Current Models of Mammalian Target of Rapamycin Complex 1 (mTORC1) Activation by Growth Factors and Amino Acids
Abstract
:1. Introduction
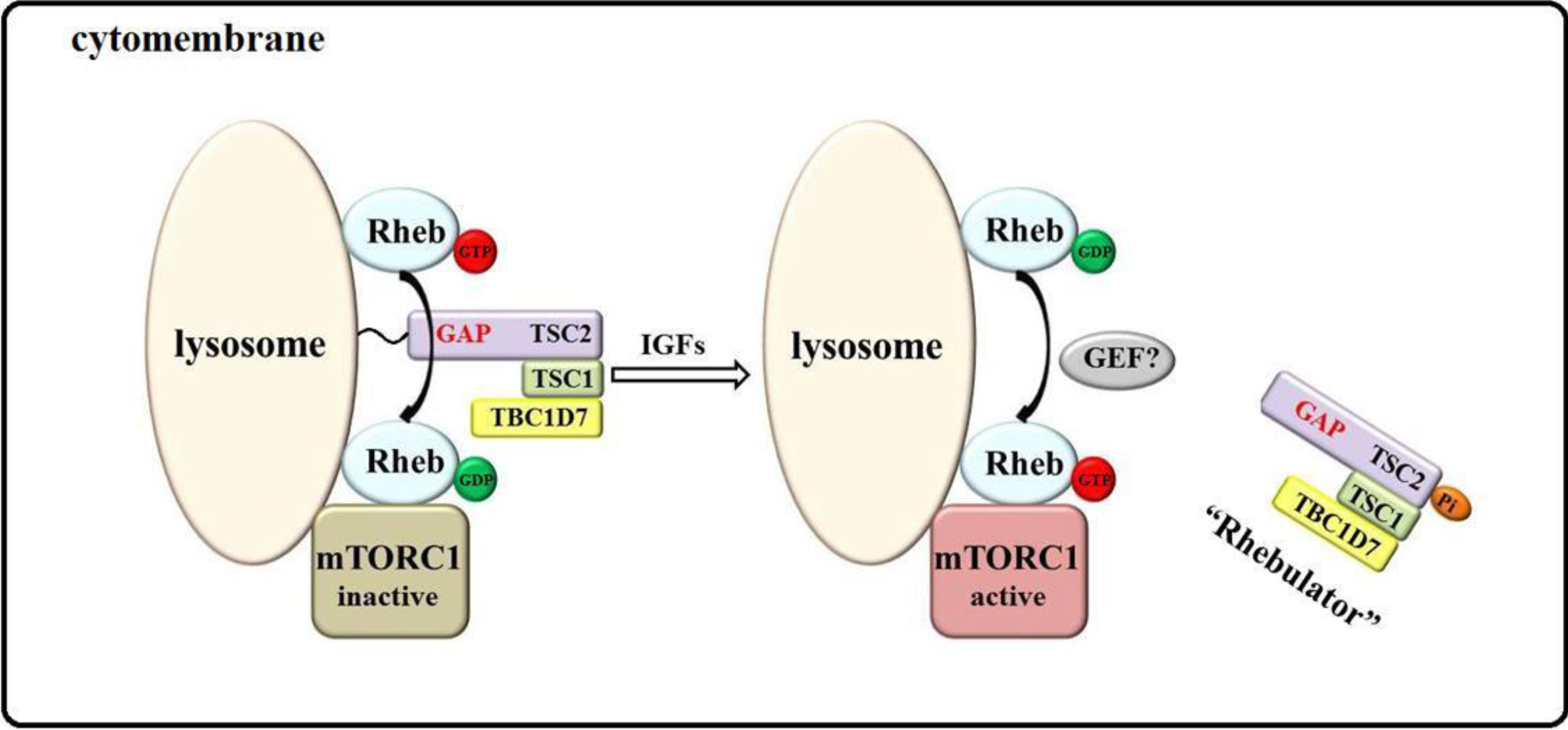
2. Model of mTORC1 Activation by Growth Factors
3. Models of mTORC1 Activation by Amino Acids
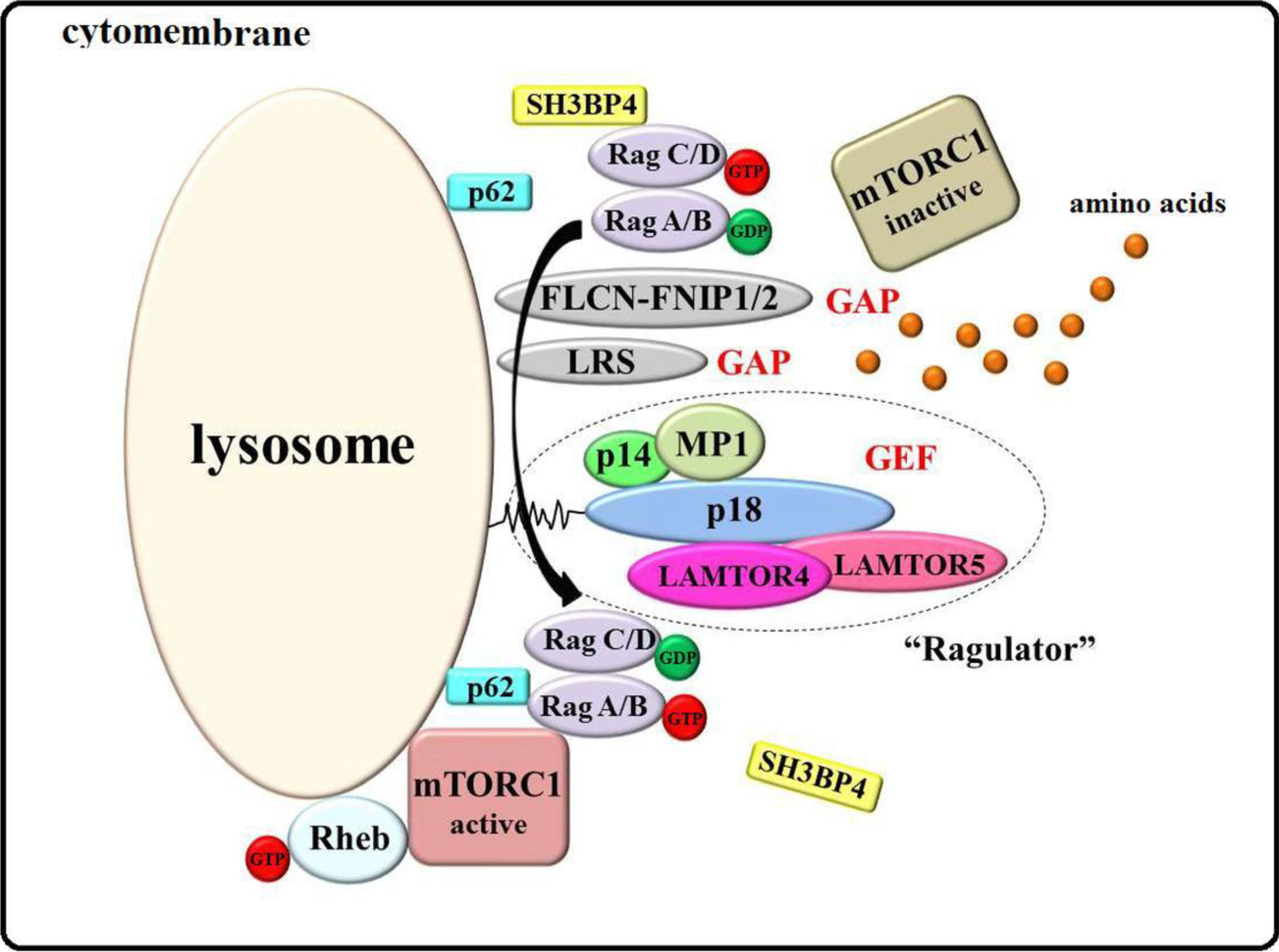
3.1. Ragulator-Rags Model
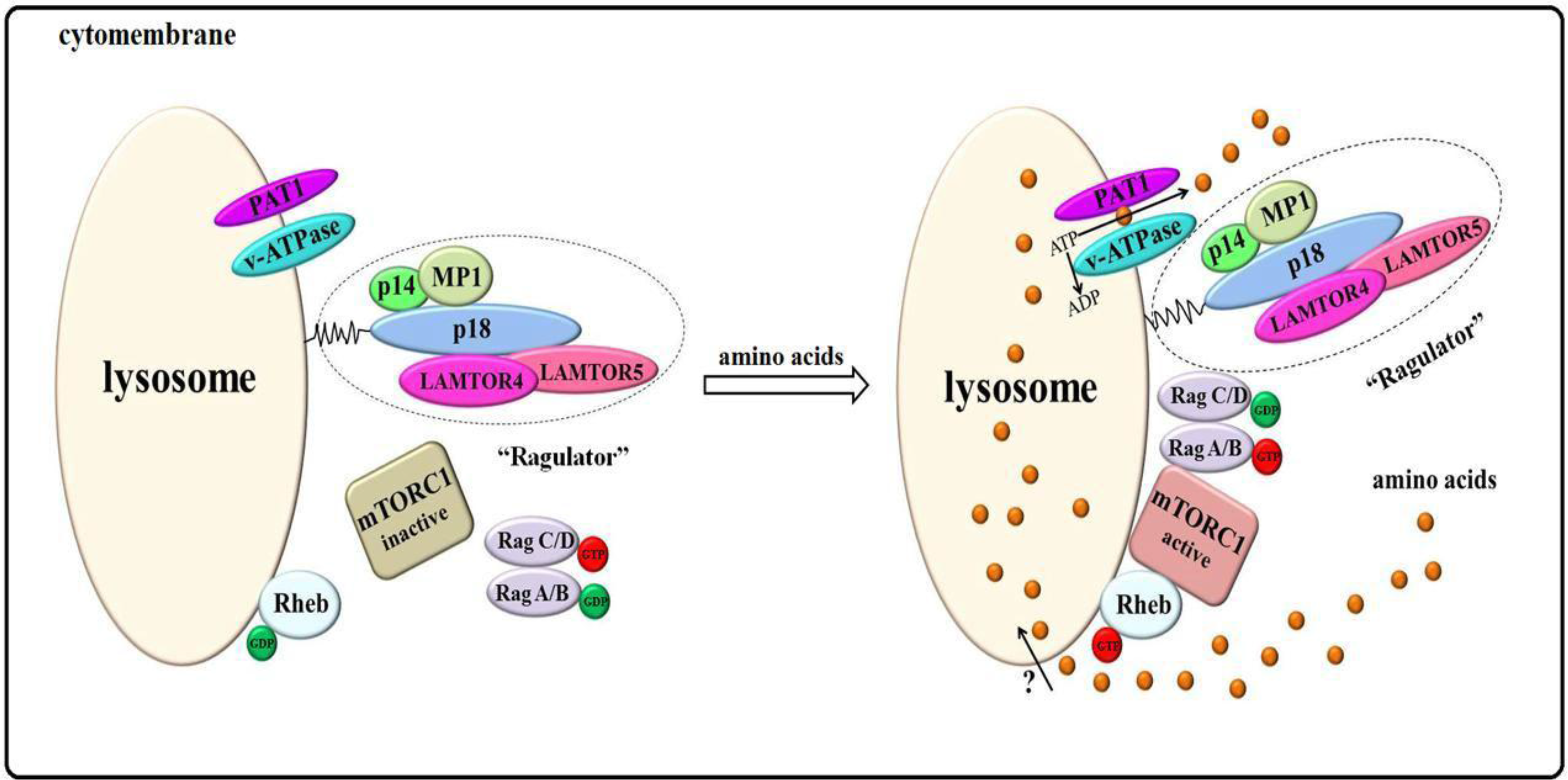
3.2. Amino Acids “inside-out” Model
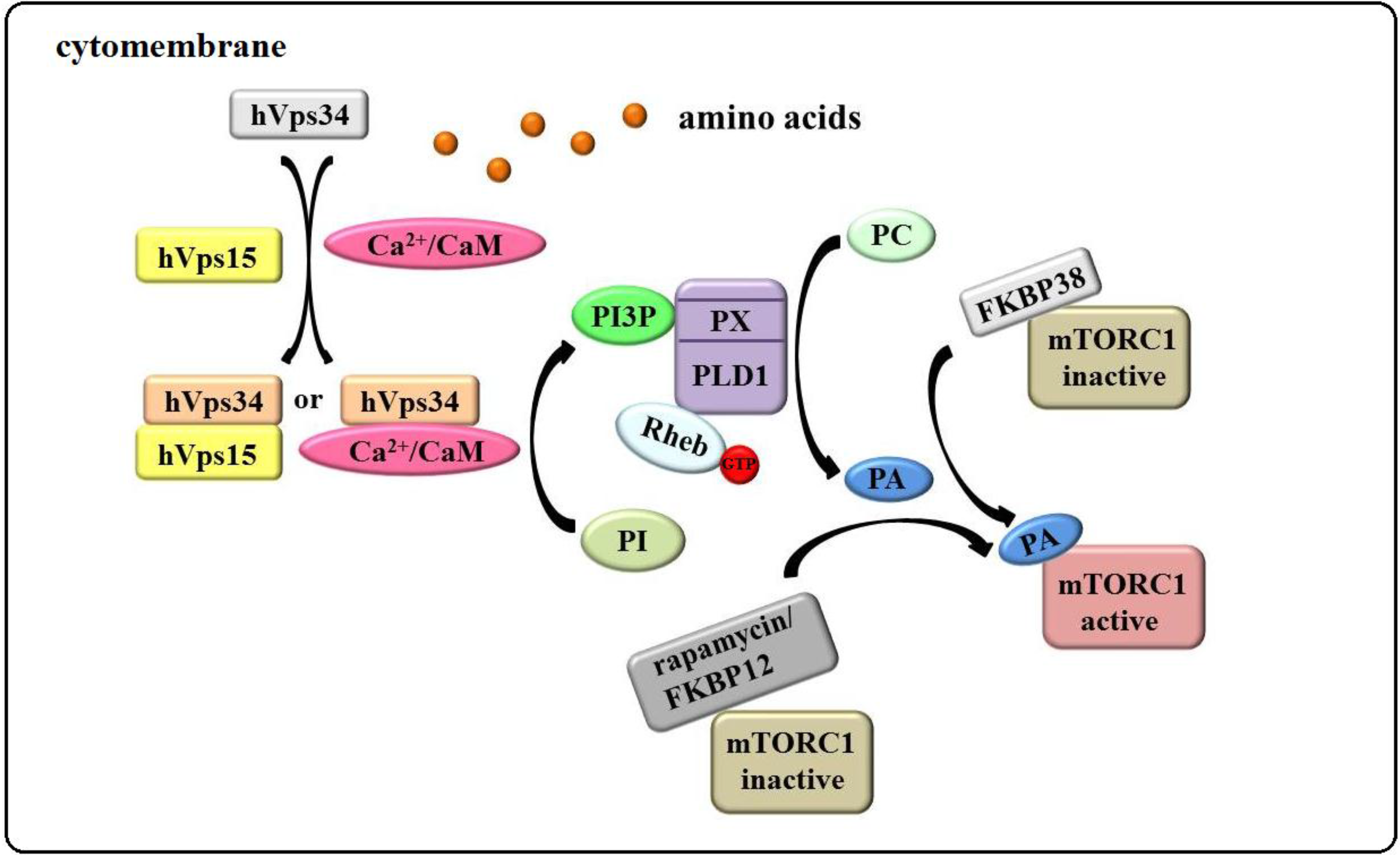
3.3. The “hVps34/Rheb-PLD1” Model
3.4. Rheb Binds to FKBP38 and mTOR Is Unleashed
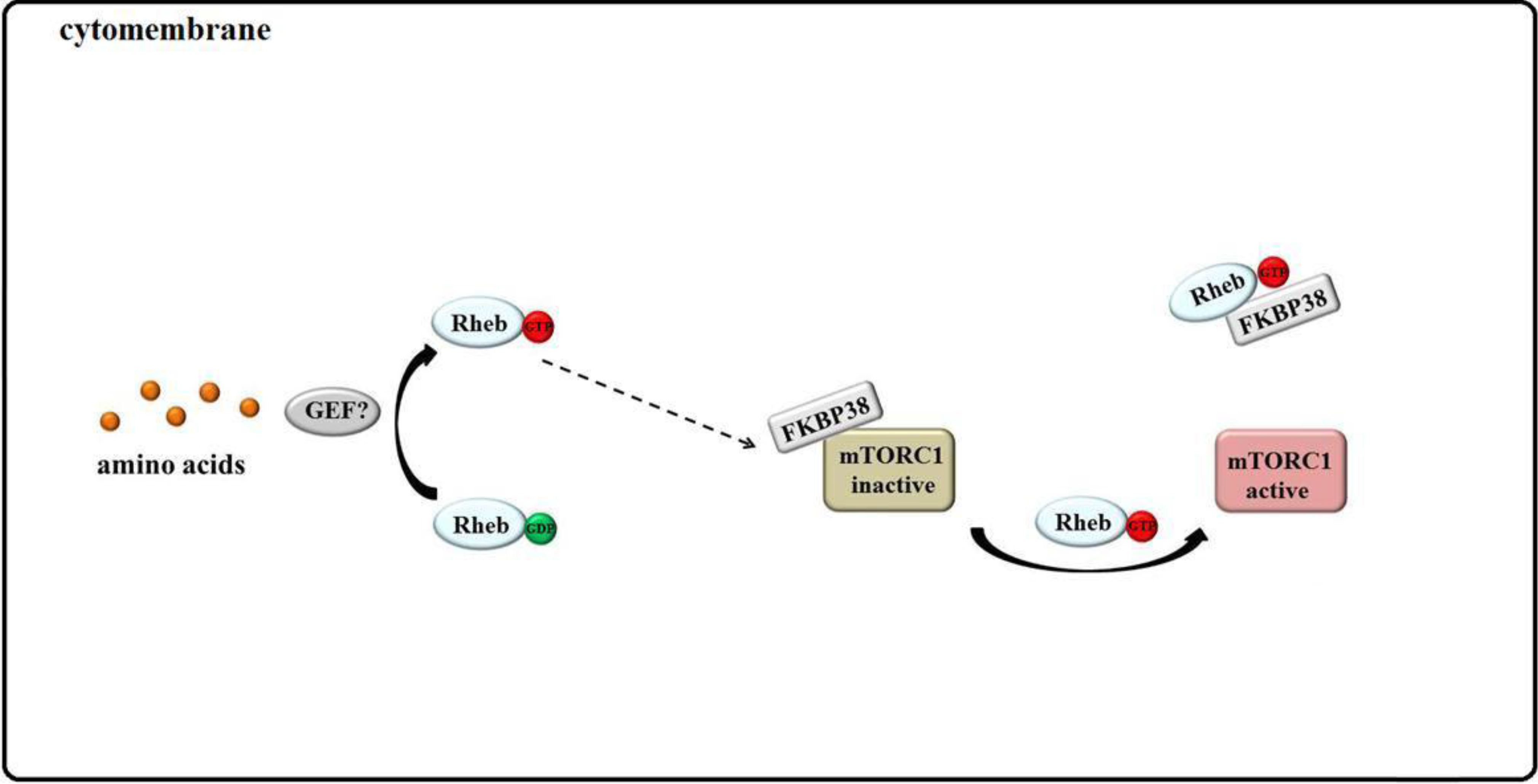
4. Conclusions
Acknowledgments
Author Contributions
Abbreviations
Conflicts of Interest
References
- Wullschleger, S.; Loewith, R.; Hall, M.N. Tor signaling in growth and metabolism. Cell 2006, 124, 471–484. [Google Scholar]
- Yang, H.; Rudge, D.G.; Koos, J.D.; Vaidialingam, B.; Yang, H.J.; Pavletich, N.P. Mtor kinase structure, mechanism and regulation. Nature 2013, 497, 217–223. [Google Scholar]
- Brown, E.J.; Albers, M.W.; Shin, T.B.; Ichikawa, K.; Keith, C.T.; Lane, W.S.; Schreiber, S.L. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 1994, 369, 756–758. [Google Scholar]
- Heitman, J.; Movva, N.R.; Hall, M.N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 1991, 253, 905–909. [Google Scholar]
- Laplante, M.; Sabatini, D.M. mTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar]
- Sabatini, D.M.; Erdjument-Bromage, H.; Lui, M.; Tempst, P.; Snyder, S.H. Raft1: A mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 1994, 78, 35–43. [Google Scholar]
- Sabers, C.J.; Martin, M.M.; Brunn, G.J.; Williams, J.M.; Dumont, F.J.; Wiederrecht, G.; Abraham, R.T. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J. Biol. Chem. 1995, 270, 815–822. [Google Scholar]
- Vezina, C.; Kudelski, A.; Sehgal, S. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J. Antibiot. 1975, 28, 721–726. [Google Scholar]
- Sekulić, A.; Hudson, C.C.; Homme, J.L.; Yin, P.; Otterness, D.M.; Karnitz, L.M.; Abraham, R.T. A direct linkage between the phosphoinositide 3-kinase-AKT signaling pathway and the mammalian target of rapamycin in mitogen-stimulated and transformed cells. Cancer Res. 2000, 60, 3504–3513. [Google Scholar]
- Sarbassov, D.D.; Ali, S.M.; Kim, D.-H.; Guertin, D.A.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004, 14, 1296–1302. [Google Scholar]
- Glidden, E.J.; Gray, L.G.; Vemuru, S.; Li, D.; Harris, T.E.; Mayo, M.W. Multiple site acetylation of rictor stimulates mammalian target of rapamycin complex 2 (mTORC2)-dependent phosphorylation of akt protein. J. Biol. Chem. 2012, 287, 581–588. [Google Scholar]
- Akcakanat, A.; Singh, G.; Hung, M.-C.; Meric-Bernstam, F. Rapamycin regulates the phosphorylation of rictor. Biochem. Biophys. Res. Commun. 2007, 362, 330–333. [Google Scholar]
- Jacinto, E.; Loewith, R.; Schmidt, A.; Lin, S.; Rüegg, M.A.; Hall, A.; Hall, M.N. Mammalian tor complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell. Biol. 2004, 6, 1122–1128. [Google Scholar]
