Effect of Hydrogen Peroxide on the Biosynthesis of Heme and Proteins: Potential Implications for the Partitioning of Glu-tRNAGlu between These Pathways
Abstract
:1. Introduction
2. Results and Discussion
2.1. Hydrogen Peroxide Inactivates Glutamyl-tRNA Synthetase 1 (GluRS1) and Reduces Heme Levels in A. ferrooxidans

2.2. Glutamyl-tRNA Reductase (GluTR) from A. ferrooxidans Is Inactivated by Hydrogen Peroxide
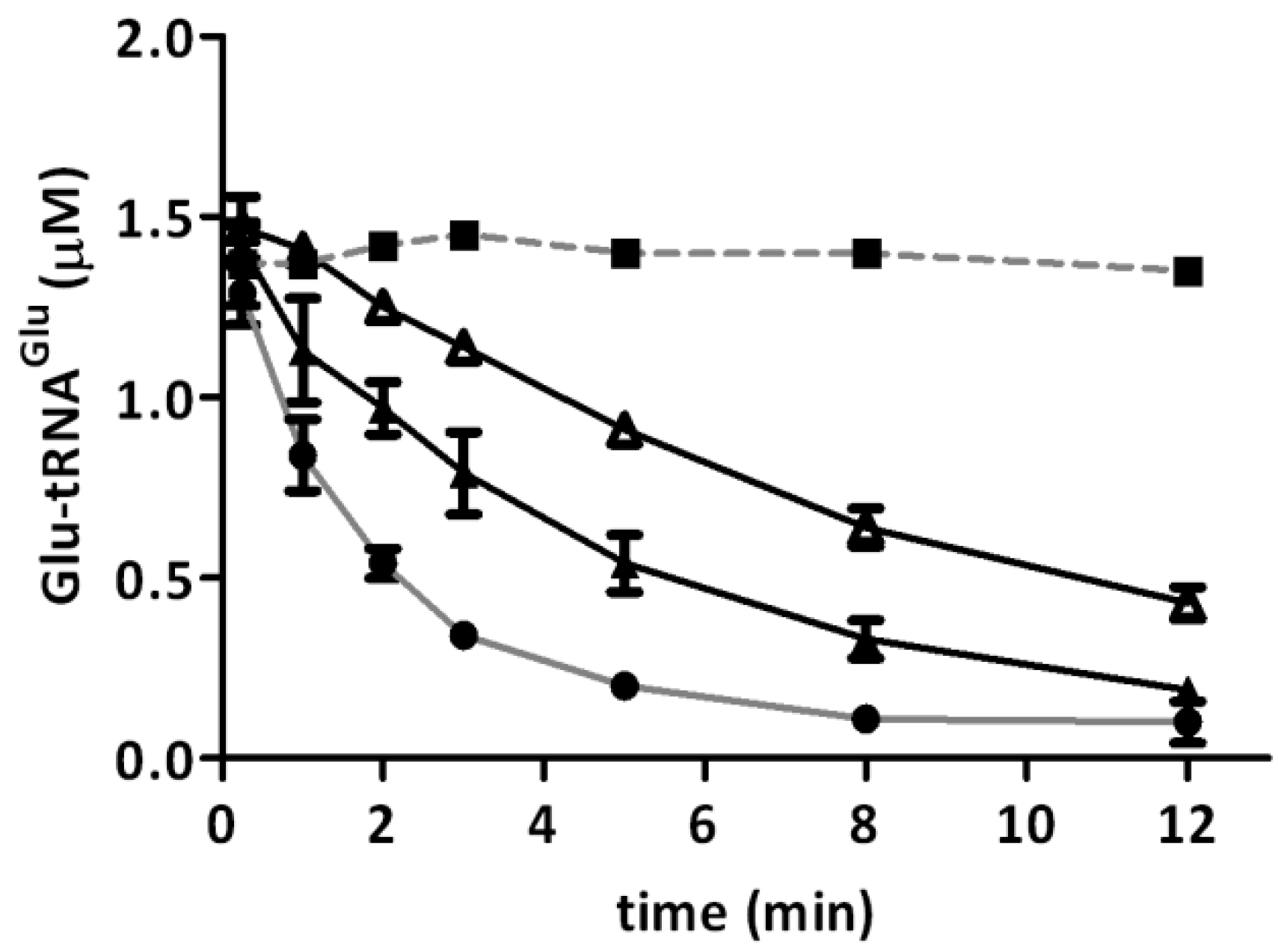
| Type of GluTR (H2O2) | GluTR (1/4) 250 µM H2O2 | GluTR (1/4) 500 µM H2O2 | GluTR (1/12) 500 µM H2O2 |
|---|---|---|---|
| kobs (s−1) | 3.18 × 10−4 ± (4.43 × 10−5) | 6.96 × 10−4 ± (4.49 × 10−5) | 4.82 × 10−4 ± (2.29 × 10−5) |
2.3. Hydrogen Peroxide Decreases the Stimulation of GluTR by Glutamate Semialdehyde 1-2 Aminomutase (GSAM)
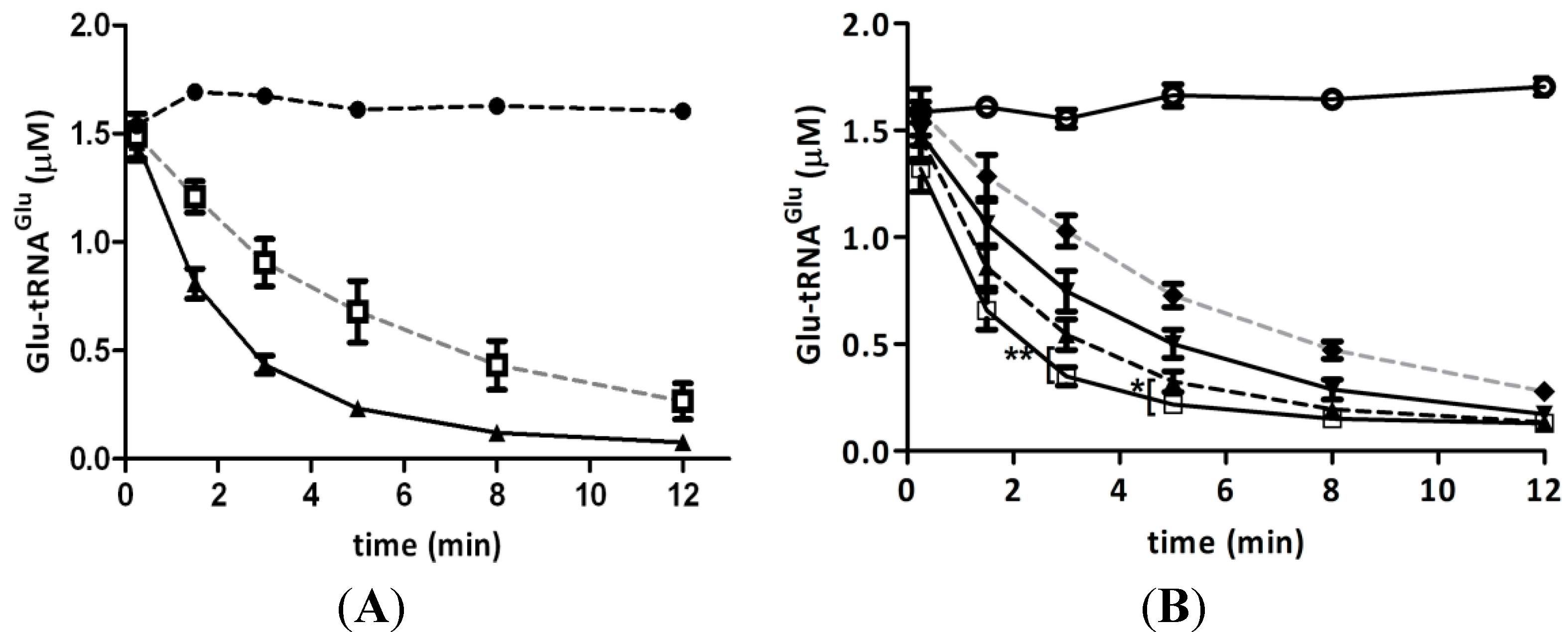
2.4. Elongation Factor Tu (EF-Tu) Is not Affected by Treatment with H2O2
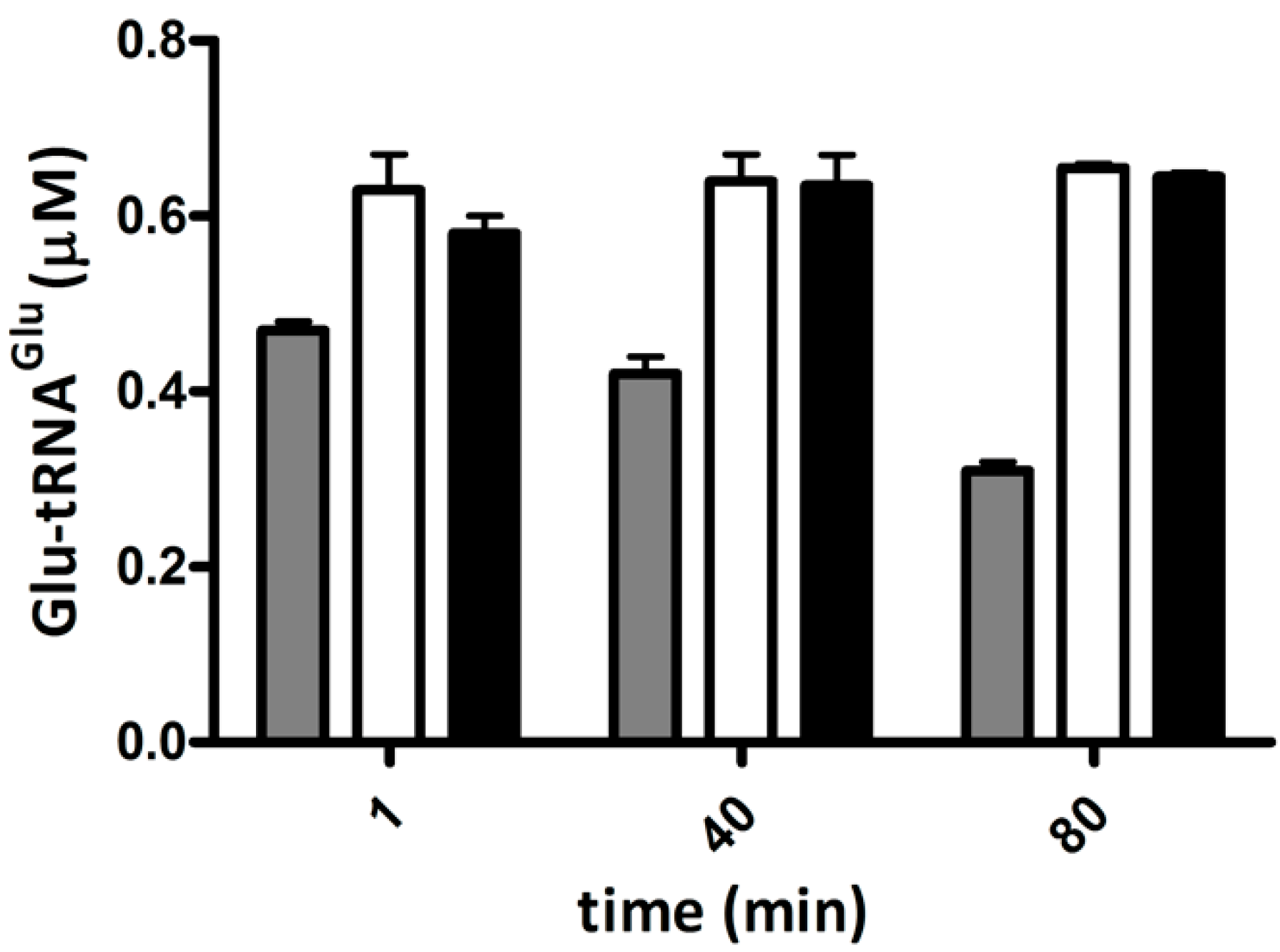
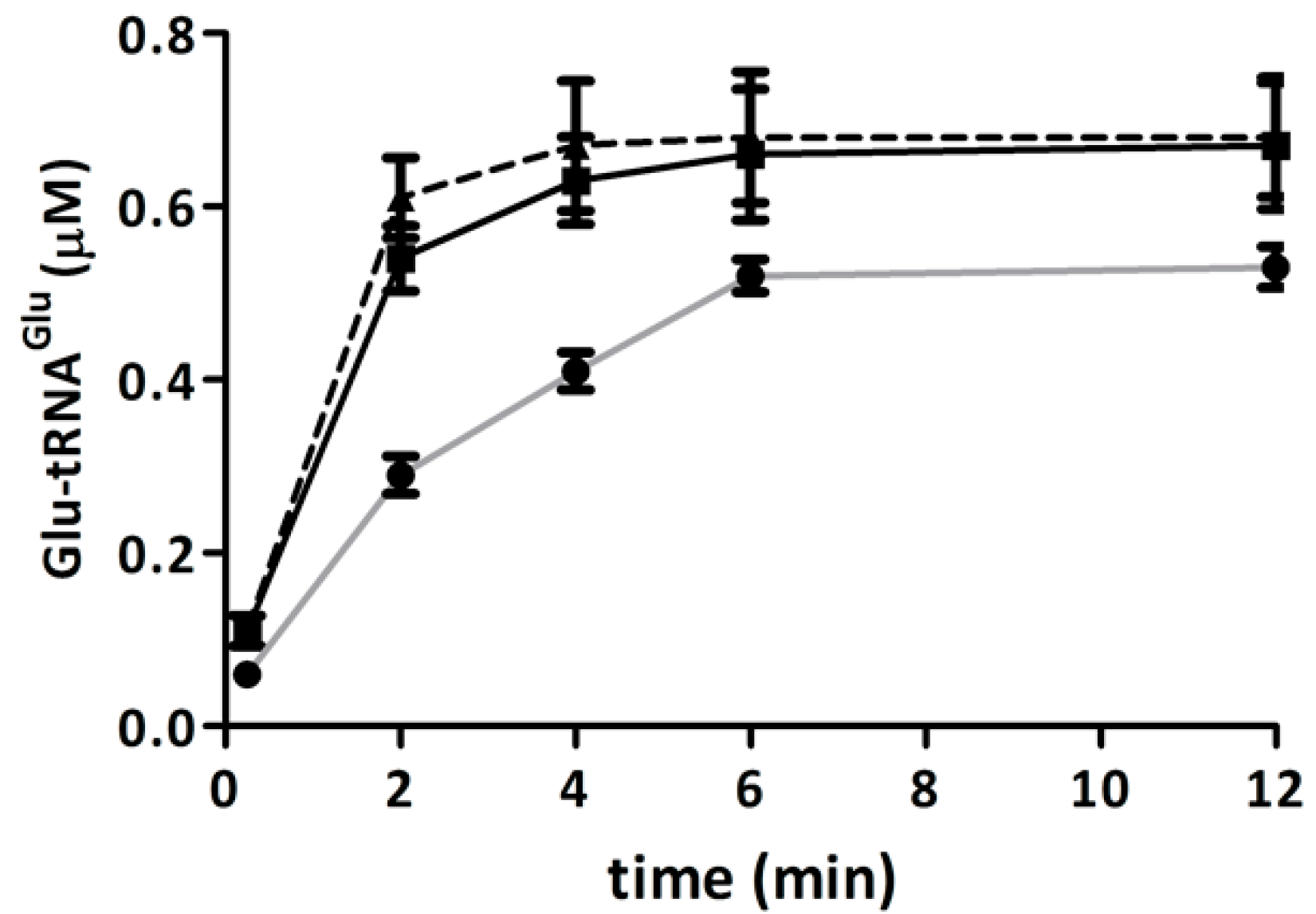
2.5. Competition of GluTR and EF-Tu for Glu-tRNAGlu Is Affected by H2O2
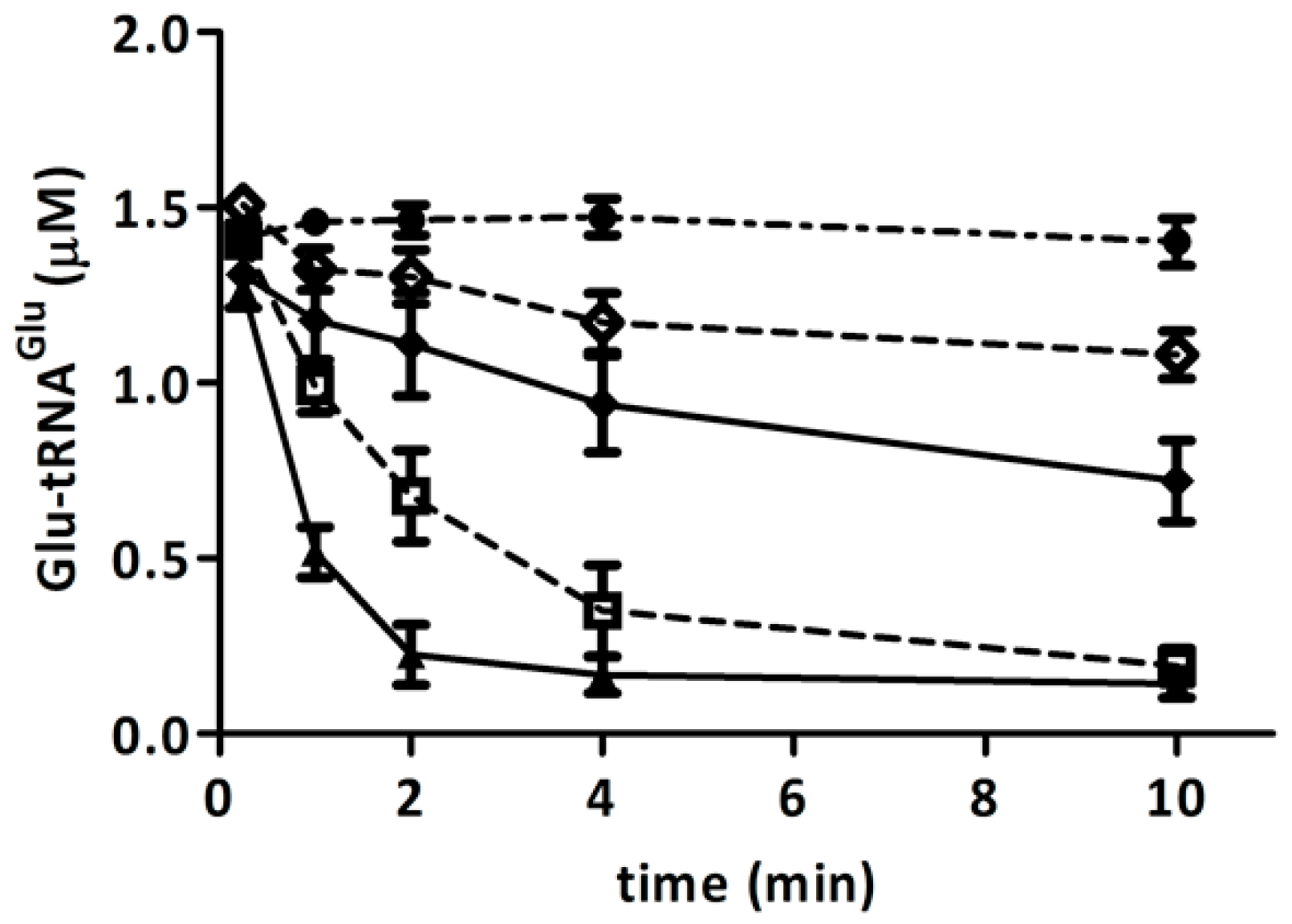
2.6. Discussion
3. Experimental Section
3.1. Bacterial Strains and Culture Conditions
3.2. Overproduction and Purification of Proteins
