Identification of a Plastid-Localized Bifunctional Nerolidol/Linalool Synthase in Relation to Linalool Biosynthesis in Young Grape Berries
Abstract
:1. Introduction
2. Results and Discussion
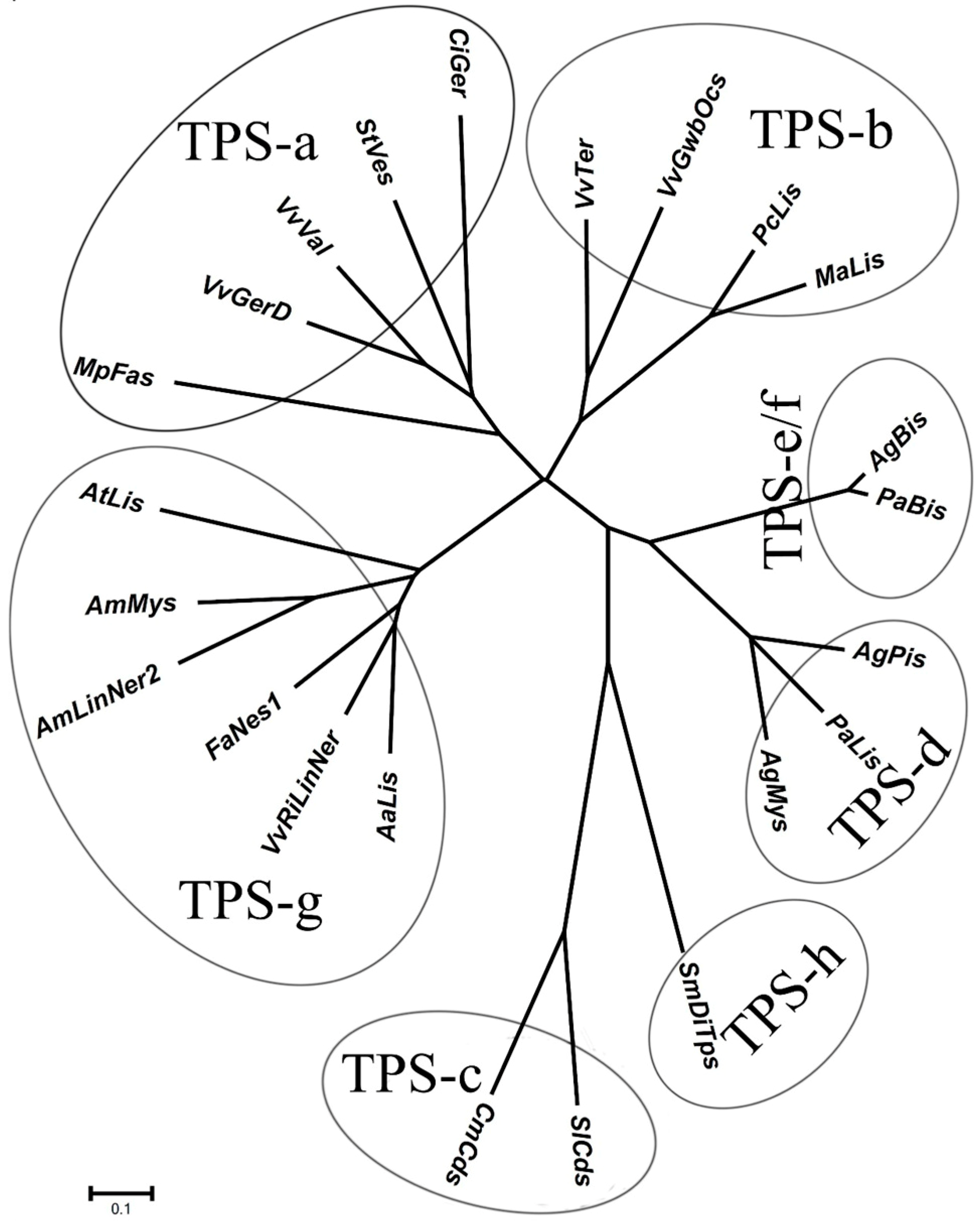
2.1. Identification of Gene Encoding 3S-Linalool Synthase from Grape Berries
| GenBank Number | Gene ID (12×) | VvTPS Gene Model † | Cloned Genes | Note ‡ |
|---|---|---|---|---|
| XM002270140.1 | GSVIVT01005272001 | VvTPS56 | VvRiLinNer (JQ062931, this study) VvPNLinNer2 (HM807392) VvCSLinNer (HM807393) | Full |
| XM002270071.1 | GSVIVT01005271001 | VvTPS65 | No gene has been cloned | Stop in first exon |
| XM002266813.1 | GSVIVT01005221001 | VvTPS54 | VvPNLinNer1(HM807391) | Full |

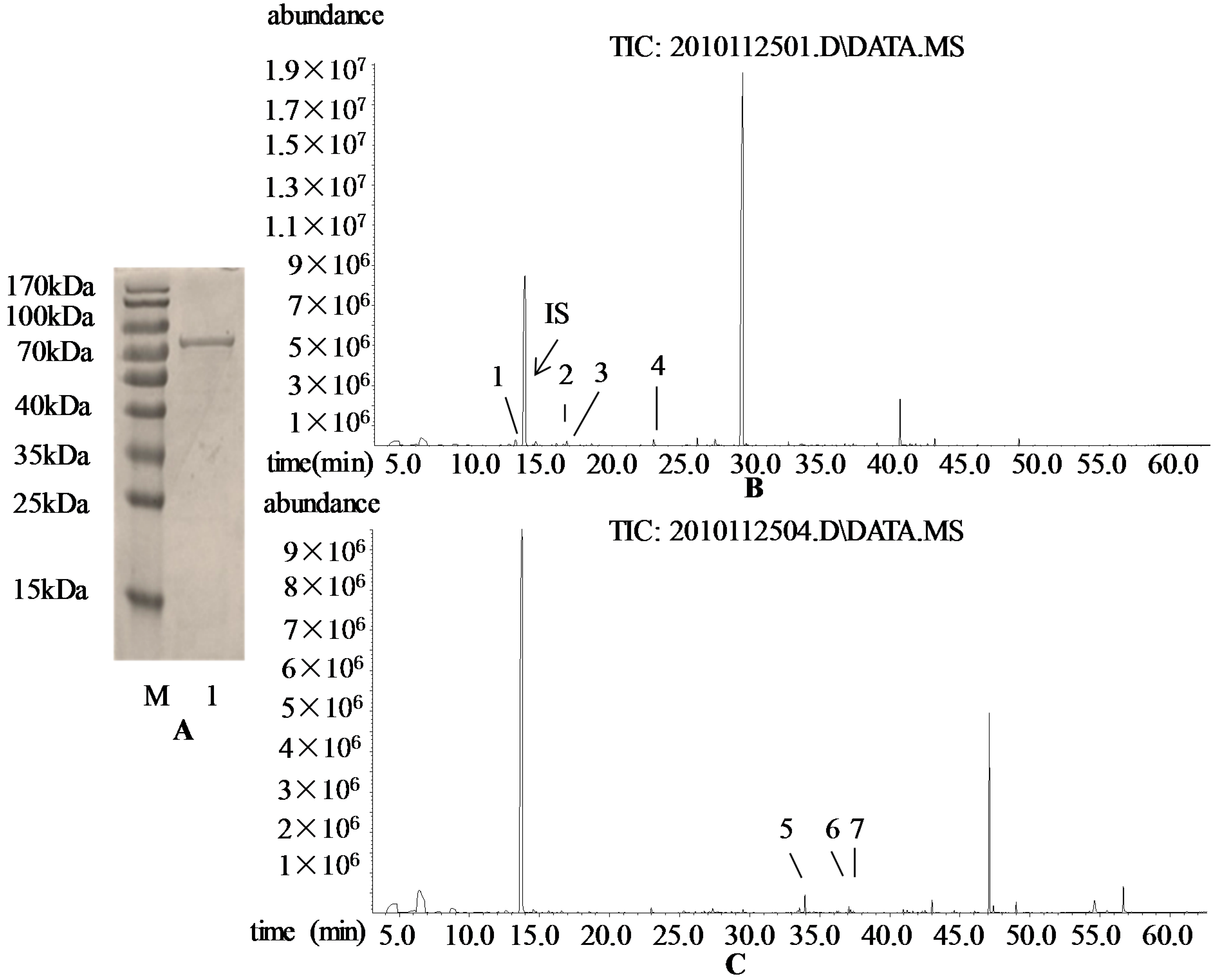
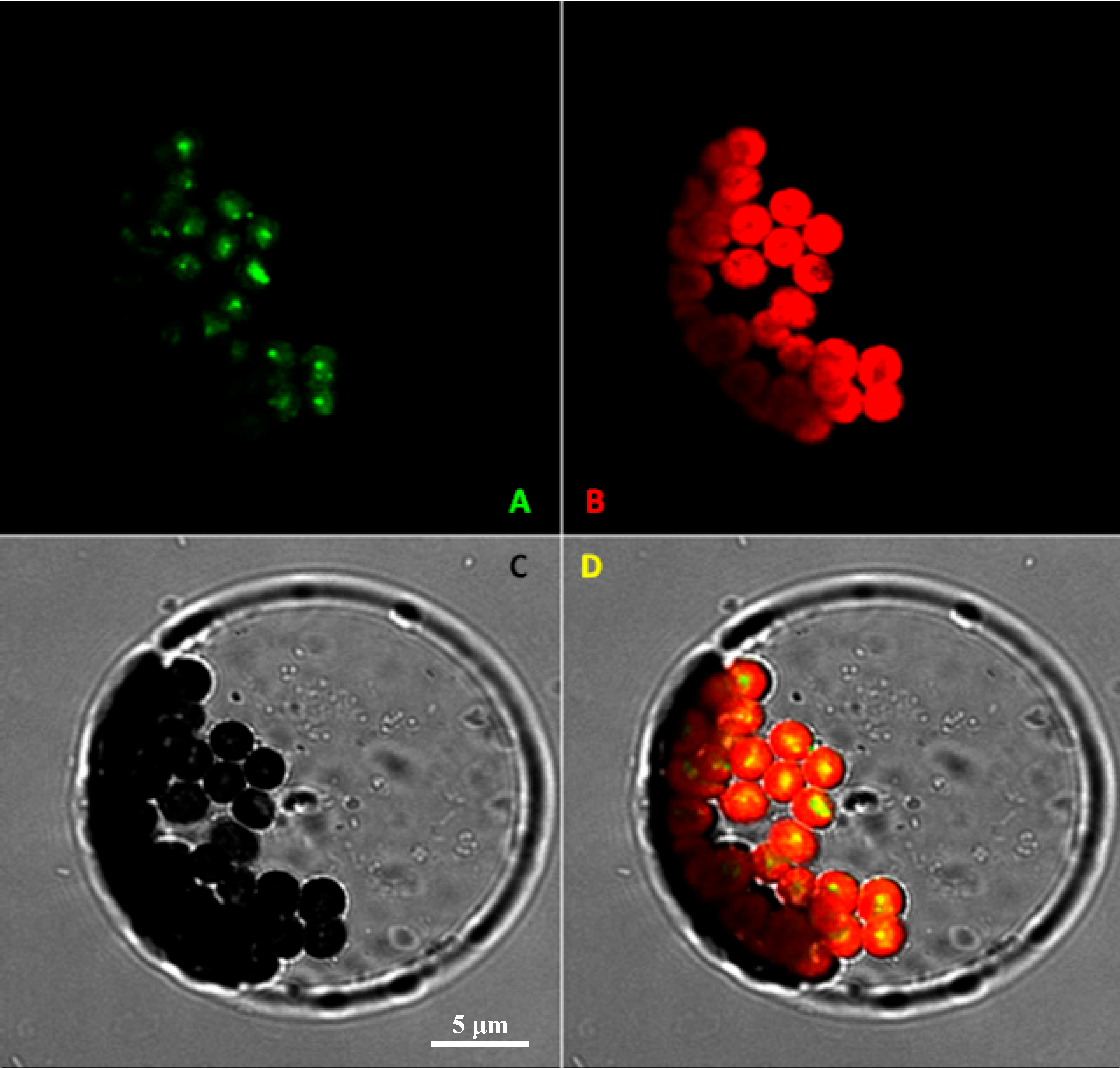
2.2. Subcellular Localization of VvRiLinNer Protein
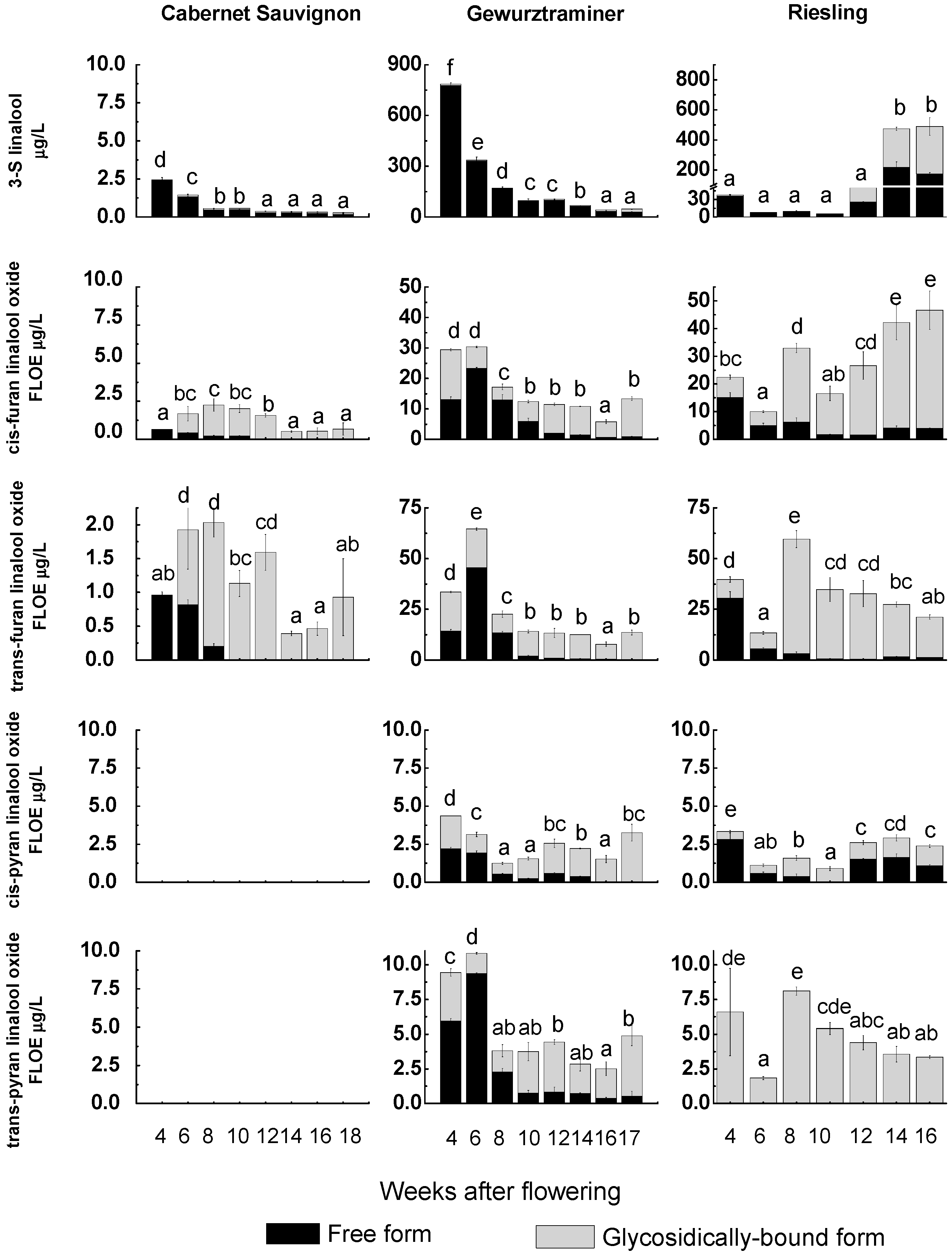
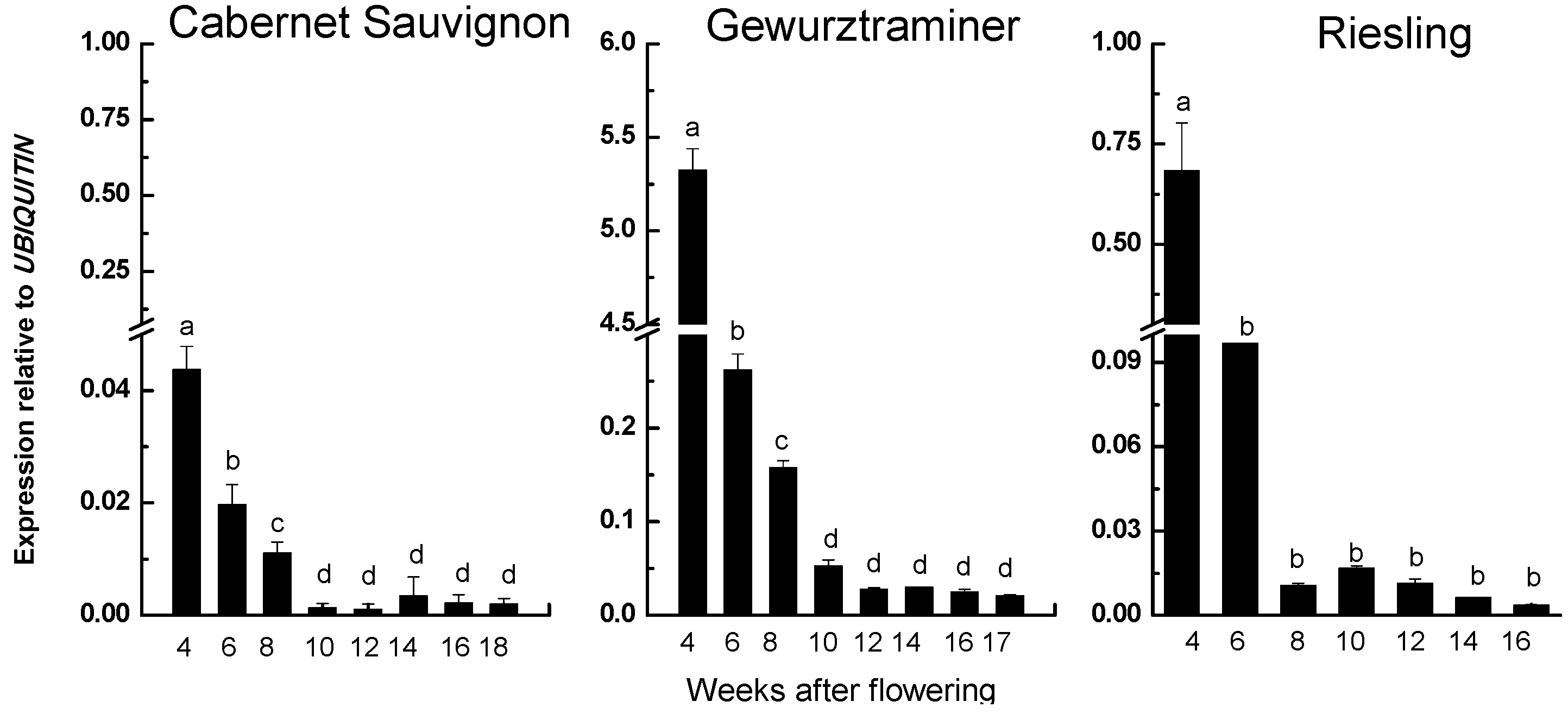
2.3. Correspondence Relationship between VvRiLinNer Expression and Linalool Accumulation in Three Grape Cultivars
3. Experimental Section
3.1. Plant Materials
3.2. Total RNA Extraction, Purification and cDNA Synthesis
3.3. Isolation of Linalool Synthase (VvRiLin) Gene from Vitis vinifera Riesling Berries
3.4. Prokaryotic Expression of VvRiLin and Purification of Recombinant Protein
3.5. Catalytic Activity of the Purified VvRiLinNer Protein
3.6. Subcellular Localization of VvRiLinNer
3.7. Analysis of VvRiLinNer Expression by Real Time PCR
3.8. Quantification of Linalool and Its Derivatives by GC–MS