- Barlow, A.; Xie, J.; Moore, C.; Campbell, S.; Shaw, J.; Nicholson, M.; Herbert, T. Rapamycin toxicity in MIN6 cells and rat and human islets is mediated by the inhibition of mTOR complex 2 (mTORC2). Diabetologia 2012, 55, 1355–1365. [Google Scholar]
- Lamming, D.W.; Ye, L.; Katajisto, P.; Goncalves, M.D.; Saitoh, M.; Stevens, D.M.; Davis, J.G.; Salmon, A.B.; Richardson, A.; Ahima, R.S. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science 2012, 335, 1638–1643. [Google Scholar]
- Loewith, R.; Jacinto, E.; Wullschleger, S.; Lorberg, A.; Crespo, J.L.; Bonenfant, D.; Oppliger, W.; Jenoe, P.; Hall, M.N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 2002, 10, 457–468. [Google Scholar]
- Sarbassov, D.D.; Ali, S.M.; Sengupta, S.; Sheen, J.-H.; Hsu, P.P.; Bagley, A.F.; Markhard, A.L.; Sabatini, D.M. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell 2006, 22, 159–168. [Google Scholar]
- Hay, N.; Sonenberg, N. Upstream and downstream of mtor. Genes Dev. 2004, 18, 1926–1945. [Google Scholar]
- Hara, K.; Maruki, Y.; Long, X.; Yoshino, K.; Oshiro, N.; Hidayat, S.; Tokunaga, C.; Avruch, J.; Yonezawa, K. Raptor, a binding partner of target of rapamycin (TOR), mediates tor action. Cell 2002, 110, 177–189. [Google Scholar]
- Kim, D.-H.; Sarbassov, D.D.; Ali, S.M.; King, J.E.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002, 110, 163–175. [Google Scholar]
- Nojima, H.; Tokunaga, C.; Eguchi, S.; Oshiro, N.; Hidayat, S.; Yoshino, K.; Hara, K.; Tanaka, N.; Avruch, J.; Yonezawa, K. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mtor substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J. Biol. Chem. 2003, 278, 15461–15464. [Google Scholar]
- Schalm, S.S.; Blenis, J. Identification of a conserved motif required for mtor signaling. Curr. Biol. 2002, 12, 632–639. [Google Scholar]
- Schalm, S.S.; Fingar, D.C.; Sabatini, D.M.; Blenis, J. Tos motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr. Biol. 2003, 13, 797–806. [Google Scholar]
- Brunn, G.J.; Hudson, C.C.; Sekulić, A.; Williams, J.M.; Hosoi, H.; Houghton, P.J.; Lawrence, J.C.; Abraham, R.T. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 1997, 277, 99–101. [Google Scholar]
- Dann, S.G.; Thomas, G. The amino acid sensitive tor pathway from yeast to mammals. FEBS Lett. 2006, 580, 2821–2829. [Google Scholar]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell. Biol. 2011, 13, 132–141. [Google Scholar]
- Kim, D.-H.; Sarbassov, D.D.; Ali, S.M.; Latek, R.R.; Guntur, K.V.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. GβL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell 2003, 11, 895–904. [Google Scholar]
- Guertin, D.A.; Stevens, D.M.; Thoreen, C.C.; Burds, A.A.; Kalaany, N.Y.; Moffat, J.; Brown, M.; Fitzgerald, K.J.; Sabatini, D.M. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev. Cell 2006, 11, 859–871. [Google Scholar]
- Laplante, M.; Sabatini, D.M. mTOR signaling at a glance. J. Cell. Sci. 2009, 122, 3589–3594. [Google Scholar]
- Kaizuka, T.; Hara, T.; Oshiro, N.; Kikkawa, U.; Yonezawa, K.; Takehana, K.; Iemura, S.; Natsume, T.; Mizushima, N. Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. J. Biol. Chem. 2010, 285, 20109–20116. [Google Scholar]
