Abstract
Nail patella syndrome (NPS) is an autosomal dominant disorder characterized by nail malformations, patellar apoplasia, or patellar hypoplasia. Mutations within the LMX1B gene are found in 85% of families with NPS; thus, this gene has been characterized as the causative gene of NPS. In this study, we identified a heterozygous microdeletion of the entire LMX1B gene using multiplex ligation-dependent probe amplification (MLPA) in a Chinese family with NPS. The determination of the deletion breakpoints by Illumina genome-wide DNA analysis beadchip showed that the deletion was located in chromosome 9q33.3 and spanned about 0.66 Mb in size. This heterozygous deletion provides strong evidence for haploinsufficiency as the pathogenic mechanism of NPS.
1. Introduction
Nail patella syndrome (NPS; OMIM 161200) is an autosomal dominant disorder characterized by nail malformations, patellar apoplasia, or patellar hypoplasia. Additional skeletal abnormalities can be present that encompass the iliac horns, produce elbow dysplasia, cause progressive nephropathy, or produce primary open angle glaucoma; thus, it is apparent that the phenotype of this disease is variable among or within families [1,2,3,4,5,6].
In 1998, Dreyer et al. [7] showed that NPS is caused by mutations of the LMX1B gene. The involvement of this gene in NPS was subsequently confirmed by other studies [8,9]. LMX1B is one of the LIM-homeodomain proteins, which encode LIM-homeodomain transcription factors involved in pattern formation during development [10,11]. Previous studies have suggested that the LMX1B gene plays a pivotal role in the development of limb, kidney, eye, nervous system, as well as other organs or systems; these abnormalities are consistent with the phenotypes of NPS disease [7,8,12,13,14,15,16,17,18].
NPS is a rare hereditary disease with the incidence roughly estimated at 1 in 50,000 live births [19]. Mutations within the LMX1B gene have been detected in approximately 85% of families with NPS [1], including missense, nonsense, frameshift, splice-site mutations, small intragenic insertions/deletions, gross insertions/deletions, and complex rearrangements [7,8,9,20,21,22,23,24,25,26,27,28,29,9,20]. While most of the mutations were present in Caucasians, only a missense mutation c.742A>G (p.R248G) within the homeodomain of LXM1B has been reported to cause NPS in a Chinese family [30]. In this study, we first present the identification of a 0.66 Mb heterozygous microdeletion encompassing entire LMX1B and flanking the MVB12B and ZBTB43 genes in a Chinese family with NPS.
2. Results
2.1. Clinical Manifestations
There were no other clinical abnormalities in the proband except for nail hypoplasia and patellar dysplasia. The nail abnormalities of the proband were prominent on both thumbs and the right index finger. They primarily manifested as nail bed shortening and longitudinal ridging; in addition, a typical triangular lunula was clearly visible in the proband’s nails (Figure 1B). Nail abnormalities of the father were subtle; they just manifested as a triangular lunula at the base of the nail. Radiographic examination results of the proband showed severe bilateral patellar dysplasia as his patella was obviously subnormal in size, while his father showed slight bilateral hypoplastic patellae that were displaced superiorly (Figure 1C). All subjects evaluated had normal renal function. There were no abnormalities of facial features, short stature, or elbow contractures in our patients, and there were no clinical abnormalities in other family members. The chromosomal analysis of the proband and his father revealed a normal male karyotype: 46, XY. Paternity was further confirmed by genotype analysis.
2.2. Genetic Analysis
Two hemizygous synonymous variants, c.441A>G (p.E147) and c.726G>C (p.S242), were detected in the proband’s father by direct DNA sequence analysis, these genetic alterations passed on to the proband’s normal elder sister as they were also identified (apparently heterozygous) in his sister (Figure 2). Notably, these two point mutations were not identified in the proband and his mother by DNA sequencing. These sequence results suggest a haploinsufficiency of LMX1B as the father’s synonymous variants were not passed on to the proband.
