Real-Time qPCR Identifies Suitable Reference Genes for Borna Disease Virus-Infected Rat Cortical Neurons
Abstract
:1. Introduction
2. Results
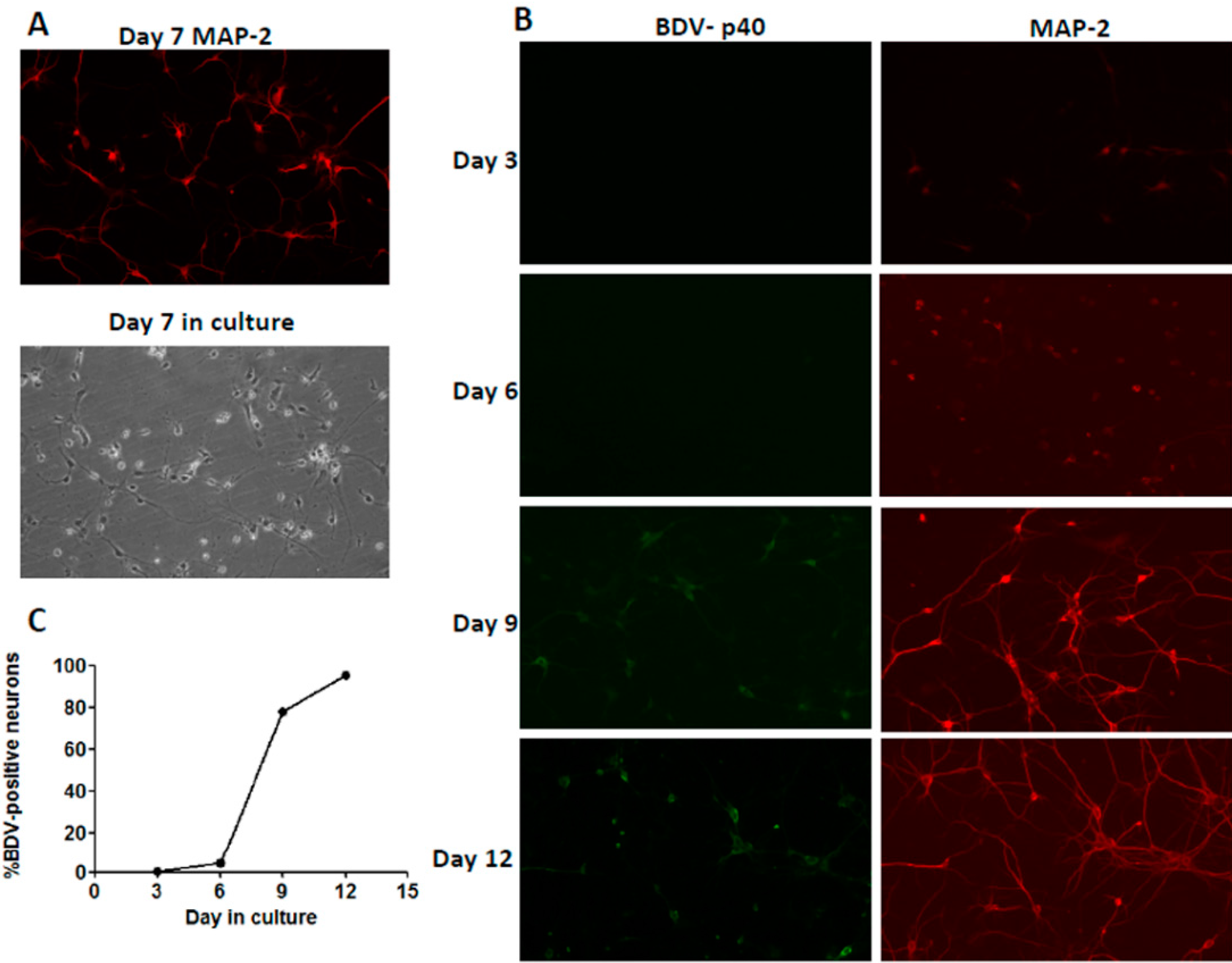
2.1. Immunofluorescence Assay
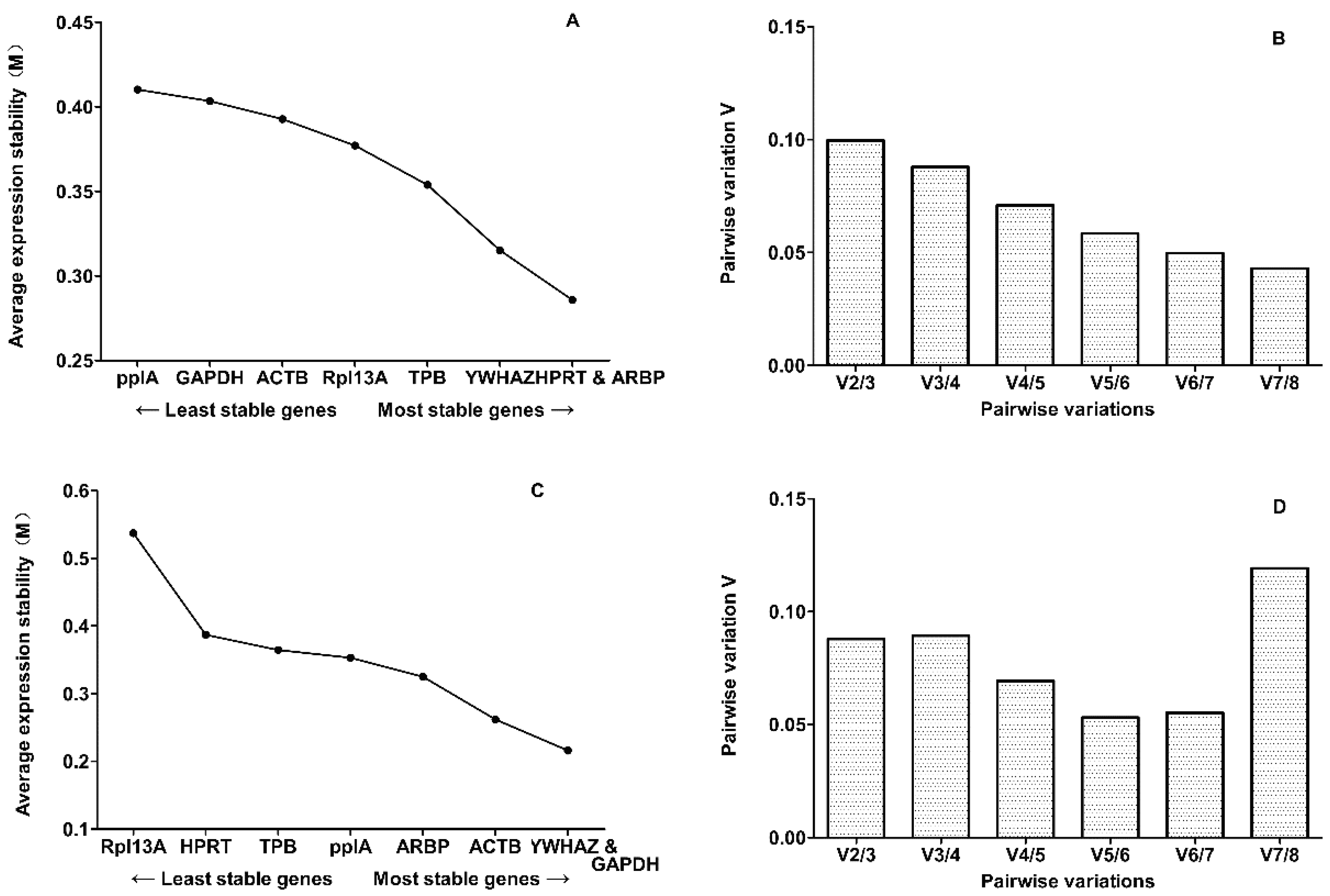
2.2. Evaluation of Expression Stability
2.3. Candidate Reference Gene Ranking
| Symbol | Accession | Name | Forward and Reverse Primer (5'-3') | Amplicon Size | Primer Efficiency | Cq Value (Average ± SD) | Reference a | Mean Relative Quantification b(9, 12 Days) |
|---|---|---|---|---|---|---|---|---|
| HPRT | NM012583 | Hypoxanthine phosphoribosyl-transferase | CTCATGGACTGATTATGGACAGGACGCAGGTCAGCAAAGAACTTATAGCC | 123 | 93 | 23.36 ± 0.78 | [10] | 0.83; 1.16 |
| YWHAZ | NM013011 | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | GATGAAGCCATTGCTGAACTTGGTCTCCTTGGGTATCCGATGTC | 117 | 100 | 19.04 ± 0.66 | [10] | 0.78; 0.91 |
| TPB | NM001004198 | TATA box binding protein | TGGGATTGTACCACAGCTCCACTCATGATGACTGCAGCAAACC | 131 | 93 | 23.94 ± 0.65 | [10] | 0.91; 1.25 |
| Rpl13A | NM173340 | Ribosomal protein L13A | GGATCCCTCCACCCTATGACACTGGTACTTCCACCCGACCTC | 132 | 94 | 18.67 ± 1.12 | [10] | 0.99; 1.28 |
| GAPDH | NM017008 | Glyceraldehyde-3-phosphate dehydrogenase | CAACTCCCTCAAGATTGTCAGCAAGGCATGGACTGTGGTCATGA | 118 | 105 | 20.35 ± 0.67 | [14] | 0.80; 0.78 |
| ACTB | NM031144 | Beta-actin | CAGGGTGTGATGGTGGGTATGGAGTTGGTGACAATGCCGTGTTC | 115 | 103 | 17.88 ± 0.85 | [15] | 1.00; 0.99 |
| PPIA | NM017101 | peptidylprolylisomerase A | GTCAACCCCACCGTGTTCTTCATCCTTTCTCCCCAGTGCTCAG | 133 | 93 | 17.06 ± 0.82 | [15] | 0.85; 0.97 |
| ARBP | NM022402 | Acidic ribosomal phosphoprotein P0 | TAGAGGGTGTCCGCAATGTGCAGTGGGAAGGTGTAGTCAGTC | 137 | 102 | 22.02 ± 0.62 | [16] | 0.81; 0.73 |
| B2M | NM012512 | Beta-2 microglobulin | CGAGACCGATGTATATGCTTGCGTCCAGATGATTCAGAGCTCCA | 114 | 92 | 18.16 ± 1.15 | [14] | 2.51; 3.24 |
| 18s rRNA | M11188 | 18s subunit ribosomal RNA | ACGGACCAGAGCGAAAGCATTGTCAATCCTGTCCGTGTCC | 310 | 107 | 18.07±0.76 | [17] | 0.72; 0.69 |
| BDNF | NM001270630 | Brain-derived neurotrophic factor | ACAGTATTAGCGAGTGGGATTGGGTAGTTCGGCATT | 213 | 96 | 25.55 ± 0.30 | a | - |