- Kanoh, J.; Yanagida, M. Tel2: A common partner of PIK‐related kinases and a link between DNA checkpoint and nutritional response? Genes Cells 2007, 12, 1301–1304. [Google Scholar]
- Kovacina, K.S.; Park, G.Y.; Bae, S.S.; Guzzetta, A.W.; Schaefer, E.; Birnbaum, M.J.; Roth, R.A. Identification of a proline-rich Akt substrate as a 14-3-3 binding partner. J. Biol. Chem. 2003, 278, 10189–10194. [Google Scholar]
- Morrison, D.K. The 14-3-3 proteins: Integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell. Biol. 2009, 19, 16–23. [Google Scholar]
- Thedieck, K.; Polak, P.; Kim, M.L.; Molle, K.D.; Cohen, A.; Jenö, P.; Arrieumerlou, C.; Hall, M.N. PRAS40 and PRR5-like protein are new mtor interactors that regulate apoptosis. PLoS One 2007, 2, e1217. [Google Scholar]
- Wang, L.; Harris, T.E.; Roth, R.A.; Lawrence, J.C. PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J. Biol. Chem. 2007, 282, 20036–20044. [Google Scholar]
- Fonseca, B.D.; Smith, E.M.; Lee, V.H.-Y.; MacKintosh, C.; Proud, C.G. Pras40 is a target for mammalian target of rapamycin complex 1 and is required for signaling downstream of this complex. J. Biol. Chem. 2007, 282, 24514–24524. [Google Scholar]
- Oshiro, N.; Takahashi, R.; Yoshino, K.; Tanimura, K.; Nakashima, A.; Eguchi, S.; Miyamoto, T.; Hara, K.; Takehana, K.; Avruch, J. The proline-rich Akt substrate of 40 kDa (pras40) is a physiological substrate of mammalian target of rapamycin complex 1. J. Biol. Chem. 2007, 282, 20329–20339. [Google Scholar]
- Sancak, Y.; Thoreen, C.C.; Peterson, T.R.; Lindquist, R.A.; Kang, S.A.; Spooner, E.; Carr, S.A.; Sabatini, D.M. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell 2007, 25, 903–915. [Google Scholar]
- Kazi, A.A.; Hong-Brown, L.; Lang, S.M.; Lang, C.H. Deptor knockdown enhances mtor activity and protein synthesis in myocytes and ameliorates disuse muscle atrophy. Mol. Med. 2011, 17, 925–936. [Google Scholar]
- Liu, M.; Wilk, S.A.; Wang, A.; Zhou, L.; Wang, R.-H.; Ogawa, W.; Deng, C.; Dong, L.Q.; Liu, F. Resveratrol inhibits mtor signaling by promoting the interaction between mTOR and DEPTOR. J. Biol. Chem. 2010, 285, 36387–36394. [Google Scholar]
- Peterson, T.R.; Laplante, M.; Thoreen, C.C.; Sancak, Y.; Kang, S.A.; Kuehl, W.M.; Gray, N.S.; Sabatini, D.M. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 2009, 137, 873–886. [Google Scholar]
- Duan, S.; Skaar, J.R.; Kuchay, S.; Toschi, A.; Kanarek, N.; Ben-Neriah, Y.; Pagano, M. mTOR generates an auto-amplification loop by triggering the βTrCP-and CK1α-dependent degradation of DEPTOR. Mol. Cell 2011, 44, 317–324. [Google Scholar]
- Gao, D.; Inuzuka, H.; Tan, M.-K.M.; Fukushima, H.; Locasale, J.W.; Liu, P.; Wan, L.; Zhai, B.; Chin, Y.R.; Shaik, S. mTOR drives its own activation viaSCF(βTrCP)-dependent degradation of the mTOR inhibitor DEPTOR. Mol. Cell 2011, 44, 290–303. [Google Scholar]
- Zhao, Y.; Xiong, X.; Sun, Y. DEPTOR, an mtor inhibitor, is a physiological substrate of SCF (βTrCP) E3 ubiquitin ligase and regulates survival and autophagy. Mol. Cell 2011, 44, 304–316. [Google Scholar]