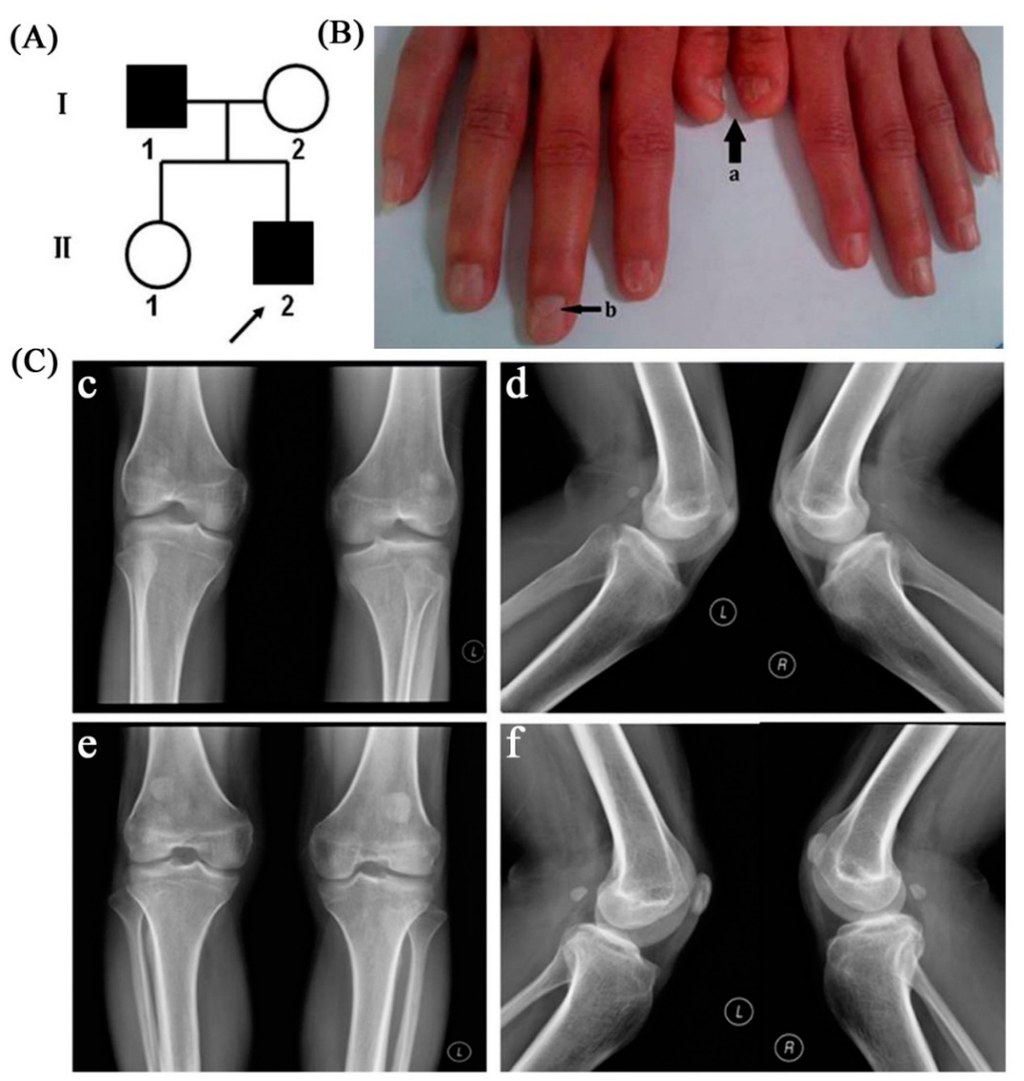
Figure 1.
(A) Chinese pedigree with nail patella syndrome; patients are indicated by solid black, denoting the proband; (B) Clinical manifestation of the proband’s nails (a: short nail bed with longitudinal ridging; b: triangular lunula at the base of the nail); (C) Radiographic examination results of patients’ knee joint. The radiographs of proband’s knee joint showed severe bilateral patellar dysplasia (c,d); The radiographs of the father showed bilateral hypoplastic, superiorly misplaced patellae (e,f).
This hypothesis was confirmed by the results of MLPA analysis, which showed a single-copy deletion of the entire LMX1B (exons 1 to 8) in the proband and his father (Figure 3 and Figure S1). MLPA failed to detect deletions in the coding sequence of LMX1B in the proband’s mother and elder sister (Figure S1). These results confirmed that haploinsufficiency of LMX1B gene was the genetic pathogenic mechanism of this NPS family.