| AldoC | NM012497 | Aldolase C, fructose bisphosphate | ACCTGGAAGGGACTCTCCTCAAAGTCACCCCTGGGACAGCT | 141 | 103 | 20.86 ± 0.77 | [18] | - |
| Genorm | 9 Days (C + V, n = 12) | 12 Days (C + V, n = 12) | ||
|---|---|---|---|---|
| Rank | Gene | M-Value | Gene | M-Value |
| 1 | ARBP | 0.2859 | GAPDH | 0.2159 |
| 2 | HPRT | 0.2859 | YWHAZ | 0.2159 |
| 3 | YWHAZ | 0.3152 | ACTB | 0.2617 |
| 4 | TPB | 0.3539 | ARBP | 0.3249 |
| 5 | Rpl13A | 0.3771 | ppIA | 0.3529 |
| 6 | ACTB | 0.3928 | TPB | 0.3645 |
| 7 | GAPDH | 0.4035 | HPRT | 0.387 |
| 8 | ppIA | 0.4103 | Rpl13A | 0.537 |
| NormFinder | 9 Days (C + V, n = 12) | 12 Days (C + V, n = 12) | ||
| Rank | Gene | Variability | Gene | Variability |
| 1 | ARBP | 0.0565 | ACTB | 0.0709 |
| 2 | ACTB | 0.0627 | YWHAZ | 0.1131 |
| 3 | TPB | 0.0713 | ppIA | 0.1363 |
| 4 | YWHAZ | 0.0796 | TPB | 0.1521 |
| 5 | HPRT | 0.0815 | HPRT | 0.1626 |
| 6 | Rpl13A | 0.0821 | GAPDH | 0.1643 |
| 7 | GAPDH | 0.0873 | ARBP | 0.199 |
| 8 | ppIA | 0.1055 | Rpl13A | 0.3008 |
| BestKeeper | 9 Days (C + V, n = 12) | 12 Days (C + V, n = 12) | ||
| Rank | Gene | (r) | Gene | (r) |
| 1 | ppIA | 0.879 | ppIA | 0.874 |
| 2 | ARBP | 0.868 | TPB | 0.809 |
| 3 | GAPDH | 0.784 | ACTB | 0.749 |
| 4 | YWHAZ | 0.72 | ARBP | 0.734 |
| 5 | HPRT | 0.703 | HPRT | 0.584 |
| 6 | ACTB | 0.7 | YWHAZ | 0.583 |
| 7 | Rpl13A | 0.527 | GAPDH | 0.511 |
| 8 | TPB | 0.497 | Rpl13A | 0.146 |
| Delta-Ct | 9 Days (C + V, n = 12) | 12 Days (C + V, n = 12) | ||
| Rank | Gene | Mean SD | Gene | Mean SD |
| 1 | ARBP | 0.34 | ACTB | 0.42 |
| 2 | TPB | 0.4 | YWHAZ | 0.45 |
| 3 | GAPDH | 0.4 | TPB | 0.45 |
| 4 | HPRT | 0.41 | GAPDH | 0.46 |
| 5 | ACTB | 0.41 | ARBP | 0.49 |
| 6 | Rpl13A | 0.42 | ppIA | 0.5 |
| 7 | ppIA | 0.42 | HPRT | 0.52 |
| 8 | YWHAZ | 0.43 | Rpl13A | 1 |
| Day 9 | |||||
| Ranking | geNorm | NormFinder | BestKeeper | Delta-Ct | Consensus |
| 1 | ARBP and HPRT | ARBP | ppIA | ARBP | ARBP |
| 2 | - | ACTB | ARBP | TPB | ACTB |
| 3 | YWHAZ | TPB | GAPDH | GAPDH | TPB |
| 4 | TPB | YWHAZ | YWHAZ | HPRT | YWHAZ |
| 5 | Rpl13A | HPRT | HPRT | ACTB | HPRT |
| 6 | ACTB | Rpl13A | ACTB | Rpl13A | GAPDH |
| 7 | GAPDH | GAPDH | Rpl13A | ppIA | RpI13A |