3.3. Preparation and Purification of tRNA Transcripts
3.4. Preparation of A. ferrooxidans Cellular Extracts
3.5. GluRS Activity
3.6. GluTR Activity
3.7. Binding of Glutamyl-tRNAGlu to EF-Tu
3.8. Protein Oxidation
3.9. Heme Measurement
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Heinemann, I.U.; Jahn, M.; Jahn, D. The biochemistry of heme biosynthesis. Arch. Biochem. Biophys. 2008, 474, 238–251. [Google Scholar] [CrossRef]
- Panek, H.; O’Brian, M.R. A whole genome view of prokaryotic haem biosynthesis. Microbiology 2002, 148, 2273–2282. [Google Scholar] [PubMed]
- Sadrzsdeh, S.; Graf, E.; Panter, S.; Hallaway, P.E.; Eaton, J.W. Hemoglobin: Abiologic fenton reagent. J. Biol. Chem. 1984, 259, 14354–14356. [Google Scholar]
- Pazos, M.; Andersen, M.; Skibsted, L.H. Heme-mediated production of free radicals via preformed lipid hydroperoxide fragmentation. J. Agric. Food Chem. 2008, 56, 11478–11484. [Google Scholar] [CrossRef] [PubMed]
- Hiraku, Y.; Kawanishi, S. Mechanism of oxidative DNA damage induced by δ-aminolevulinic acid in the presence of copper ion. Cancer Res. 1996, 56, 1786–1793. [Google Scholar] [PubMed]
- Fotinos, N.; Convert, M.; Piffaretti, J.; Gurny, R.; Lange, N. Effects on Gram-negative and Gram-positive bacteria mediated by 5-aminolevulinic acid and 5-aminolevulinic acid derivatives. Antimicrob. Agents Chemother. 2008, 52, 1366–1373. [Google Scholar] [CrossRef] [PubMed]