3.9. Statistics Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mateo, J.; Jiménez, M. Monoterpenes in grape juice and wines. J. Chromatogr. 2000, 881, 557–567. [Google Scholar] [CrossRef]
- Fenoll, J.; Manso, A.; Hellin, P.; Ruiz, L.; Flores, P. Changes in the aromatic composition of the Vitis vinifera grape muscat hamburg during ripening. Food Chem. 2009, 114, 420–428. [Google Scholar] [CrossRef]
- Caven-Quantrill, D.J.; Buglass, A.J. Seasonal variation of flavour content of English vineyard grapes, determined by stir-bar sorptive extraction-gas chromatography-mass spectrometry. Flavour Fragr. J. 2008, 23, 239–248. [Google Scholar] [CrossRef]
- Duchêne, E.; Legras, J.L.; Karst, F.; Merdinoglu, D.; Claudel, P.; Jaegli, N.; Pelsy, F. Variation of linalool and geraniol content within two pairs of aromatic and non-aromatic grapevine clones. Aust. J. Grape Wine Res. 2009, 15, 120–130. [Google Scholar] [CrossRef]
- Ebeler, S.E. Analytical chemistry: Unlocking the secrets of wine flavor. Food Rev. Int. 2001, 17, 45–64. [Google Scholar] [CrossRef]
- Skinkis, P.A.; Bordelon, B.P.; Wood, K.V. Comparison of monoterpene constituents in Traminette, Gewürztraminer, and Riesling winegrapes. Am. J. Enol. Vitic. 2008, 59, 440–445. [Google Scholar]
- Pichersky, E.; Noel, J.P.; Dudareva, N. Biosynthesis of plant volatiles: Nature’s diversity and ingenuity. Science 2006, 311, 808–811. [Google Scholar] [CrossRef] [PubMed]