- Laplante, M.; Sabatini, D.M. An emerging role of mTOR in lipid biosynthesis. Curr. Biol. 2009, 19, R1046–R1052. [Google Scholar]
- Dibble, C.C.; Elis, W.; Menon, S.; Qin, W.; Klekota, J.; Asara, J.M.; Finan, P.M.; Kwiatkowski, D.J.; Murphy, L.O.; Manning, B.D. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol. Cell 2012, 47, 535–546. [Google Scholar]
- Inoki, K.; Ouyang, H.; Zhu, T.; Lindvall, C.; Wang, Y.; Zhang, X.; Yang, Q.; Bennett, C.; Harada, Y.; Stankunas, K. TSC2 integrates wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 2006, 126, 955–968. [Google Scholar]
- Ma, L.; Chen, Z.; Erdjument-Bromage, H.; Tempst, P.; Pandolfi, P.P. Phosphorylation and functional inactivation of TSC2 by Erk: Implications for tuberous sclerosisand cancer pathogenesis. Cell 2005, 121, 179–193. [Google Scholar]
- Potter, C.J.; Pedraza, L.G.; Xu, T. Akt regulates growth by directly phosphorylating Tsc2. Nat. Cell. Biol. 2002, 4, 658–665. [Google Scholar]
- Roux, P.P.; Ballif, B.A.; Anjum, R.; Gygi, S.P.; Blenis, J. Tumor-promoting phorbol esters and activated ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal s6 kinase. Proc. Natl. Acad. Sci. USA 2004, 101, 13489–13494. [Google Scholar]
- Capo-Chichi, J.M.; Tcherkezian, J.; Hamdan, F.F.; Décarie, J.C.; Dobrzeniecka, S.; Patry, L.; Nadon, M.A.; Mucha, B.E.; Major, P.; Shevell, M. Disruption of TBC1D7, a subunit of the TSC1-TSC 2 protein complex, in intellectual disability and megalencephaly. J. Med. Genet. 2013, 50, 740–744. [Google Scholar]
- Yamagata, K.; Sanders, L.K.; Kaufmann, W.E.; Yee, W.; Barnes, C.A.; Nathans, D.; Worley, P.F. rheb, a growth factor-and synaptic activity-regulated gene, encodes a novel Ras-related protein. J. Biol. Chem. 1994, 269, 16333–16339. [Google Scholar]
- Bai, X.; Ma, D.; Liu, A.; Shen, X.; Wang, Q.J.; Liu, Y.; Jiang, Y. Rheb activates mTOR by antagonizing its endogenous inhibitor, FKBP38. Science 2007, 318, 977–980. [Google Scholar]
- Zhang, Y.; Gao, X.; Saucedo, L.J.; Ru, B.; Edgar, B.A.; Pan, D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat. Cell. Biol. 2003, 5, 578–581. [Google Scholar]
- Jacobs, B.L.; You, J.S.; Frey, J.W.; Goodman, C.A.; Gundermann, D.M.; Hornberger, T.A. Eccentric contractions increase TSC2 phosphorylation and alter the targeting of TSC 2 and mTOR to the lysosome. J. Physiol. 2013, 591, 4611–4620. [Google Scholar]
- Cai, S.L.; Tee, A.R.; Short, J.D.; Bergeron, J.M.; Kim, J.; Shen, J.; Guo, R.; Johnson, C.L.; Kiguchi, K.; Walker, C.L. Activity of TSC2 is inhibited by akt-mediated phosphorylation and membrane partitioning. J. Cell. Biol. 2006, 173, 279–289. [Google Scholar]
- Miyazaki, M.; McCarthy, J.J.; Esser, K.A. Insulin like growth factor‐1‐induced phosphorylation and altered distribution of tuberous sclerosis complex (TSC)1/TSC2 in C2C12 myotubes. FEBS J. 2010, 277, 2180–2191. [Google Scholar]
- Menon, S.; Dibble, C.C.; Talbott, G.; Hoxhaj, G.; Valvezan, A.J.; Takahashi, H.; Cantley, L.C.; Manning, B.D. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 2014, 156, 771–785. [Google Scholar]