- Nakayashiki, T.; Nishimura, K.; Tanaka, R.; Inokuchi, H. Partial inhibition of protein synthesis accelerates the synthesis of porphyrin in heme-deficient mutants of Escherichia coli. Mol. Gen. Genet. 1995, 249, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Francklyn, C.S.; Minajigi, A. tRNA as an active chemical scaffold for diverse chemical transformations. FEBS Lett. 2010, 584, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Levicán, G.; Katz, A.; de Armas, M.; Nuñez, H.; Orellana, O. Regulation of a glutamyl-tRNA synthetase by the heme status. Proc. Natl. Acad. Sci. USA 2007, 104, 3135–3140. [Google Scholar] [CrossRef] [PubMed]
- Yarzabal, A.; Brasseur, G.; Bonnefoy, V. Cytochromes c of Acidithiobacillus ferrooxidans. FEMS Microbiol. Lett. 2002, 209, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Salazar, J.C.; Ahel, I.; Orellana, O.; Tumbula-Hansen, D.; Krieger, R.; Daniels, L.; Söll, D. Coevolution of an aminoacyl-tRNA synthetase with its tRNA substrates. Proc. Natl. Acad. Sci. USA 2003, 100, 13863–13868. [Google Scholar] [CrossRef] [PubMed]
- Levicán, G.; Katz, A.; Valenzuela, P.; Soll, D.; Orellana, O. A tRNAGlu that uncouples protein and tetrapyrrole biosynthesis. FEBS Lett. 2005, 579, 6383–6387. [Google Scholar] [CrossRef] [PubMed]
- Katz, A.; Banerjee, R.; de Armas, M.; Ibba, M; Orellana, O. Redox status affects the catalytic activity of glutamyl-tRNA synthetase. Biochem. Biophys. Res. Commun. 2010, 398, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Brathwaite, O.; Cosloy, S; Russell, C.S. 5-Aminolevulinic acid synthesis in Escherichia coli. J. Bacteriol. 1989, 171, 2547–2552. [Google Scholar] [PubMed]
- Kang, Z.; Wang, Y.; Gu, P.; Wang, Q.; Qi, Q. Engineering Escherichia coli for efficient production of 5-aminolevulinicacid from glucose. Metab. Eng. 2011, 13, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Schobert, M.; Jahn, D. Regulation of heme biosynthesis in non-phototrophic bacteria. J. Mol. Microbiol. Biotechnol. 2002, 4, 287–294. [Google Scholar] [PubMed]
- De Armas, M.; Levicán, G.; Katz, A.; Moser, J.; Jahn, D.; Orellana, O. Cellular levels of heme affect the activity of dimeric glutamyl-tRNA reductase. Biochem. Biophys. Res. Commun. 2011, 405, 134–139. [Google Scholar] [CrossRef] [PubMed]
