Microarray-Based Gene Expression Profiling to Elucidate Effectiveness of Fermented Codonopsis lanceolata in Mice
Abstract
:1. Introduction
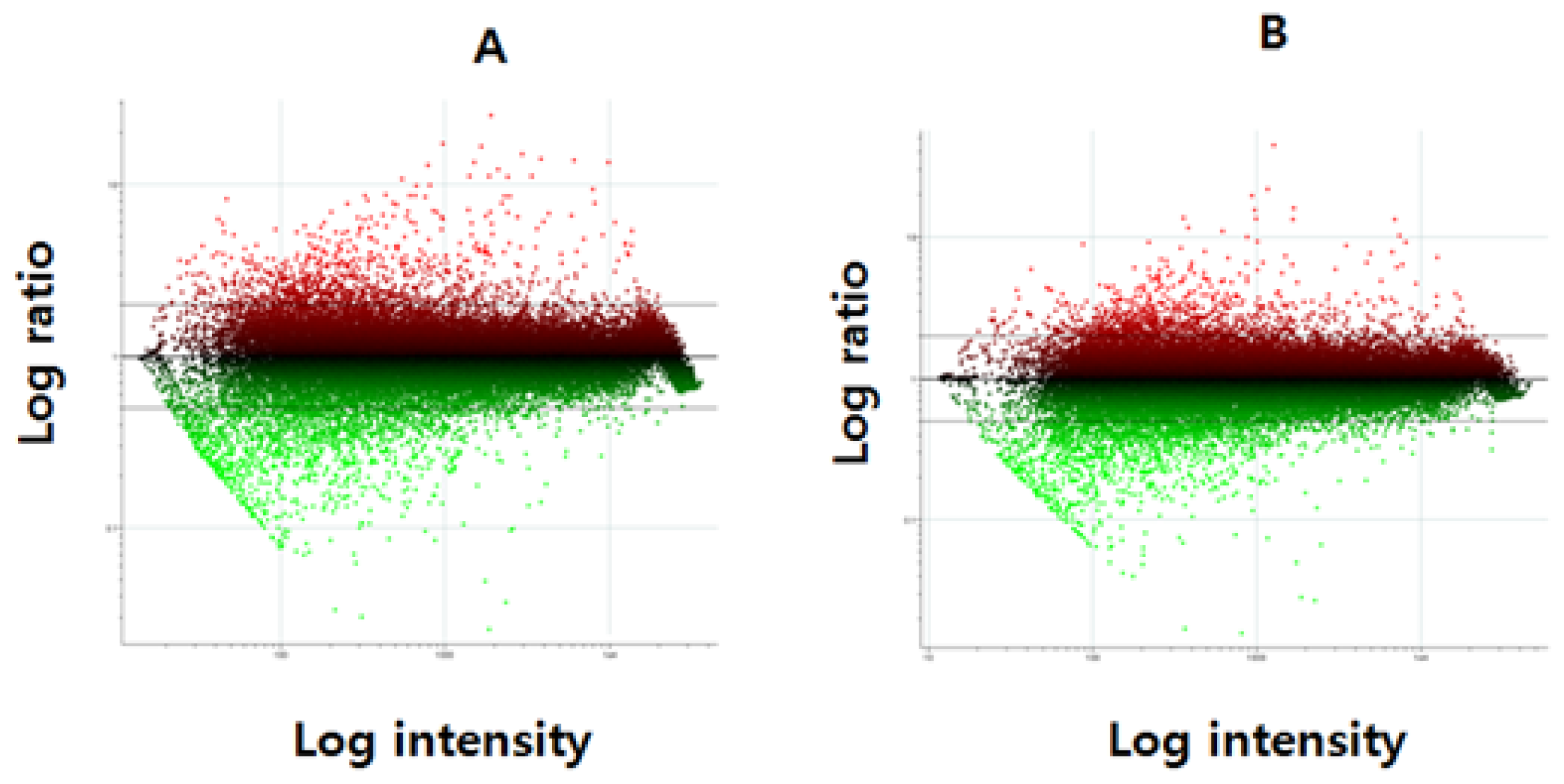
2. Results and Discussion
3. Experimental Section
3.1. Sample Preparation
3.2. Animals and Feeding Protocols
3.3. RNA Extraction and Purification
3.4. Oligonucleotide Microarray Assay
3.5. Microarray Data Analysis
3.6. Statistical Analysis
4. Conclusions
Conflicts of Interest
References
- Mitchell, M.; Armstrong, D.T.; Robker, R.L.; Norman, R.J. Adipokines: Implications for female fertility and obesity. Reproduction 2005, 130, 583–597. [Google Scholar]
- Kunitomi, M.; Wada, J.; Takahashi, K.; Tsuchiyama, Y.; Mimura, Y.; Hida, K.; Miyatake, N.; Fujii, M.; Kira, S.; Shikata, K.; et al. Relationship between reduced serum IGF-I levels and accumulation of visceral fat in Japanese men. Int. J. Obes. Relat. Metab. Disord 2002, 26, 361–369. [Google Scholar]
- Fujita, H.; Fujishima, H.; Koshimura, J.; Hosoba, M.; Yoshioka, N.; Shimotomai, T.; Morii, T.; Narita, T.; Kakei, M.; Ito, S. Effects of antidiabetic treatment with metformin and insulin on serum and adipose tissue adiponectin levels in db/db mice. Endocr. J 2005, 52, 427–433. [Google Scholar]
- Maeda, N.; Shimomura, I.; Kishida, K.; Nishizawa, H.; Matsuda, M.; Nagaretani, H.; Furuyama, N.; Kondo, H.; Takahashi, M.; Arita, Y.; et al. Diet-induced insulin resistance in mice lacking adiponectin/ACRP30. Nat. Med 2002, 8, 731–737. [Google Scholar]
- Schena, M.; Shalon, D.; Davis, R.W.; Brown, P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995, 270, 467–470. [Google Scholar]
- Blanchard, A.P.; Kaiser, R.J.; Hood, L.E. High-density oligonucleotide arrays. Biosensors Bioelectron 1996, 11, 687–690. [Google Scholar]
- Boyer, J.; Allen, W.L.; McLean, E.G.; Wilsor, P.M.; McCulla, A.; Moore, S.; Longley, D.B.; Cadas, C.; Johnston, P.G. Phamacogenomic identification of novel determinants of reponse to chemotheraphy in colon cancer. Cancer Res 2006, 66, 2765–2777. [Google Scholar]