The complete genome analysis beadchip from Illumina was used to determine the breakpoints of the segmental deletion. The evaluation indicated a heterozygous deletion spanning from 128,952,700 to 129,613,085 in 9q33.3, which demonstrated the deletion to be 0.66 Mb in size [31]. This segmental deletion included the whole LMX1B gene, encoding a LIM-homeobox transcription factor as being the causative gene of NPS. In addition, it contained MVB12B and ZBTB43 genes, which locate in the up and downstream of LMX1B, respectively (Figure 4).
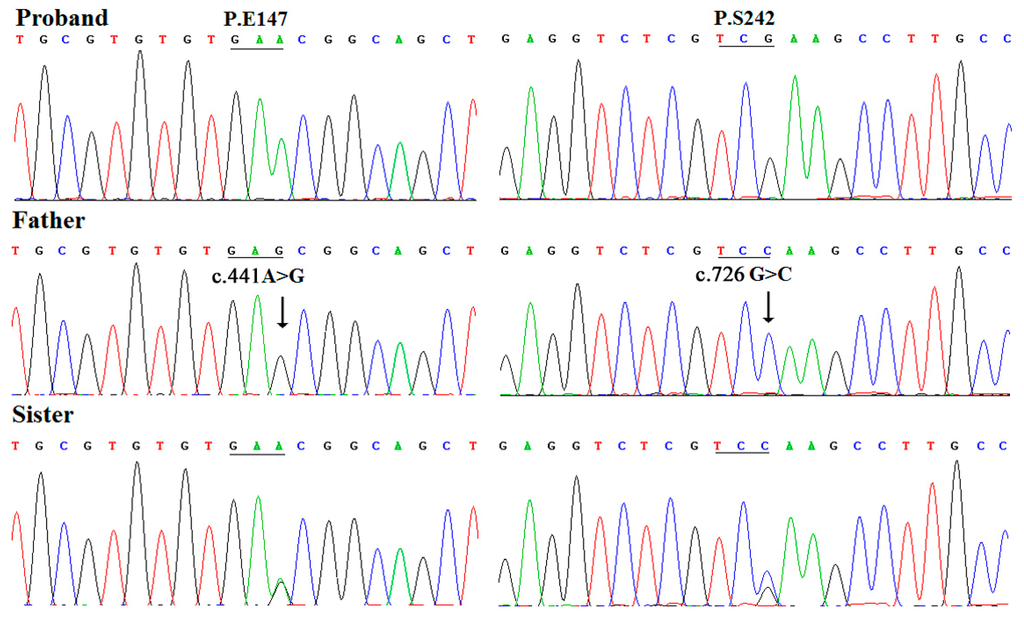
Figure 2.
Chromatography of synonymous mutations of LMX1B gene in the family. The proband, his father, and elder sister, were wild type, homozygous at the 441 locus and heterozygous at the 726 locus.
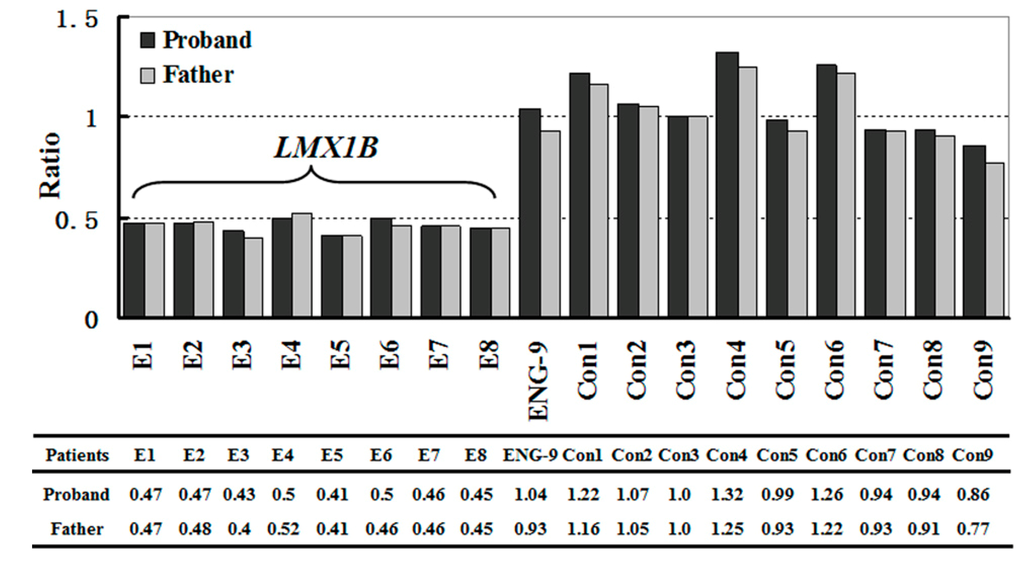
Figure 3.