- Luer, C.; Schauer, S.; Mobius, K.; Schulze, J.; Schubert, W.; Heinz, D.W.; Jahn, D.; Moser, J. Complex Formation between glutamyl-tRNA reductase and glutamate-1-semialdehyde 2,1-aminomutase in Escherichia coli during the initial reactions of porphyrin biosynthesis. J. Biol. Chem. 2005, 280, 18568–18572. [Google Scholar] [CrossRef] [PubMed]
- Nogaj, L.A.; Beale, S.I. Physical and kinetic interactions between glutamyl-tRNA reductase and glutamate-1-semialdehyde aminotransferase of Chlamydomonas reinhardtii. J. Biol. Chem. 2005, 280, 24301–24307. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Perona, J.; Ryu, K.; Francklyn, C.; Hou, Y. Distinct kinetic mechanisms of the two classes of aminoacyl-tRNA synthetases. J. Mol. Biol. 2006, 361, 300–311. [Google Scholar] [CrossRef] [PubMed]
- Kaspy, I.; Rotem, E.; Weiss, N.; Ronin, I.; Balaban, N.Q.; Glaser, G. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat. Commun. 2013, 4, 3001. [Google Scholar] [CrossRef] [PubMed]
- Aarti, D.; Tanaka, R.; Ito, H.; Tanaka, A. High light inhibits chlorophyll biosynthesis at the level of 5-aminolevulinate synthesis during de-etiolation in cucumber (Cucumis sativus) cotyledons. Photochem. Photobiol. 2007, 83, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Elgrably-Weiss, M.; Park, S.; Schlosser-Silverman, E.; Rosenshine, I.; Imlay, J.; Altuvia, S. A Salmonella enterica serovar typhimurium hemA mutant is highly susceptible to oxidative DNA damage. J. Bacteriol. 2002, 184, 3774–3784. [Google Scholar] [CrossRef] [PubMed]