| 8 | ppIA | ppIA | TPB | YWHAZ | ppIA |
| Day 12 | |||||
| Ranking | geNorm | NormFinder | BestKeeper | Delta-Ct | Consensus |
| 1 | GAPDH and YWHAZ | ACTB | ppIA | ACTB | ACTB |
| 2 | - | YWHAZ | TPB | YWHAZ | YWHAZ |
| 3 | ACTB | ppIA | ACTB | TPB | GAPDH |
| 4 | ARBP | TPB | ARBP | GAPDH | TPB |
| 5 | ppIA | HPRT | HPRT | ARBP | ARBP |
| 6 | TPB | GAPDH | YWHAZ | ppIA | ppIA |
| 7 | HPRT | ARBP | GAPDH | HPRT | HPRT |
| 8 | Rpl13A | Rpl13A | Rpl13A | Rpl13A | RpI13A |
| Two Time Points | |||||
| Ranking | Day 9 | Day 12 | Consensus | ||
| 1 | ARBP | ACTB | ACTB | ||
| 2 | ACTB | YWHAZ | ARBP | ||
| 3 | TPB | GAPDH | TPB | ||
| 4 | YWHAZ | TPB | YWHAZ | ||
| 5 | HPRT | ARBP | GAPDH | ||
| 6 | GAPDH | ppIA | HPRT | ||
| 7 | RpI13A | HPRT | ppIA | ||
| 8 | ppIA | RpI13A | RpI13A | ||
2.4. Assessment of the Validity
| Reference Genes at Day 12 | BDNF | AldoC |
|---|---|---|
| (p-value, n) | (p-value, n) | |
| ACTB and YWHAZ | 0.74 | 0.086 |
| (0.002, n = 12) | (0.612, n = 12) | |
| RpI13A | 1.37 | 1.95 |
| (0.075, n = 12) | (0.873, n = 12) |
3. Discussion
4. Experimental Section
4.1. Primary Culture of Neurons and Viral Infections
4.2. Immunofluorescence
4.3. RNA Isolation and Reverse Transcription
4.4. Quantitative Real-Time PCR
4.5. Data Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ludwig, H.; Bode, L.; Gosztonyi, G. Borna disease: A persistent virus infection of the central nervous system. Prog. Med. Virol. 1988, 35, 107–151. [Google Scholar]
- Lipkin, W.I.; Briese, T.; Bornaviridae. Fields Virology, 5th ed.; Knipe, D., Howley, P., Griffin, D., Lamb, R., Martin, M., Roizman, B., Straus, S., Eds.; Lippincott Williams and Wilkins: Philadelphia, PA, USA, 2007; Volume II, pp. 1829–1851. [Google Scholar]
- Kinnunen, P.M.; Palva, A.; Vaheri, A.; Vapalahti, O. Epidemiology and host spectrum of Borna disease virus infections. J. Gen. Virol. 2013, 94, 247–262. [Google Scholar]
- Zhang, L.; Wang, X.; Zhan, Q.; Wang, Z.; Xu, M.; Zhu, D.; He, F.; Liu, X.; Huang, R.; Li, D.; et al. Evidence for natural Borna disease virus infection in healthy domestic animals in three areas of western China. Arch. Virol. 2014, 159, 1941–1949. [Google Scholar]
