecular Sciences, 15(7), 11578-11596. https://doi.org/10.3390/ijms150711578