- Li, Y.; Inoki, K.; Guan, K.L. Biochemical and functional characterizations of small GTPase Rheb and TSC2 GAP activity. Mol. Cell. Biol. 2004, 24, 7965–7975. [Google Scholar]
- Amson, R.; Pece, S.; Marine, J.C.; Fiore, P.P.D.; Telerman, A. TPT1/TCTP-regulated pathways in phenotypic reprogramming. Trends. Cell. Biol. 2012, 23, 37–46. [Google Scholar]
- Hsu, Y.C.; Chern, J.J.; Cai, Y.; Liu, M.; Choi, K.W. Drosophila TCTP is essential for growth and proliferation through regulation of dRheb GTPase. Nature 2007, 445, 785–788. [Google Scholar]
- Polak, P.; Hall, M.N. mTOR and the control of whole body metabolism. Curr. Opin. Cell. Biol. 2009, 21, 209–218. [Google Scholar]
- Rehmann, H.; Brüning, M.; Berghaus, C.; Schwarten, M.; Köhler, K.; Stocker, H.; Stoll, R.; Zwartkruis, F.; Wittinghofer, A. Biochemical characterisation of TCTP questions its function as a guanine nucleotide exchange factor for Rheb. FEBS Lett. 2008, 582, 3005–3010. [Google Scholar]
- Lee, M.N.; Koh, A.; Park, D.; Jang, J.-H.; Kwak, D.; Jeon, H.; Kim, J.; Choi, E.-J.; Jeong, H.; Suh, P.-G. Deacetylated αβ-tubulin acts as a positive regulator of Rheb GTPase through increasing its GTP-loading. Cell. Signal. 2012, 25, 535–551. [Google Scholar]
- Maeurer, C.; Holland, S.; Pierre, S.; Potstada, W.; Scholich, K. Sphingosine-1-phosphate induced mTOR-activation is mediated by the E3-ubiquitin ligase PAM. Cell. Signal. 2009, 21, 293–300. [Google Scholar]
- Sancak, Y.; Bar-Peled, L.; Zoncu, R.; Markhard, A.L.; Nada, S.; Sabatini, D.M. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 2010, 141, 290–303. [Google Scholar]
- Bar-Peled, L.; Schweitzer, L.D.; Zoncu, R.; Sabatini, D.M. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 2012, 150, 1196–1208. [Google Scholar]
- Sancak, Y.; Sabatini, D.M. Rag proteins regulate amino-acid-induced mTORC1 signalling. Biochem. Soc. Trans. 2009, 37, 289–290. [Google Scholar]
- Schürmann, A.; Brauers, A.; Maßmann, S.; Becker, W.; Joost, H.-G. Cloning of a novel family of mammalian GTP-binding proteins (RagA, RagBs, RagB1) with remote similarity to the Ras-related GTPases. J. Biol. Chem. 1995, 270, 28982–28988. [Google Scholar]
- Sekiguchi, T.; Hirose, E.; Nakashima, N.; Ii, M.; Nishimoto, T. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J. Biol. Chem. 2001, 276, 7246–7257. [Google Scholar]
- Sancak, Y.; Peterson, T.R.; Shaul, Y.D.; Lindquist, R.A.; Thoreen, C.C.; Bar-Peled, L.; Sabatini, D.M. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008, 320, 1496–1501. [Google Scholar]
- Hirose, E.; Nakashima, N.; Sekiguchi, T.; Nishimoto, T. RagA is a functional homologue of S. Cerevisiae Gtr1p involved in the Ran/Gsp1-GTPase pathway. J. Cell. Sci. 1998, 111, 11–21. [Google Scholar]
- Hong, S.-B.; Oh, H.; Valera, V.A.; Baba, M.; Schmidt, L.S.; Linehan, W.M. Inactivation of the FLCN tumor suppressor gene induces TFE3 transcriptional activity by increasing its nuclear localization. PLoS One 2010, 5, e15793. [Google Scholar]