- Zhang, L.; Xu, M.M.; Zeng, L.; Liu, S.; Liu, X.; Wang, X.; Li, D.; Huang, R.Z.; Zhao, L.B.; Zhan, Q.L.; et al. Evidence for Borna disease virus infection in neuropsychiatric patients in three western China provinces. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 621–627. [Google Scholar]
- Ludwig, H.; Bode, L. Borna disease virus: New aspects on infection, disease, diagnosis and epidemiology. Rev. Sci. Tech. 2000, 19, 259–288. [Google Scholar]
- Hornig, M.; Solbrig, M.; Horscroft, N.; Weissenbock, H.; Lipkin, W.I. Borna disease virus infection of adult and neonatal rats: Models for neuropsychiatric disease. Curr. Top. Microbiol. Immunol. 2001, 253, 157–177. [Google Scholar]
- Gosztonyi, G. Natural and experimental Borna disease virus infections—Neuropathology and pathogenetic considerations. APMIS 2008, 116, 53–57. [Google Scholar]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes. Immun. 2005, 6, 279–284. [Google Scholar]
- Nelissen, K.; Smeets, K.; Mulder, M.; Hendriks, J.J.; Ameloot, M. Selection of reference genes for gene expression studies in rat oligodendrocytes using quantitative real time PCR. J. Neurosci. Methods 2010, 187, 78–83. [Google Scholar]
- Vandesompele, J.; de Preter, K.; Pattyn, F.; Poppe, B.; van Roy, N.; de Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3. [Google Scholar] [CrossRef]
- Vesentini, N.; Barsanti, C.; Martino, A.; Kusmic, C.; Ripoli, A.; Rossi, A.; L’Abbate, A. Selection of reference genes in different myocardial regions of an in vivo ischemia/reperfusion rat model for normalization of antioxidant gene expression. BMC Res. 2012, 5. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar]
- Langnaese, K.; John, R.; Schweizer, H.; Ebmeyer, U.; Keilhoff, G. Selection of reference genes for quantitative real-time PCR in a rat asphyxial cardiac arrest model. BMC Mol. Biol. 2008, 9. [Google Scholar] [CrossRef]
- Kim, I.; Yang, D.; Tang, X.; Carroll, J.L. Reference gene validation for qPCR in rat carotid body during postnatal development. BMC Res. Notes 2011, 4. [Google Scholar] [CrossRef]