- Zeller, T.; Moskvin, O.V.; Li, K.; Klug, G.; Gomelsky, M. Transcriptome and physiological responses to hydrogen peroxide of the facultatively phototrophic bacterium Rhodobacter sphaeroides. J. Bacteriol. 2005, 187, 7232–7242. [Google Scholar] [CrossRef] [PubMed]
- Faulkner, M.J.; Ma, Z.; Fuangthong, M.; Helmann, J. Derepression of the Bacillus subtilis PerR peroxide stress response leads to iron deficiency. J. Bacteriol. 2012, 194, 1226–1235. [Google Scholar] [CrossRef] [PubMed]
- Sampson, J.R.; Uhlenbeck, O.C. Biochemical and physical characterization of an unmodified yeast phenylalanine. Proc. Natl. Acad. Sci. USA 1988, 85, 1033–1037. [Google Scholar] [CrossRef] [PubMed]
- Roy, H.; Becker, H.D.; Mazauric, M.; Kern, D. Structural elements defining elongation factor Tu mediated suppression of codon ambiguity. Nucleic Acids Res. 2007, 35, 3420–3430. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.; Ran So, B.; Yadavalli, S.S.; Roy, H.; Shoji, S.; Fredrick, K.; Musier-Forsyth, K.; Ibba, M. Resampling and editing of mischarged tRNA prior to translation elongation. Mol. Cell 2000, 33, 654–660. [Google Scholar] [CrossRef]