- Durán, R.V.; Hall, M.N. Leucyl-tRNA synthetase: Double duty in amino acid sensing. Cell. Res. 2012, 22, 1207–1209. [Google Scholar]
- Han, J.M.; Jeong, S.J.; Park, M.C.; Kim, G.; Kwon, N.H.; Kim, H.K.; Ha, S.H.; Ryu, S.H.; Kim, S. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 2012, 149, 410–424. [Google Scholar]
- Hsieh, A. mTOR: The master regulator. Cell 2012, 149, 955–957. [Google Scholar]
- Kim, S.G.; Buel, G.R.; Blenis, J. Nutrient regulation of the mTOR complex 1 signaling pathway. Mol. Cells 2013, 35, 1–11. [Google Scholar]
- Kim, Y.M.; Kim, D.H. dRAGging amino acid-mTORC1 signaling by SH3BP4. Mol. Cells 2013, 35, 1–6. [Google Scholar]
- Kim, Y.M.; Stone, M.; Hwang, T.H.; Kim, Y.G.; Dunlevy, J.R.; Griffin, T.J.; Kim, D.H. SH3BP4 is a negative regulator of amino acid-Rag GTPase-mTORC1 signaling. Mol. Cell 2012, 46, 833–846. [Google Scholar]
- Shaw, R.J. GATORS take a bite out of mTOR. Science 2013, 340, 1056–1057. [Google Scholar]
- Bar-Peled, L.; Chantranupong, L.; Cherniack, A.D.; Chen, W.W.; Ottina, K.A.; Grabiner, B.C.; Spear, E.D.; Carter, S.L.; Meyerson, M.; Sabatini, D.M. A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 2013, 340, 1100–1106. [Google Scholar]
- Duran, A.; Amanchy, R.; Linares, J.F.; Joshi, J.; Abu-Baker, S.; Porollo, A.; Hansen, M.; Moscat, J.; Diaz-Meco, M.T. P62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol. Cell 2011, 44, 134–146. [Google Scholar]
- Bar-Peled, L.; Sabatini, D.M. Snapshot: mTORC1 signaling at the lysosomal surface. Cell 2012, 151, 1390–1390.e1. [Google Scholar]
- Jewell, J.L.; Guan, K.-L. Nutrient signaling to mTOR and cell growth. Trends Biochem. Sci. 2013, 38, 233–242. [Google Scholar]
- Yang, H.; Gong, R.; Xu, Y. Control of cell growth: Rag GTPases in activation of TORC1. Cell. Mol. Life Sci. 2013, 70, 2873–2875. [Google Scholar]
- Demetriades, C.; Doumpas, N.; Teleman, A.A. Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell. 2014, 156, 786–799. [Google Scholar]
- Zoncu, R.; Bar-Peled, L.; Efeyan, A.; Wang, S.; Sancak, Y.; Sabatini, D.M. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science 2011, 334, 678–683. [Google Scholar]
- Efeyan, A.; Zoncu, R.; Sabatini, D.M. Amino acids and mTORC1: From lysosomes to disease. Trends Mol. Med. 2012, 18, 524–533. [Google Scholar]
- Sagné, C.; Agulhon, C.; Ravassard, P.; Darmon, M.; Hamon, M.; El Mestikawy, S.; Gasnier, B.; Giros, B. Identification and characterization of a lysosomal transporter for small neutral amino acids. Proc. Natl. Acad. Sci. USA 2001, 98, 7206–7211. [Google Scholar]
- Goberdhan, D.C.; Meredith, D.; Boyd, C.R.; Wilson, C. PAT-related amino acid transporters regulate growth via a novel mechanism that does not require bulk transport of amino acids. Development 2005, 132, 2365–2375. [Google Scholar]
- Heublein, S.; Kazi, S.; Ögmundsdóttir, M.; Attwood, E.; Kala, S.; Boyd, C.; Wilson, C.; Goberdhan, D. Proton-assisted amino-acid transporters are conserved regulators of proliferation and amino-acid-dependent mTORC1 activation. Oncogene 2010, 29, 4068–4079. [Google Scholar]