- Hemmerlin, A.; Harwood, J.L.; Bach, T.J. A raison d’êtrefor two distinct pathways in the early steps of plant isoprenoid biosynthesis? Prog. Lipid Res. 2012, 51, 95–148. [Google Scholar] [CrossRef] [PubMed]
- Dudareva, N.; Negre, F.; Nagegowda, D.A.; Orlova, I. Plant volatiles: Recent advances and future perspectives. Crit. Rev. Plant Sci. 2006, 25, 417–440. [Google Scholar] [CrossRef]
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011, 66, 212–229. [Google Scholar] [CrossRef] [PubMed]
- Bohlmann, J.; Meyer-Gauen, G.; Croteau, R. Plant terpenoid synthases: Molecular biology and phylogenetic analysis. Proc. Natl. Acad. Sci. USA 1998, 95, 4126–4133. [Google Scholar] [CrossRef] [PubMed]
- Dudareva, N.; Martin, D.; Kish, C.M.; Kolosova, N.; Gorenstein, N.; Fäldt, J.; Miller, B.; Bohlmann, J. (E)-β-Ocimene and myrcene synthase genes of floral scent biosynthesis in snapdragon: Function and expression of three terpene synthase genes of a new terpene synthase subfamily. Plant Cell 2003, 15, 1227–1241. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, A.; Giri, A.P.; Verstappen, F.W.; Bertea, C.M.; Sevenier, R.; Sun, Z.; Jongsma, M.A.; Schwab, W.; Bouwmeester, H.J. Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell 2004, 16, 3110–3131. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.; Aubourg, S.; Schouwey, M.; Daviet, L.; Schalk, M.; Toub, O.; Lund, S.; Bohlmann, J. Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, flcDNA cloning, and enzyme assays. BMC Plant Biol. 2010, 10, 226. [Google Scholar] [CrossRef] [PubMed]