- Kim, S.Y.; Kim, H.S.; Kim, S.H.; Kim, H.S.; Su, I.S.; Chung, S.Y. Effect of the feeding Platycodon grandiflorum and Codonopsis lancedata on the fatty acid composition of serum and liver in rats. J. Korean Soc. Food Nutr 1993, 22, 524–530. [Google Scholar]
- Park, S.J.; Park, D.S.; Kim, S.S.; He, X.; Ahn, J.H.; Yoon, W.B.; Lee, H.Y. The effect of fermented codonopsis lanceolata on the memory impairment of mice. J. Korean Soc. Food Nutr 2010, 39, 1691–1694. [Google Scholar]
- Lee, K.T.; Choi, J.; Jung, W.T.; Nam, J.H.; Jung, H.J.; Park, H.J. Structure of a new echinocystic acid bisdesmoside isolated from Codonopsis lanceolata roots and the cytotoxic activity of prosapogenins. J. Agric. Food Chem 2002, 50, 4190–4193. [Google Scholar]
- Lee, K.W.; Jung, H.J.; Park, H.J.; Kim, D.G.; Lee, J.Y.; Lee, K.T. Beta-d-xylopyranosyl-(1-->3)-beta-d-glucuronopyranosyl echinocystic acid isolated from the roots of Codonopsis lanceolata induces caspase-dependent apoptosis in human acute promyelocytic leukemia HL-60 cells. Biol. Pharm. Bull 2005, 28, 854–859. [Google Scholar]
- He, Z.; Zhou, J. Empirical evaluation of a new method for calculating signal-to-noise ratio for microarray data analysis. Appl. Environ. Microbiol 2008, 74, 2957–2966. [Google Scholar]
- Sergeev, N.; Volokhov, D.; Chizhikov, V.; Rasooly, A. Simultaneous analysis of multiple staphylococcal enterotoxin genes by an oligonucleotide microarray assay. J. Clin. Microbiol 1998, 42, 2134–2143. [Google Scholar]
- He, Z.; Wu, L.; Li, X.; Fields, M.W.; Zhou, J. Empirical establishment of oligonucleotide probe design criteria. Appl. Environ. Microbiol 2005, 71, 3753–3760. [Google Scholar]
- Pfaffl, M.W.; Meyer, H.H.D.; Sauerwein, H. Quantification of the insulin like growth factor-1 mRNA: Development and validation of an internally standardized competitive reverse transcription-polymerase chain reaction. Exp. Clin. Endocrinol. Diabetes 1998, 106, 502–512. [Google Scholar]
- The Gene Ontology Consortium. Available online: http://www.geneontology.org/index.shtml (accessed on 11 January 2013).