- Ögmundsdóttir, M.H.; Heublein, S.; Kazi, S.; Reynolds, B.; Visvalingam, S.M.; Shaw, M.K.; Goberdhan, D.C. Proton-assisted amino acid transporter PAT1 complexes with Rag GTPases and activates TORC1 on late endosomal and lysosomal membranes. PLoS One 2012, 7, e36616. [Google Scholar]
- Ruivo, R.; Bellenchi, G.C.; Chen, X.; Zifarelli, G.; Sagné, C.; Debacker, C.; Pusch, M.; Supplisson, S.; Gasnier, B. Mechanism of proton/substrate coupling in the heptahelical lysosomal transporter cystinosin. Proc. Natl. Acad. Sci. USA 2012, 109, E210–E217. [Google Scholar]
- Byfield, M.P.; Murray, J.T.; Backer, J.M. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J. Biol. Chem. 2005, 280, 33076–33082. [Google Scholar]
- Nobukuni, T.; Joaquin, M.; Roccio, M.; Dann, S.G.; Kim, S.Y.; Gulati, P.; Byfield, M.P.; Backer, J.M.; Natt, F.; Bos, J.L. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc. Natl. Acad. Sci. USA 2005, 102, 14238–14243. [Google Scholar]
- Nobukuni, T.; Kozma, S.C.; Thomas, G. hVps34, an ancient player, enters a growing game: mTOR Complex1/S6K1 signaling. Curr. Opin. Cell. Biol. 2007, 19, 135–141. [Google Scholar]
- Zhou, X.; Takatoh, J.; Wang, F. The mammalian class 3 PI3K (PIK3C3) is required for early embryogenesis and cell proliferation. PLoS One 2011, 6, e16358. [Google Scholar]
- Gulati, P.; Gaspers, L.D.; Dann, S.G.; Joaquin, M.; Nobukuni, T.; Natt, F.; Kozma, S.C.; Thomas, A.P.; Thomas, G. Amino acids activate mTOR complex 1 via Ca2+/CaM signaling to hVps34. Cell. MeTable 2008, 7, 456–465. [Google Scholar]
- Yan, Y.; Flinn, R.; Wu, H.; Schnur, R.; Backer, J. hVps15, but not Ca2+/CaM, is required for the activity and regulation of hVps34 in mammalian cells. Biochem. J. 2009, 417, 747–755. [Google Scholar]
- Hodgkin, M.N.; Masson, M.R.; Powner, D.; Saqib, K.M.; Ponting, C.P.; Wakelam, M.J. Phospholipase D regulation and localisation is dependent upon a phosphatidylinositol 4,5-bisphosphate-specific PH domain. Curr. Biol. 2000, 10, 43–46. [Google Scholar]
- Stahelin, R.V.; Ananthanarayanan, B.; Blatner, N.R.; Singh, S.; Bruzik, K.S.; Murray, D.; Cho, W. Mechanism of membrane binding of the phospholipase D1 PX domain. J. Biol. Chem. 2004, 279, 54918–54926. [Google Scholar]
- Vanhaesebroeck, B.; Leevers, S.J.; Ahmadi, K.; Timms, J.; Katso, R.; Driscoll, P.C.; Woscholski, R.; Parker, P.J.; Waterfield, M.D. Synthesis and function of 3-phosphorylated inositol lipids. Annu. Rev. Biochem. 2001, 70, 535–602. [Google Scholar]
- Volinia, S.; Dhand, R.; Vanhaesebroeck, B.; MacDougall, L.; Stein, R.; Zvelebil, M.; Domin, J.; Panaretou, C.; Waterfield, M. A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. EMBO J. 1995, 14, 3339–3348. [Google Scholar]
- Chen, J.; Fang, Y. A novel pathway regulating the mammalian target of rapamycin (mTOR) signaling. Biochem. Pharmacol. 2002, 64, 1071–1077. [Google Scholar]
- Fang, Y.; Vilella-Bach, M.; Bachmann, R.; Flanigan, A.; Chen, J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Sci. Signal. 2001, 294, 1942–1950. [Google Scholar]
- Foster, D.A. Regulation of mTOR by phosphatidic acid? Cancer Res. 2007, 67, 1–4. [Google Scholar]