- Nagegowda, D.A.; Gutensohn, M.; Wilkerson, C.G.; Dudareva, N. Two nearly identical terpene synthases catalyze the formation of nerolidol and linalool in snapdragon flowers. Plant J. 2008, 55, 224–239. [Google Scholar] [CrossRef]
- Luan, F.; Wüst, M. Differential incorporation of 1-deoxy-d-xylulose into (3S)-linalool and geraniol in grape berry exocarp and mesocarp. Phytochemistry 2002, 60, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Dudareva, N.; Cseke, L.; Blanc, V.M.; Pichersky, E. Evolution of floral scent in clarkia: Novel patterns of S-linalool synthase gene expression in the c. Breweri flower. Plant Cell 1996, 8, 1137–1148. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Tholl, D.; D’Auria, J.C.; Farooq, A.; Pichersky, E.; Gershenzon, J. Biosynthesis and emission of terpenoid volatiles from arabidopsis flowers. Plant Cell 2003, 15, 481–494. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.S.; Köllner, T.G.; Wiggins, G.; Grant, J.; Degenhardt, J.; Chen, F. Molecular and genomic basis of volatile-mediated indirect defense against insects in rice. Plant J. 2008, 55, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.L.; Lee, Y.R.; Huang, W.K.; Chang, S.T.; Chu, F.H. Characterization of S-(+)-linalool synthase from several provenances of Cinnamomum osmophloeum. Tree Genet. Genomes 2014, 10, 75–86. [Google Scholar] [CrossRef]
- Nagegowda, D.A. Plant volatile terpenoid metabolism: Biosynthetic genes, transcriptional regulation and subcellular compartmentation. FEBS Lett. 2010, 584, 2965–2973. [Google Scholar] [CrossRef] [PubMed]