- Zhao, C.; Zha, Y.; Wu, X.; Chen, L.; Shi, J.; Cui, L. The quantification of ADAMTS4 and 8 expression and selection of reference genes for quantitative real-time PCR analysis in myocardial infarction. Biomed. Pharmacother. 2011, 65, 555–559. [Google Scholar]
- Bonefeld, B.E.; Elfving, B.; Wegener, G. Reference genes for normalization: A study of rat brain tissue. Synapse 2008, 62, 302–309. [Google Scholar]
- Williams, B.L.; Yaddanapudi, K.; Hornig, M.; Lipkin, W.I. Spatiotemporal analysis of purkinje cell degeneration relative to parasagittal expression domains in a model of neonatal viral infection. J. Virol. 2007, 81, 2675–2687. [Google Scholar]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar]
- Liu, X.; Zhang, L.; Cheng, K.; Wang, X.; Ren, G.; Xie, P. Identification of suitable plasma-based reference genes for miRNAome analysis of major depressive disorder. J. Affect. Disord. 2014, 163, 133–139. [Google Scholar]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7. [Google Scholar] [CrossRef]
- Lewin, G.R.; Barde, Y.A. Physiology of the neurotrophins. Annu. Rev. Neurosci. 1996, 19, 289–317. [Google Scholar]
- Huang, E.J.; Reichardt, L.F. Neurotrophins: Roles in neuronal development and function. Annu. Rev. Neurosci. 2001, 24, 677–736. [Google Scholar]
- McAllister, A.K.; Katz, L.C.; Lo, D.C. Neurotrophins and synaptic plasticity. Annu. Rev. Neurosci. 1999, 22, 295–318. [Google Scholar]
- Ahn, A.H.; Dziennis, S.; Hawkes, R.; Herrup, K. The cloning of zebrin II reveals its identity with aldolase C. Development 1994, 120, 2081–2090. [Google Scholar]
- Carbone, K.M.; Rubin, S.A.; Nishino, Y.; Pletnikov, M.V. Borna disease: Virus-induced neurobehavioral disease pathogenesis. Curr. Opin. Microbiol. 2001, 4, 467–475. [Google Scholar]
- Wensman, J.J.; Ilback, C.; Hjertstrom, E.; Blomstrom, A.L.; Gustavsson, M.H.; Jaderlund, K.H.; Strom-Holst, B.; Belak, S.; Berg, A.L.; Berg, M. Expression of interferon gamma in the brain of cats with natural Borna disease virus infection. Vet. Immunol. Immunopathol. 2011, 141, 162–167. [Google Scholar]
- Jehle, C.; Herpfer, I.; Rauer, M.; Schwemmle, M.; Sauder, C. Identification of differentially expressed genes in brains of newborn Borna disease virus-infected rats in the absence of inflammation. Arch. Virol. 2003, 148, 45–63. [Google Scholar]