- Choi, J.; Chen, J.; Schreiber, S.L.; Clardy, J. Structure of the FKBP12-rapamycin complex interacting with binding domain of human FRAP. Science 1996, 273, 239–242. [Google Scholar]
- Yip, C.K.; Murata, K.; Walz, T.; Sabatini, D.M.; Kang, S.A. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol. Cell. 2010, 38, 768–774. [Google Scholar]
- Sun, Y.; Fang, Y.; Yoon, M.-S.; Zhang, C.; Roccio, M.; Zwartkruis, F.; Armstrong, M.; Brown, H.; Chen, J. Phospholipase D1 is an effector of Rheb in the mTOR pathway. Proc. Natl. Acad. Sci. USA 2008, 105, 8286–8291. [Google Scholar]
- Proud, C.G. mTOR, unleashed. Science 2007, 318, 926–927. [Google Scholar]
- Dunlop, E.A.; Dodd, K.M.; Seymour, L.A.; Tee, A.R. Mammalian target of rapamycin complex 1-mediated phosphorylation of eukaryotic initiation factor 4E-binding protein 1 requires multiple protein-protein interactions for substrate recognition. Cell. Signal. 2009, 21, 1073–1084. [Google Scholar]
- Wang, X.; Fonseca, B.D.; Tang, H.; Liu, R.; Elia, A.; Clemens, M.J.; Bommer, U.A.; Proud, C.G. Re-evaluating the roles of proposed modulators of mammalian target of rapamycin complex 1 (mTORC1) signaling. J. Biol. Chem. 2008, 283, 30482–30492. [Google Scholar]
- Maehama, T.; Tanaka, M.; Nishina, H.; Murakami, M.; Kanaho, Y.; Hanada, K. RalA functions as an indispensable signal mediator for the nutrient-sensing system. J. Biol. Chem. 2008, 283, 35053–35059. [Google Scholar]
- Sato, T.; Nakashima, A.; Guo, L.; Tamanoi, F. Specific activation of mTORC1 by Rheb G-protein in vitro involves enhanced recruitment of its substrate protein. J. Biol. Chem. 2009, 284, 12783–12791. [Google Scholar]
- Uhlenbrock, K.; Weiwad, M.; Wetzker, R.; Fischer, G.; Wittinghofer, A.; Rubio, I. Reassessment of the role of FKBP38 in the Rheb/mTORC1 pathway. FEBS Lett. 2009, 583, 965–970. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, X.; Liang, Y.; He, Q.; Yao, R.; Bao, W.; Bao, L.; Wang, Y.; Wang, Z. Current Models of Mammalian Target of Rapamycin Complex 1 (mTORC1) Activation by Growth Factors and Amino Acids. Int. J. Mol. Sci. 2014, 15, 20753-20769. https://doi.org/10.3390/ijms151120753
Zheng X, Liang Y, He Q, Yao R, Bao W, Bao L, Wang Y, Wang Z. Current Models of Mammalian Target of Rapamycin Complex 1 (mTORC1) Activation by Growth Factors and Amino Acids. International Journal of Molecular Sciences. 2014; 15(11):20753-20769. https://doi.org/10.3390/ijms151120753
Chicago/Turabian StyleZheng, Xu, Yan Liang, Qiburi He, Ruiyuan Yao, Wenlei Bao, Lili Bao, Yanfeng Wang, and Zhigang Wang. 2014. "Current Models of Mammalian Target of Rapamycin Complex 1 (mTORC1) Activation by Growth Factors and Amino Acids" International Journal of Molecular Sciences 15, no. 11: 20753-20769. https://doi.org/10.3390/ijms151120753
APA StyleZheng, X., Liang, Y., He, Q., Yao, R., Bao, W., Bao, L., Wang, Y., & Wang, Z. (2014). Current Models of Mammalian Target of Rapamycin Complex 1 (mTORC1) Activation by Growth Factors and Amino Acids. International Journal of Molecular Sciences, 15(11), 20753-20769. https://doi.org/10.3390/ijms151120753