- May, B.; Lange, B.M.; Wüst, M. Biosynthesis of sesquiterpenes in grape berry exocarp of Vitis vinifera L.: Evidence for a transport of farnesyl diphosphate precursors from plastids to the cytosol. Phytochemistry 2013, 95, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yauk, Y.K.; Nieuwenhuizen, N.J.; Matich, A.J.; Wang, M.Y.; Perez, R.L.; Atkinson, R.G.; Beuning, L.L. Characterisation of an (S)-linalool synthase from kiwifruit (Actinidia arguta) that catalyses the first committed step in the production of floral lilac compounds. Funct. Plant Biol. 2010, 37, 232–243. [Google Scholar] [CrossRef]
- Aubourg, S.; Lecharny, A.; Bohlmann, J. Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol. Genet. Genomics 2002, 267, 730–745. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.M.; Bohlmann, J. Identification of vitis vinifera (−)-α-terpineol synthase by in silico screening of full-length cdna ests and functional characterization of recombinant terpene synthase. Phytochemistry 2004, 65, 1223–1229. [Google Scholar] [CrossRef] [PubMed]
- Lücker, J.; Bowen, P.; Bohlmann, J. Vitis vinifera terpenoid cyclases: Functional identification of two sesquiterpene synthase cDNAs encoding (+)-valencene synthase and (−)-germacrene D synthase and expression of mono- and sesquiterpene synthases in grapevine flowers and berries. Phytochemistry 2004, 65, 2649–2659. [Google Scholar] [CrossRef] [PubMed]