Results of MLPA analysis. A single-copy deletion of the entire LMX1B was detected in the proband and his father.
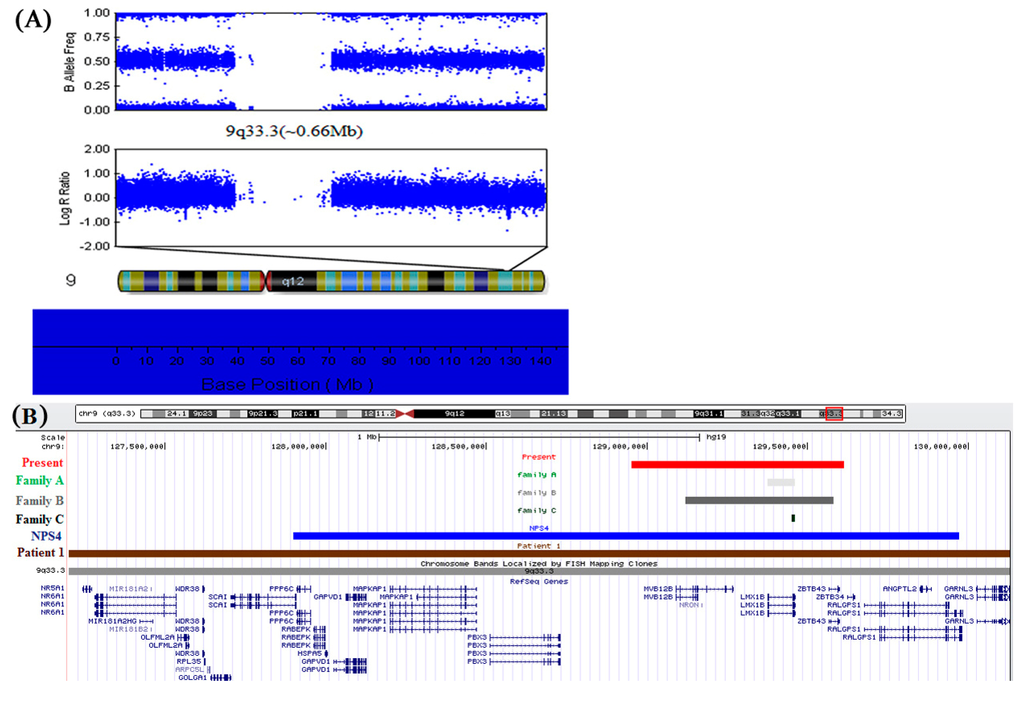
Figure 4.
(A) The complete genome analysis of the proband. A 0.66 Mb deletion in chromosome band 9q33.3, between 128,952,700 and 129,613,085 bp, which was detected by using an Illumina genome-wide DNA analysis beadchip; (B) Enlargement of the 9q33.3–q34.11 region from the UCSC genome browser shows a comparison between the deleted segments, cytogenetics bands and RefSeq genes. Patient 1 from Schlaubitz et al. [32]: brown bar. Patients of families A, B, and C from Bongers et al. [25]: light gray to black bars. Patient NPS4 from Marini et al. [22]: blue bar. Patient in present study: red bar.
3. Discussion
In this NPS family, patients displayed only nail and patellar dysplasia; no other clinical abnormalities were observed in the family. The nail abnormalities of the proband are prominent on both thumbs and the right index finger, primarily manifesting as nail bed shortening and longitudinal ridging; in addition, typical triangular lunula was clearly visible in the proband’s nails. Nail abnormalities of the father were subtle, just manifesting as a triangular lunula at the base of the nail. Radiographic examination of the proband showed severe bilateral patellar dysplasia, as his patellae were obviously subnormal in size, while his father showed slightly bilateral hypoplastic, higher than normal misplaced patella. The presence and severity of different NPS manifestations showed high variability at the individual, intrafamilial, and interfamilial levels. In addition to typical nail dysplasia and patellar apoplasia/hypoplasia, this disease could also manifest as elbow dysplasia, iliac horns, muscle dystrophy, progressive nephropathy, primary open angle glaucoma, attention deficit hyperactivity disorder, and symptoms of depressive disorder [1,2,3,4,33,34]. The phenotypic expression of NPS varies widely within and among families. This might be due to variable penetrance; however, other endogenous or environmental modifier factors could also be involved in the pathogenesis of this disease.