- Kuross, S.A.; Rank, B.H.; Hebbel, R.P. Excess heme in sickle erythrocyte inside-out membranes: Possible role in thiol oxidation. Blood 1988, 4, 876–882. [Google Scholar]
- Flora, J.S.; Mittal, M.; Mehta, A. Heavy metal induced oxidative stress & its possible reversal by chelation therapy. Indian J. Med. Res. 2008, 128, 501–523. [Google Scholar] [PubMed]
- </i>Stohs, S.J.; Bagchi, D. Oxidative mechanisms in the toxicity of metal ions. Free Radic. Biol. Med. 1995, 18, 321–336. [Google Scholar] [CrossRef] [PubMed]
- López-Archilla, A.I.; Marin, I.; Amils, R. Microbial community composition and ecology of an acidic aquatic environment: The tinto river, Spain. Microb. Ecol. 2001, 41, 20–35. [Google Scholar] [PubMed]
- Cortés, A.; Flores, R.; Norambuena, J.; Cardenas, J.P.; Quatrini, R.; Orellana, O.; Levicán, G. Comparative study of redox stress response in the acidophilic bacteria Leptospirillum ferriphilum and Acidithiobacillus ferrooxidans. In Biohydrometallurgy, Biotech Key to Unlock Mineral Resources Value; Qiu, G., Jiang, T., Qin, W., Liu, X., Yang, Y., Wang, H., Eds.; Central South University Press: Changsha, China, 2011; pp. 354–357. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farah, C.; Levicán, G.; Ibba, M.; Orellana, O. Effect of Hydrogen Peroxide on the Biosynthesis of Heme and Proteins: Potential Implications for the Partitioning of Glu-tRNAGlu between These Pathways. Int. J. Mol. Sci. 2014, 15, 23011-23023. https://doi.org/10.3390/ijms151223011
Farah C, Levicán G, Ibba M, Orellana O. Effect of Hydrogen Peroxide on the Biosynthesis of Heme and Proteins: Potential Implications for the Partitioning of Glu-tRNAGlu between These Pathways. International Journal of Molecular Sciences. 2014; 15(12):23011-23023. https://doi.org/10.3390/ijms151223011
Chicago/Turabian StyleFarah, Carolina, Gloria Levicán, Michael Ibba, and Omar Orellana. 2014. "Effect of Hydrogen Peroxide on the Biosynthesis of Heme and Proteins: Potential Implications for the Partitioning of Glu-tRNAGlu between These Pathways" International Journal of Molecular Sciences 15, no. 12: 23011-23023. https://doi.org/10.3390/ijms151223011
APA StyleFarah, C., Levicán, G., Ibba, M., & Orellana, O. (2014). Effect of Hydrogen Peroxide on the Biosynthesis of Heme and Proteins: Potential Implications for the Partitioning of Glu-tRNAGlu between These Pathways. International Journal of Molecular Sciences, 15(12), 23011-23023. https://doi.org/10.3390/ijms151223011




