MicroRNA-103 Promotes Colorectal Cancer by Targeting Tumor Suppressor DICER and PTEN
Abstract
:1. Introduction
2. Results and Discussion
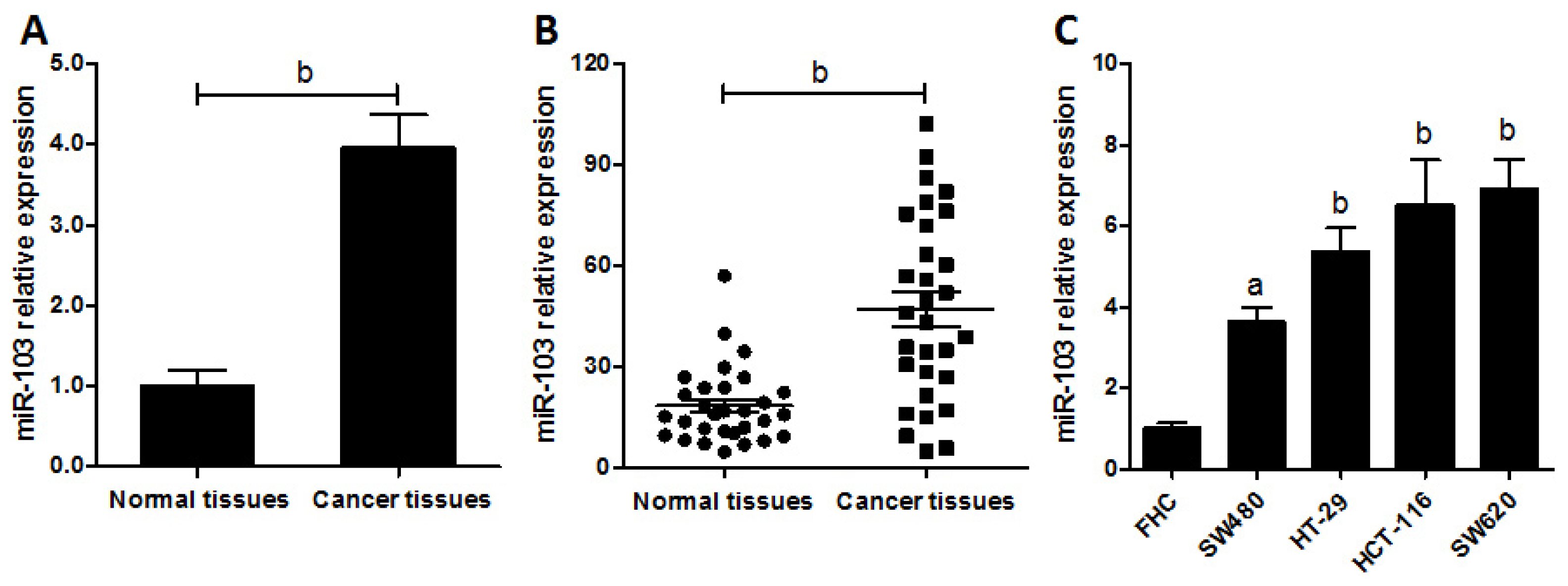
2.1. miR-103 Is Up-Regulated in Colorectal Cancer Patients
2.2. miR-103 Directly Promotes Colorectal Carcinoma Cell Proliferation and Migration
2.3. DICER and PTEN Are Direct Target Genes of miR-103
2.4. miR-103 Promotes Cell Proliferation and Migration by Repressing DICER and PTEN
2.5. Repressed miR-103 Expression Inhibits Cancer Cell Growth in Vivo
3. Experimental Section
3.1. Materials
3.2. Colorectal Carcinoma Patients and Tissue Specimens
3.3. Cell Culture and Transfection
3.4. Cell Growth Assay
3.5. Colony Formation Assay
3.6. RNA Isolation and Real-Time PCR (Polymerase Chain Reaction)
3.7. Western Blot Assay
3.8. Vector Construction and Luciferase Reporter Assays
3.9. Cell Cycle Analysis
3.10. Cell Migration and Invasion Assay
3.11. Colorectal Carcinoma Cell Xenografts Growth
3.12. Statistical Analysis
4. Conclusions
Acknowledgments
Conflicts of Interest
- Author ContributionsL.G. and X.-D.F. conceived and designed the experiments; L.G. B.S. B.G. and Z.W. performed the experiments; C.Q. and X.-D.F. analyzed the data; L.G. F.W. and X.-D.F. wrote the paper; all the authors have read and approved the final manuscript to be published.