In the present study, two synonymous variants, c.441A>G (p.E147) and c.726G>C (p.S242), apparently hemizygous, were detected in the proband’s father and passed on to his elder sister without NPS. It is worth noting that these genetic alterations were not found in the proband and his mother by direct DNA sequence analysis. These results suggest that these synonymous substitutions could be single-nucleotide polymorphisms rather than pathogenic mutations and have no correlation with NPS [29]. Recently, the same synonymous mutation, c.726G>C (p.S242) of LMX1B, has been reported in a Korean Family with NPS [23]; the author could not demonstrate any segregation of this synonymous mutation with NPS. Our findings indicate that this genetic alteration of LMX1B was not pathogenic for this NPS family; thus, there must be other pathogenic mechanism for the observed phenomenon in this Korean family.
An increasing number of studies have attempted to elucidate the molecular pathogenic mechanism of NPS. In 1998, Dreyer et al. [7] demonstrated that NPS is the result of mutations within the LMX1B gene. Concurrently, Chen et al. [17] showed that LMX1B−/− mice exhibited limb and kidney defects similar to NPS. Moreover, Vollrath et al. [8] identified four mutations within LMX1B in four unrelated families with NPS and open-angle glaucoma (OAG). Since then, a large number of LMX1B mutations have been reported; however, no correlation in the range of severity of NPS symptoms has been reported among patients with missense, nonsense, frameshift, or splice mutations; furthermore, those with entire/partial gene deletions, strongly support haploinsufficiency for LMX1B as the mechanism of NPS [1,25]. This assumption is supported by the lack of any dominant-negative effect detected by in vitro experiments studying missense and truncation LMX1B mutations [30,35,36]. A study of LMX1B+/− mice showed diminished compensatory renal growth compared to the kidneys of LMX1B+/+ mice in which renal damage was induced by unilateral nephrectomy [18]. This result further supports the assumption that a critical dosage of LMX1B is critical for normal kidney development. The majority of mutations that have been identified are point mutations. Recently, Bongers et al. [25] identified two entire LMX1B gene deletions and one smaller partial LMX1B deletion (exons 3 to 8) in a series of eight unrelated Dutch families with classical features of NPS (Figure 4B and Table 1). Their finding strongly confirmed that loss of function is the main pathogenic mechanism of NPS in human. Marini et al. [22] and Schlaubitz et al. [32] identified two entire LMX1B gene deletions on chromosome 9q33.3–34.11 involving large regions (~2 and ~3.07 Mb) by using array-CGH (Figure 4B and Table 1). In addition to signs of NPS, both patients had facial anomalies, club feet, genital anomalies, and mental retardation. It is possible that other genes (except for LMX1B) deleted in these families could contribute to the etiopathogenesis of facial anomalies, club feet, genital anomalies, and mental retardation that were observed in these patients. In present study, a 0.66 Mb heterozygous microdeletion was identified in chromosome band 9q33.3 (128,952,700~129,613,085), encompassing the entire LMX1B and flanking MVB12B and ZBTB43 genes in a Chinese family. This 0.66 Mb heterozygous deletion was first reported in NPS patients. In 2008, Bongers et al. [25] identified three different deletions in a series of eight unrelated families with classical features of NPS in whom no pathogenic LMX1B mutation was found by sequence analysis, as shown in Figure 4B, a deletion of exons 3–8 of LMX1B was found in family C, Further determination of the size of the genomic microdeletions revealed a deletion of the whole LMX1B gene in family A, whereas a deletion of the entire LMX1B and flanking FAM125B and ZNF297B genes was shown in family B which was similar to that of our patients [25]. However, it is uncertain whether these two deletions are identical because the location of the probes Bongers et al. [25] used were different than ours. The deletion was about approximately 0.44 Mb in length according to their probes’ position. Moreover, Bongers et al. [25] reported families revealed renal and extrarenal symptoms while our patients displayed only nail and patellar dysplasia. Despite this difference, our research can still further confirm the deletion of entire LMX1B as the pathogenic mechanism underlying NPS.