- Koster-Patzlaff, C.; Hosseini, S.M.; Reuss, B. Layer specific changes of astroglial gap junctions in the rat cerebellar cortex by persistent Borna Disease Virus infection. Brain Res. 2008, 1219, 143–158. [Google Scholar]
- Southall, M.D.; Li, T.; Gharibova, L.S.; Pei, Y.; Nicol, G.D.; Travers, J.B. Activation of epidermal vanilloid receptor-1 induces release of proinflammatory mediators in human keratinocytes. J. Pharmacol. Exp. Ther. 2003, 304, 217–222. [Google Scholar]
- Pihur, V.; Datta, S.; Datta, S. RankAggreg, an R package for weighted rank aggregation. BMC Bioinform. 2009, 10. [Google Scholar] [CrossRef]
- Bellon, A. New genes associated with schizophrenia in neurite formation: A review of cell culture experiments. Mol. Psychiatry 2007, 12, 620–629. [Google Scholar]
- Binder, L.I.; Frankfurter, A.; Kim, H.; Caceres, A.; Payne, M.R.; Rebhun, L.I. Heterogeneity of microtubule-associated protein 2 during rat brain development. Proc. Natl. Acad. Sci. USA 1984, 81, 5613–5617. [Google Scholar]
- Bajramovic, J.J.; Munter, S.; Syan, S.; Nehrbass, U.; Brahic, M.; Gonzalez-Dunia, D. Borna disease virus glycoprotein is required for viral dissemination in neurons. J. Virol. 2003, 77, 12222–12231. [Google Scholar]
- Wang, G.P.; Xu, C.S. Reference gene selection for real-time RT-PCR in eight kinds of rat regenerating hepatic cells. Mol. Biotechnol. 2010, 46, 49–57. [Google Scholar]
- Lilly, S.T.; Drummond, R.S.; Pearson, M.N.; MacDiarmid, R.M. Identification and validation of reference genes for normalization of transcripts from virus-infected Arabidopsis thaliana. Mol. Plant Microbe Interact. 2011, 24, 294–304. [Google Scholar]
- Bode, L.; Ludwig, H. Borna disease virus infection, a human mental-health risk. Clin. Microbiol. Rev. 2003, 16, 534–545. [Google Scholar]
- Bode, L.; Durrwald, R.; Rantam, F.A.; Ferszt, R.; Ludwig, H. First isolates of infectious human Borna disease virus from patients with mood disorders. Mol. Psychiatry 1996, 1, 200–212. [Google Scholar]
- De la Torre, J.C.; Bode, L.; Durrwald, R.; Cubitt, B.; Ludwig, H. Sequence characterization of human Borna disease virus. Virus Res. 1996, 44, 33–44. [Google Scholar]