References
- Jemal, A.; Ward, E.; Hao, Y.; Thun, M. Trends in the leading causes of death in the United States, 1970–2002. JAMA 2005, 294, 1255–1259. [Google Scholar]
- Jafri, S.H.; Mills, G. Lifestyle modification in colorectal cancer patients: An integrative oncology approach. Future Oncol 2013, 9, 207–218. [Google Scholar]
- Bjerrum, J.T.; Nielsen, O.H.; Wang, Y.L.; Olsen, J. Technology insight: Metabonomics in gastroenterology-basic principles and potential clinical applications. Nat. Clin. Pr. Gastroenterol. Hepatol 2008, 5, 332–343. [Google Scholar]
- Astin, M.; Griffin, T.; Neal, R.D.; Rose, P.; Hamilton, W. The diagnostic value of symptoms for colorectal cancer in primary care: A systematic review. Br. J. Gen. Pr 2011, 61, e231–e243. [Google Scholar]
- Hristova, N.R.; Tagscherer, K.E.; Fassl, A.; Kopitz, J.; Roth, W. Notch1-dependent regulation of p27 determines cell fate in colorectal cancer. Int. J. Oncol 2013, 43, 1967–1975. [Google Scholar]
- Sung, M.K.; Yeon, J.Y.; Park, S.Y.; Park, J.H.; Choi, M.S. Obesity-induced metabolic stresses in breast and colon cancer. Ann. N. Y. Acad. Sci 2011, 1229, 61–68. [Google Scholar]
- Coghlin, C.; Murray, G.I. Progress in the identification of plasma biomarkers of colorectal cancer. Proteomics 2013, 13, 2227–2228. [Google Scholar]
- Chen, K.; Rajewsky, N. The evolution of gene regulation by transcription factors and microRNAs. Nat. Rev. Genet 2007, 8, 93–103. [Google Scholar]
- Du, T.; Zamore, P.D. Beginning to understand microRNA function. Cell Res 2007, 17, 661–663. [Google Scholar]
- Cao, J.; Shen, Y.; Zhu, L.; Xu, Y.; Zhou, Y.; Wu, Z.; Li, Y.; Yan, X.; Zhu, X. miR-129-3p controls cilia assembly by regulating CP110 and actin dynamics. Nat. Cell Biol 2012, 14, 697–706. [Google Scholar]
- Nakasa, T.; Miyaki, S.; Okubo, A.; Hashimoto, M.; Nishida, K.; Ochi, M.; Asahara, H. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum 2008, 58, 1284–1292. [Google Scholar]
- Chan, E.K.; Satoh, M.; Pauley, K.M. Contrast in aberrant microRNA expression in systemic lupus erythematosus and rheumatoid arthritis: Is microRNA-146 all we need? Arthritis Rheum 2009, 60, 912–915. [Google Scholar]
- Chen, C.Z. MicroRNAs as oncogenes and tumor suppressors. N. Engl. J. Med 2005, 353, 1768–1771. [Google Scholar]
- Hammond, S.M. MicroRNAs as oncogenes. Curr. Opin. Genet. Dev 2006, 16, 4–9. [Google Scholar]
- Zhang, B.; Chen, H.; Zhang, L.; Dakhova, O.; Zhang, Y.; Lewis, M.T.; Creighton, C.J.; Ittmann, M.M.; Xin, L. A dosage-dependent pleiotropic role of Dicer in prostate cancer growth and metastasis. Oncogene 2013. [Google Scholar] [CrossRef]
- Tamura, M.; Gu, J.; Matsumoto, K.; Aota, S.; Parsons, R.; Yamada, K.M. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science 1998, 280, 1614–1617. [Google Scholar]
- Maiques, O.; Santacana, M.; Valls, J.; Pallares, J.; Mirantes, C.; Gatius, S.; Garcia Dios, D.A.; Amant, F.; Pedersen, H.C.; Dolcet, X.; et al. Optimal protocol for PTEN immunostaining; role of analytical and preanalytical variables in PTEN staining in normal and neoplastic endometrial, breast, and prostatic tissues. Hum. Pathol 2014, 45, 522–532. [Google Scholar]
- Nyakern, M.; Cappellini, A.; Mantovani, I.; Martelli, A.M. Synergistic induction of apoptosis in human leukemia T cells by the Akt inhibitor perifosine and etoposide through activation of intrinsic and Fas-mediated extrinsic cell death pathways. Mol. Cancer Ther 2006, 5, 1559–1570. [Google Scholar]
- Zhang, L.; Liu, X.; Jin, H.; Guo, X.; Xia, L.; Chen, Z.; Bai, M.; Liu, J.; Shang, X.; Wu, K.; et al. miR-206 inhibits gastric cancer proliferation in part by repressing cyclinD2. Cancer Lett 2013, 332, 94–101. [Google Scholar]