- Soler, E.; Feron, G.; Clastre, M.; Dargent, R.; Gleizes, M.; Ambid, C. Evidence for a geranyl-diphosphate synthase located within the plastids of Vitis vinifera L. Cultivated in vitro. Planta 1992, 187, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Sallaud, C.; Rontein, D.; Onillon, S.; Jabès, F.; Duffé, P.; Giacalone, C.; Thoraval, S.; Escoffier, C.; Herbette, G.; Leonhardt, N. A novel pathway for sesquiterpene biosynthesis from Z,Z-farnesyl pyrophosphate in the wild tomato solanum habrochaites. Plant Cell 2009, 21, 301–317. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.M.; Chiang, A.; Lund, S.T.; Bohlmann, J. Biosynthesis of wine aroma: Transcript profiles of hydroxymethylbutenyl diphosphate reductase, geranyl diphosphate synthase, and linalool/nerolidol synthase parallel monoterpenol glycoside accumulation in Gewürztraminer grapes. Planta 2012, 236, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Kalua, C.M.; Boss, P.K. Evolution of volatile compounds during the development of cabernet sauvignon grapes (Vitis viniferaL.). J. Agric. Food Chem. 2009, 57, 3818–3830. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.; Strauss, C.R.; Williams, P.J. Changes in free and glycosidically bound monoterpenes in developing muscat grapes. J. Agric. Food Chem. 1984, 32, 919–924. [Google Scholar] [CrossRef]
- Ebeler, S.E.; Thorngate, J.H. Wine chemistry and flavor: Looking into the crystal glass. J. Agric. Food Chem. 2009, 57, 8098–8108. [Google Scholar] [CrossRef] [PubMed]