Table 1.
LMX1B Deletions Reported.
| Deletion | Size | Phenotype | Reference |
|---|---|---|---|
| Entire LMX1B | ~0.66 Mb | NPS | Present study |
| Entire LMX1B | ~82 Kb | NPS | Family A [25] |
| Entire LMX1B | ~0.44 Mb | NPS | Family B [25] |
| Partial LMX1B (exon 3–8) | ~5.4 Kb | NPS | Family C [25] |
| Entire LMX1B | ~2 Mb | NPS, facial anomalies, club feet, mental retardation, genital anomalies | NPS4 [22] |
| Entire LMX1B | ~3.07 Mb | NPS, facial anomalies, club feet, mental retardation, genital anomalies | Patient 1 [32] |
4. Experimental Section
4.1. Subjects and Clinical Evaluation
This is a small family comprised of four members (Figure 1A). The proband is a 27-year-old-man who presented at our genetic clinic for nail hypoplasia. The proband’s father is also affected, while his mother and elder sister are normal. Detailed history and physical examination were carried out. Knee joints of the patients were assessed by radiographic examination. Cytogenetic analysis was performed to exclude a karyotype abnormality. Renal function was assessed by urinalysis and blood tests.
4.2. Sequencing of Genomic DNA
Genomic DNA was extracted from peripheral blood leukocytes using a DNA extraction kit (Watson Biotechnologies Inc., Shanghai, China), after obtaining informed consent. This experiment was approved by the ethical committee. Exons 1–8 of LMX1B were screened for mutations by DNA sequencing. Briefly, genomic DNA was amplified by PCR using the pair of primers (Table 2). PCR amplification was performed in 25 μL reaction volumes, containing 50 ng genomic DNA, 1× PCR buffer, 2× GC buffer, and 1 μM of each dNTP, as well as 1.5 μM·MgCl2 and 0.5 U Taq DNA polymerase (Takara, Dalian, China). After an initial denaturation at 94 °C for 5 min, the reactions were amplified for 35 cycles with denaturation at 94 °C for 45 s annealing at 61–68 °C for 45 s, and extension at 72 °C for 1 min; this was followed by a final extension at 72 °C for 10 min. DNA fragments were purified and subsequently sequenced and analyzed by the ABI PRISM 3730 DNA Analyzer (Applied Biosystems by Life Technologies., Carlsbad, CA, USA). The sequence data were analyzed by aligning with the reference sequences in NCBI (NC_000009 for LMX1B) using the DNAStar 5.0 (DNAStar., Madison, WI, USA) and BioEdit (Micro Focus., London, UK) software. Mutations or polymorphisms were identified according to the reference sequences.

Table 2.
Primer Sequences for LMX1B Amplification from Human Genomic DNA.
| Exon | Sense Primer | Antisense Primer | Product Size (bp) | Reference |
|---|---|---|---|---|
| 1 | TGACAAGCAGGTGACAGAGGA | CTGGCGATCACTCCAGGAGT | 558 | [5] |
| 2 | CCGAGGACTGGGACGGACTA | CTCTCGGAACCCTTGGAGCT | 513 | [5] |
| 3 | GGCAGGAGTGGCCTCTG | TCCAGGACACCCCAGCAAC | 359 | [6] |
| 4 + 5 + 6 | CCACGGCAGGTGTCAACAGA | GATGGCCTTGGTGGAAGGCT | 1005 | [5] |
| 7 + 8 | CTGAGCCTGGAGGAGGAGCT | GGGCACCGTATGGCTGT | 1115 | [5,7] |
4.3. Multiplex Ligation-Dependent Probe Amplification (MLPA) Analysis
MLPA analysis was performed on the family members and two normal controls to identify large gene deletions or duplications in the LMX1B gene using the SALSA MLPA kit (P289-A2 LMX1B; MRC Holland, Amsterdam, The Netherlands). The P289-A2 LMX1B probemix contains 18 MLPA probes, including 8 probes for all exons of the LMX1B gene (exons 1–5, 6a, 7a and 8), 1 probe for ENG gene located on 9q34 and 9 reference probes, which were added to detect several different autosomal chromosomal locations. Hybridization, ligation, and amplification were performed according to the manufacturer’s protocol. Amplification products were detected using an ABI PRISM 3730 DNA Analyzer (Applied Biosystems by Life Technologies., Carlsbad, CA, USA) with LIZ500 (Applied Biosystems) as an internal size standard. The raw data were analyzed by using Coffalyser MLPA data analysis software (MRC Holland., Amsterdam, The Netherlands).