- Wang, X.; Yue, P.; Kim, Y.A.; Fu, H.; Khuri, F.R.; Sun, S.Y. Enhancing mammalian target of rapamycin (mTOR)-targeted cancer therapy by preventing mTOR/raptor inhibition-initiated, mTOR/rictor-independent Akt activation. Cancer Res 2008, 68, 7409–7418. [Google Scholar]
- Xu, C.X.; Li, Y.; Yue, P.; Owonikoko, T.K.; Ramalingam, S.S.; Khuri, F.R.; Sun, S.Y. The combination of RAD001 and NVP-BEZ235 exerts synergistic anticancer activity against non-small cell lung cancer in vitro and in vivo. PLoS One 2011, 6, e20899. [Google Scholar]
- Xue, X.; Cao, A.T.; Cao, X.; Yao, S.; Carlson, E.D.; Soong, L.; Liu, C.G.; Liu, X.; Liu, Z.; Duck, L.W.; et al. Downregulation of microRNA-107 in intestinal CD11c, myeloid cells in response to microbiota and proinflammatory cytokines increases IL-23p19 expression. Eur. J. Immunol 2014, 44, 673–682. [Google Scholar]
- Bjaanaes, M.M.; Halvorsen, R.; Solberg, S.; Jorgensen, L.; Dragani, T.A.; Galvan, A.; Colombo, F.; Anderlini, M.; Pastorino, U.; Kure, E.; et al. Unique microRNA-profiles in EGFR-mutated lung adenocarcinomas. Int. J. Cancer 2014. [Google Scholar] [CrossRef]
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011, 474, 649–653. [Google Scholar]
- Ellis, K.L.; Cameron, V.A.; Troughton, R.W.; Frampton, C.M.; Ellmers, L.J.; Richards, A.M. Circulating microRNAs as candidate markers to distinguish heart failure in breathless patients. Eur. J. Heart Fail 2013, 15, 1138–1147. [Google Scholar]
- Yu, D.; Zhou, H.; Xun, Q.; Xu, X.; Ling, J.; Hu, Y. MicroRNA-103 regulates the growth and invasion of endometrial cancer cells through the downregulation of tissue inhibitor of metalloproteinase 3. Oncol. Lett 2012, 3, 1221–1226. [Google Scholar]
- Chen, H.Y.; Lin, Y.M.; Chung, H.C.; Lang, Y.D.; Lin, C.J.; Huang, J.; Wang, W.C.; Lin, F.M.; Chen, Z.; Huang, H.D.; et al. miR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Cancer Res 2012, 72, 3631–3641. [Google Scholar]
- Hong, Z.; Feng, Z.; Sai, Z.; Tao, S. PER3, a novel target of miR-103, plays a suppressive role in colorectal cancer in vitro. BMB Rep. 2014. pii 2514. [Google Scholar]
- Zhu, J.; Chen, L.; Zou, L.; Yang, P.; Wu, R.; Mao, Y.; Zhou, H.; Li, R.; Wang, K.; Wang, W.; et al. miR-20b, -21, and -130b inhibit PTEN expression resulting in B7-H1 over-expression in advanced colorectal cancer. Hum. Immunol 2014, 75, 348–353. [Google Scholar]





© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Geng, L.; Sun, B.; Gao, B.; Wang, Z.; Quan, C.; Wei, F.; Fang, X.-D. MicroRNA-103 Promotes Colorectal Cancer by Targeting Tumor Suppressor DICER and PTEN. Int. J. Mol. Sci. 2014, 15, 8458-8472. https://doi.org/10.3390/ijms15058458
Geng L, Sun B, Gao B, Wang Z, Quan C, Wei F, Fang X-D. MicroRNA-103 Promotes Colorectal Cancer by Targeting Tumor Suppressor DICER and PTEN. International Journal of Molecular Sciences. 2014; 15(5):8458-8472. https://doi.org/10.3390/ijms15058458
Chicago/Turabian StyleGeng, Li, Bing Sun, Bo Gao, Zheng Wang, Cheng Quan, Feng Wei, and Xue-Dong Fang. 2014. "MicroRNA-103 Promotes Colorectal Cancer by Targeting Tumor Suppressor DICER and PTEN" International Journal of Molecular Sciences 15, no. 5: 8458-8472. https://doi.org/10.3390/ijms15058458
APA StyleGeng, L., Sun, B., Gao, B., Wang, Z., Quan, C., Wei, F., & Fang, X.-D. (2014). MicroRNA-103 Promotes Colorectal Cancer by Targeting Tumor Suppressor DICER and PTEN. International Journal of Molecular Sciences, 15(5), 8458-8472. https://doi.org/10.3390/ijms15058458