- Ebang-Oke, J.; Billerbeck, G.D.; Ambid, C.; le Quéré, J.; Étiévant, P. Temporal expression of the lis gene from vitis vinifera L., cv. Muscat de frontignan. In Flavour Research at the Dawn of the Twenty-first Century. Proceedings of the 10th Weurman Flavour Research Symposium, Beaune, France, 25–28 June 2002; Editions Tec & Doc, Lavoisier Intercept: London, UK, 2003; pp. 321–325. [Google Scholar]
- Matarese, F.; Scalabrelli, G.; D’Onofrio, C. Analysis of the expression of terpene synthase genes in relation to aroma content in two aromatic Vitis vinifera varieties. Funct. Plant Biol. 2013, 40, 552–565. [Google Scholar] [CrossRef]
- He, F.; Fang, X.X.; Hu, M.; Pan, Q.H.; Shi, Y.; Duan, C.Q. Preparation and biological application of antibodies against leucoanthocyanidin reductase and anthocyanidin reductase from grape berry. Vitis J. Grapevine Res. 2009, 48, 69–75. [Google Scholar]
- Grimplet, J.; Cramer, G.R.; Dickerson, J.A.; Mathiason, K.; van Hemert, J.; Fennell, A.Y. Vitisnet: “Omics” integration through grapevine molecular networks. PLoS One 2009, 4, e8365. [Google Scholar] [CrossRef] [PubMed]
- Sandra, D.; Franck, S.; Jean-Philippe, T.; Cécile, G.; Véronique, B.; Philippe, G.; Séverine, G.; Philippe, L.; Jean-Charles, L.; Alain, L. Exploration of plant genomes in the FLAGdb++ environment. Plant Methods 2011, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.-M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar] [CrossRef] [PubMed]
- ChloroP. Available online: http://www.cbs.dtu.dk/services/ChloroP (accessed on 10 July 2010).