- BioCarta. Available online: http://www.biocarta.com (accessed on 10 January 2013).
- GenMAPP. Available online: http://www.genmapp.org (accessed on 24 January 2013).
- DAVID. Available online: http://david.abcc.ncifcrf.gov (accessed on 2 February 2013).
- Medline Databases. Available online: http://www.ncbi.nlm.nih.gov (accessed on 3 February 2013).

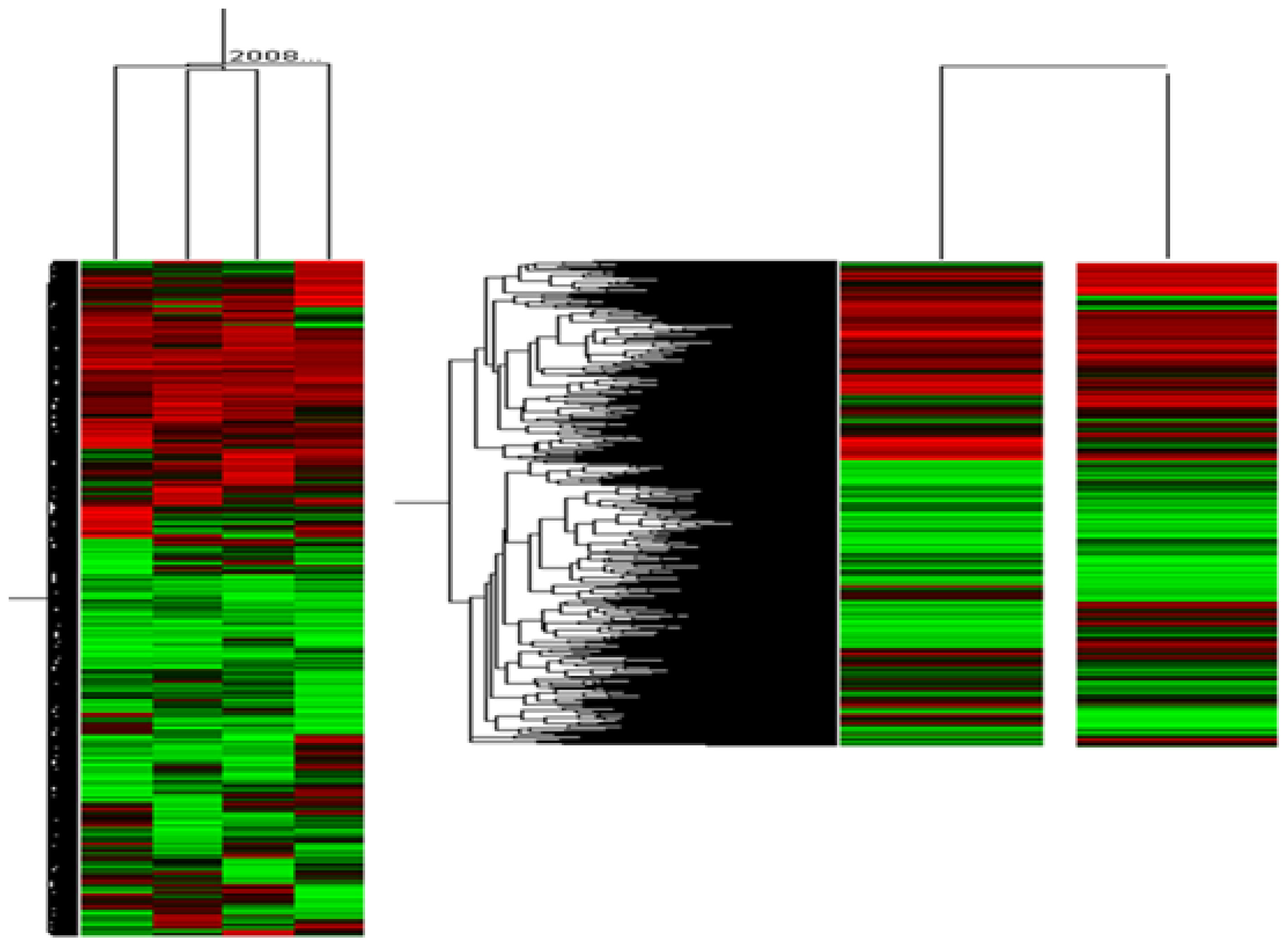
| Up-regulated gene (>2.0-fold) | Down-regulated gene (<0.5-fold) | ||
|---|---|---|---|
| Name | Fold | Name | Fold |
| Cytochrome P450, family 4, subfamily a. polypeptide 14 (Cyp4a14) | 14.28 ± 0.39 | Thyroid hormone responsive SPOT14 homolog (Rattus) | 0.11 ± 0.40 |
| Deleted in malignant brain tumors 1 (Dmbt1) | 12.47 ± 0.13 | Fatty acid syhthase | 0.18 ± 0.27 |
| Homolog to homo sapiens Cytochrome P450 4A11 precursor | 10.24 ± 0.90 | Transcribed locus | 0.21 ± 0.26 |
| PREDICTED: hypothetical protein LOC76487 [Mus musculus] | 8.74 ± 0.72 | Farnesyl diphosphate synthetase (Fdps) | 0.22 ± 0.49 |
| Metallothionein 2 | 8.42 ± 0.50 | Mid1 interacting protein 1 (gastrulation specific G12-like (zebrafish)) | 0.24 ± 0.44 |
| Deleted in malignant brain tumors 1 | 8.01 ± 0.45 | Sterol regulatory element binding protein 1 (Srebp1) | 0.24 ± 0.39 |
| Cytochrome P450, family 4, subfamily a. polypeptide 10 | 7.91 ± 0.44 | NDA(P) dependent steroid dehydrog | 0.25 ± 1.47 |