4.4. Whole Genome Copy Number Analysis
The IlluminaHumanOmniZhongHua-8 BeadChip (Illumina Inc., San Diego, CA, USA) was further used to determine the size of the sequence deletion in chromosome 9. The test was performed at Hunan Jiahui Genetics Hospital, Changsha, China. Experiments were conducted according to manufacturer’s protocol. Briefly, ~200 ng DNA was amplified, fragmented and hybridized onto the beadchip. After labeling, the beadchip was scanned using an Illumina BeadArray™ Reader (Illumina Inc., San Diego, CA, USA). Data were analyzed using the GenomeStudio software package (Illumina Inc., San Diego, CA, USA).
5. Conclusions
In this study, we identified a 0.66 Mb heterozygous microdeletion in chromosome band 9q33.3 encompassing the entire LMX1B gene and flanking MVB12B and ZBTB43 genes in a Chinese family. This is the first report of a 0.66 Mb heterozygous microdeletion containing an entire LMX1B in NPS patients, which further confirmed the hypothesis that haploinsufficiency of LMX1B is the principal pathogenic mechanism of NPS in human.
Supplementary Files
Supplementary File 1Acknowledgments
We would like to thank the members of the family for their kind cooperation in this study.
Author Contributions
Rong He and Yanyan Zhao conceived and designed the experiments; Shujuan Jiang, Jiubin Zhang and Dan Huang performed the experiments; Yuanyuan Zhang and Yinzhao Wang analyzed the data; Xiaoliang Liu contributed reagents/materials/analysis tools; and Shujuan Jiang wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- McIntosh, I.; Dunston, J.A.; Liu, L.; Hoover-Fong, J.E.; Sweeney, E. Nail patella syndrome revisited: 50 years after linkage. Ann. Hum. Genet. 2005, 69, 349–363. [Google Scholar] [CrossRef]
- Hawkins, C.F.; Smith, O.E. Renal dysplasia in a family with multiple hereditary abnormalities including iliac horns. Lancet 1950, 1, 803–808. [Google Scholar] [CrossRef] [PubMed]
- Mimiwati, Z.; Mackey, D.A.; Craig, J.E.; Mackinnon, J.R.; Rait, J.L.; Liebelt, J.E.; Ayala-Lugo, R.; Vollrath, D.; Richards, J.E. Nail patella syndrome and its association with glaucoma: A review of eight families. Br. J. Ophthalmol. 2006, 90, 1505–1509. [Google Scholar] [CrossRef] [PubMed]
- Milla, E.; Hernan, I.; Gamundi, M.J.; Martinez-Gimeno, M.; Carballo, M. Novel LMX1B mutation in familial Nail patella syndrome with variable expression of open angle glaucoma. Mol. Vis. 2007, 13, 639–648. [Google Scholar] [PubMed]
- Lee, B.H.; Cho, T.J.; Choi, H.J.; Kang, H.K.; Lim, I.S.; Park, Y.H.; Ha, I.S.; Choi, Y.; Cheong, H.I. Clinico-genetic study of nail patella syndrome. J. Korean Med. Sci. 2009, 24, S82–S86. [Google Scholar]
- Romero, P.; Sanhueza, F.; Lopez, P.; Reyes, L.; Herrera, L. c.194 A>C (Q65P) mutation in the LMX1B gene in patients with nail patella syndrome associated with glaucoma. Mol. Vis. 2011, 17, 1929–1939. [Google Scholar] [PubMed]
- Dreyer, S.D.; Zhou, G.; Baldini, A.; Winterpacht, A.; Zabel, B.; Cole, W.; Johnson, R.L.; Lee, B. Mutations in LMX1B cause abnormal skeletal patterning and renal dysplasia in nail patella syndrome. Nat. Genet. 1998, 19, 47–50. [Google Scholar] [CrossRef] [PubMed]