- TargetP. Available online: http://www.cbs.dtu.dk/services/TargetP (accessed on 10 July 2010).
- SignalP. Available online: http://www.cbs.dtu.dk/services/SignalP (accessed on 10 July 2010).
- DFCI Grape gene index. Available online: http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=grape (accessed on 20 June 2010).
- Fäldt, J.; Martin, D.; Miller, B.; Rawat, S.; Bohlmann, J. Traumatic resin defense in norway spruce (Picea abies): Methyl jasmonate-induced terpene synthase gene expression, and cDNA cloning and functional characterization of (+)-3-carene synthase. Plant Mol. Biol. 2003, 51, 119–133. [Google Scholar] [CrossRef] [PubMed]
- The Map of p-EZS-NL Vector. Available online: https://deepgreen.dpb.carnegiescience.edu (accessed on 1 August 2010).
- Fan, R.C.; Peng, C.C.; Xu, Y.H.; Wang, X.F.; Li, Y.; Shang, Y.; Du, S.Y.; Zhao, R.; Zhang, X.Y.; Zhang, L.Y.; et al. Apple sucrose transporter SUT1 and sorbitol transporter SOT6 interact with cytochrome b5 to regulate their affinity for substrate sugars. Plant Physiol. 2009, 150, 1880–1901. [Google Scholar] [CrossRef] [PubMed]
- Sheen, J. A Transient Expression Assay Using Arabidopsis Mesophyll Protoplasts. 2002. Protocols of Jen Sheen’s Lab. Available online: http://molbio.mgh.harvard.edu/sheenweb/protocols_reg.html (accessed on 29 October 2009).
- Marshall, O.J. Perlprimer: Cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics 2004, 20, 2471–2472. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Z.; Li, X.X.; Zhu, B.Q.; Wen, Y.Q.; Duan, C.Q.; Pan, Q.H. Molecular characterization and expression analysis on two isogenes encoding 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase in grapes. Mol. Biol. Rep. 2011, 38, 4739–4747. [Google Scholar] [CrossRef] [PubMed]
- Loscos, N.; Hernandez-Orte, P.; Cacho, J.; Ferreira, V. Evolution of the aroma composition of wines supplemented with grape flavour precursors from different varietals during accelerated wine ageing. Food Chem. 2010, 120, 205–216. [Google Scholar] [CrossRef]
- Wu, Y.; Pan, Q.; Qu, W.; Duan, C. Comparison of volatile profiles of nine litchi (litchi chinensis sonn.) cultivars from southern china. J. Agric. Food Chem. 2009, 57, 9676–9681. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, B.-Q.; Cai, J.; Wang, Z.-Q.; Xu, X.-Q.; Duan, C.-Q.; Pan, Q.-H. Identification of a Plastid-Localized Bifunctional Nerolidol/Linalool Synthase in Relation to Linalool Biosynthesis in Young Grape Berries. Int. J. Mol. Sci. 2014, 15, 21992-22010. https://doi.org/10.3390/ijms151221992
Zhu B-Q, Cai J, Wang Z-Q, Xu X-Q, Duan C-Q, Pan Q-H. Identification of a Plastid-Localized Bifunctional Nerolidol/Linalool Synthase in Relation to Linalool Biosynthesis in Young Grape Berries. International Journal of Molecular Sciences. 2014; 15(12):21992-22010. https://doi.org/10.3390/ijms151221992
Chicago/Turabian StyleZhu, Bao-Qing, Jian Cai, Zhi-Qun Wang, Xiao-Qing Xu, Chang-Qing Duan, and Qiu-Hong Pan. 2014. "Identification of a Plastid-Localized Bifunctional Nerolidol/Linalool Synthase in Relation to Linalool Biosynthesis in Young Grape Berries" International Journal of Molecular Sciences 15, no. 12: 21992-22010. https://doi.org/10.3390/ijms151221992
APA StyleZhu, B.-Q., Cai, J., Wang, Z.-Q., Xu, X.-Q., Duan, C.-Q., & Pan, Q.-H. (2014). Identification of a Plastid-Localized Bifunctional Nerolidol/Linalool Synthase in Relation to Linalool Biosynthesis in Young Grape Berries. International Journal of Molecular Sciences, 15(12), 21992-22010. https://doi.org/10.3390/ijms151221992