| Metallothionein 1 | 7.41 ± 0.96 | Transmembrane inner ear (Tmie) | 0.28 ± 0.84 |
| RIKEN cDNA A930041I02 gene (A930041I02Rik) | 7.12 ± 0.50 | Amino carboxymuconate semialdehyde decarboxylase (Acmsd) | 0.31 ± 0.47 |
| Metallothionein 2 | 6.82 ± 0.28 | ||
© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Choi, W.Y.; Kim, J.S.; Park, S.J.; Ma, C.J.; Lee, H.Y. Microarray-Based Gene Expression Profiling to Elucidate Effectiveness of Fermented Codonopsis lanceolata in Mice. Int. J. Mol. Sci. 2014, 15, 5907-5915. https://doi.org/10.3390/ijms15045907
Choi WY, Kim JS, Park SJ, Ma CJ, Lee HY. Microarray-Based Gene Expression Profiling to Elucidate Effectiveness of Fermented Codonopsis lanceolata in Mice. International Journal of Molecular Sciences. 2014; 15(4):5907-5915. https://doi.org/10.3390/ijms15045907
Chicago/Turabian StyleChoi, Woon Yong, Ji Seon Kim, Sung Jin Park, Choong Je Ma, and Hyeon Yong Lee. 2014. "Microarray-Based Gene Expression Profiling to Elucidate Effectiveness of Fermented Codonopsis lanceolata in Mice" International Journal of Molecular Sciences 15, no. 4: 5907-5915. https://doi.org/10.3390/ijms15045907
APA StyleChoi, W. Y., Kim, J. S., Park, S. J., Ma, C. J., & Lee, H. Y. (2014). Microarray-Based Gene Expression Profiling to Elucidate Effectiveness of Fermented Codonopsis lanceolata in Mice. International Journal of Molecular Sciences, 15(4), 5907-5915. https://doi.org/10.3390/ijms15045907