- Suberbielle, E.; Stella, A.; Pont, F.; Monnet, C.; Mouton, E.; Lamouroux, L.; Monsarrat, B.; Gonzalez-Dunia, D. Proteomic analysis reveals selective impediment of neuronal remodeling upon Borna disease virus infection. J. Virol. 2008, 82, 12265–12279. [Google Scholar]
- Huang, R.; Gao, H.; Zhang, L.; Jia, J.; Liu, X.; Zheng, P.; Ma, L.; Li, W.; Deng, J.; Wang, X.; et al. Borna disease virus infection perturbs energy metabolites and amino acids in cultured human oligodendroglia cells. PLoS One 2012, 7, e44665. [Google Scholar]
- Ludwig, H.; Furuya, K.; Bode, L.; Klein, N.; Durrwald, R.; Lee, D.S. Biology and neurobiology of Borna disease viruses (BDV), defined by antibodies, neutralizability and their pathogenic potential. Arch. Virol. Suppl. 1993, 7, 111–133. [Google Scholar]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Liu, S.; Zhang, L.; You, H.; Huang, R.; Sun, L.; He, P.; Chen, S.; Zhang, H.; Xie, P. Real-Time qPCR Identifies Suitable Reference Genes for Borna Disease Virus-Infected Rat Cortical Neurons. Int. J. Mol. Sci. 2014, 15, 21825-21839. https://doi.org/10.3390/ijms151221825
Zhang L, Liu S, Zhang L, You H, Huang R, Sun L, He P, Chen S, Zhang H, Xie P. Real-Time qPCR Identifies Suitable Reference Genes for Borna Disease Virus-Infected Rat Cortical Neurons. International Journal of Molecular Sciences. 2014; 15(12):21825-21839. https://doi.org/10.3390/ijms151221825
Chicago/Turabian StyleZhang, Lujun, Siwen Liu, Liang Zhang, Hongmin You, Rongzhong Huang, Lin Sun, Peng He, Shigang Chen, Hong Zhang, and Peng Xie. 2014. "Real-Time qPCR Identifies Suitable Reference Genes for Borna Disease Virus-Infected Rat Cortical Neurons" International Journal of Molecular Sciences 15, no. 12: 21825-21839. https://doi.org/10.3390/ijms151221825
APA StyleZhang, L., Liu, S., Zhang, L., You, H., Huang, R., Sun, L., He, P., Chen, S., Zhang, H., & Xie, P. (2014). Real-Time qPCR Identifies Suitable Reference Genes for Borna Disease Virus-Infected Rat Cortical Neurons. International Journal of Molecular Sciences, 15(12), 21825-21839. https://doi.org/10.3390/ijms151221825