- Vollrath, D.; Jaramillo-Babb, V.L.; Clough, M.V.; McIntosh, I.; Scott, K.M.; Lichter, P.R.; Richards, J.E. Loss-of-function mutations in the LIM-homeodomain gene, LMX1B, in nail patella syndrome. Hum. Mol. Genet. 1998, 7, 1091–1098. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, I.; Dreyer, S.D.; Clough, M.V.; Dunston, J.A.; Eyaid, W.; Roig, C.M.; Montgomery, T.; Ala-Mello, S.; Kaitila, I.; Winterpacht, A.; et al. Mutation analysis of LMX1B gene in nail patella syndrome patients. Am. J. Hum. Genet. 1998, 63, 1651–1658. [Google Scholar]
- Iannotti, C.A.; Inoue, H.; Bernal, E.; Aoki, M.; Liu, L.; Donis-Keller, H.; German, M.S.; Permutt, M.A. Identification of a human LMX1 (LMX1.1)-related gene, LMX1.2: Tissue-specific expression and linkage mapping on chromosome 9. Genomics 1997, 46, 520–524. [Google Scholar]
- Curtiss, J.; Heilig, J.S. DeLIMiting development. Bioessays 1998, 20, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Song, N.N.; Xiu, J.B.; Huang, Y.; Chen, J.Y.; Zhang, L.; Gutknecht, L.; Lesch, K.P.; Li, H.; Ding, Y.Q. Adult raphe-specific deletion of LMX1b leads to central serotonin deficiency. PLoS One 2011, 6, e15998. [Google Scholar]
- Vogel, A.; Rodriguez, C.; Warnken, W.; Izpisua, B.J. Dorsal cell fate specified by chick Lmx1 during vertebrate limb development. Nature 1995, 378, 716–720. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.Q.; Yin, J.; Kania, A.; Zhao, Z.Q.; Johnson, R.L.; Chen, Z.F. LMX1b controls the differentiation and migration of the superficial dorsal horn neurons of the spinal cord. Development 2004, 131, 3693–3703. [Google Scholar] [CrossRef] [PubMed]
- Rohr, C.; Prestel, J.; Heidet, L.; Hosser, H.; Kriz, W.; Johnson, R.L.; Antignac, C.; Witzgall, R. The LIM-homeodomain transcription factor LMX1b plays a crucial role in podocytes. J. Clin. Investig. 2002, 109, 1073–1082. [Google Scholar] [CrossRef] [PubMed]
- Xiang, C.; Zhang, K.H.; Yin, J.; Arends, J.J.; Erzurumlu, R.S.; Jacquin, M.F.; Chen, Z.F. The transcription factor, LMX1b, is necessary for the development of the principal trigeminal nucleus-based lemniscal pathway. Mol. Cell. Neurosci. 2010, 44, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Lun, Y.; Ovchinnikov, D.; Kokubo, H.; Oberg, K.C.; Pepicelli, C.V.; Gan, L.; Lee, B.; Johnson, R.L. Limb and kidney defects in LMX1b mutant mice suggest an involvement of LMX1B in human nail patella syndrome. Nat. Genet. 1998, 19, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Endele, S.; Klein, S.; Richter, S.; Molter, T.; Amann, K.; Klanke, B.; Witzgall, R.; Johnson, R.L.; Hilgers, K.F.; Winterpacht, A. Renal phenotype in heterozygous LMX1b knockout mice (LMX1b+/−) after unilateral nephrectomy. Transgen. Res. 2007, 16, 723–729. [Google Scholar] [CrossRef]
- Sweeney, E.; Fryer, A.; Mountford, R.; Green, A.; McIntosh, I. Nail patella syndrome: A review of the phenotype aided by developmental biology. J. Med. Genet. 2003, 40, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Clough, M.V.; Hamlington, J.D.; McIntosh, I. Restricted distribution of loss-of-function mutations within the LMX1B genes of nail patella syndrome patients. Hum. Mutat. 1999, 14, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Oshimo, T.; Fukai, K.; Higashi, N.; Kitano, T.; Imai, Y.; Shintaku, H.; Ishii, M. A novel LMX1B nonsense mutation in a family with nail patella syndrome. J. Dermatol. Sci. 2008, 52, 57–60. [Google Scholar] [PubMed]
- Marini, M.; Bocciardi, R.; Gimelli, S.; di Duca, M.; Divizia, M.T.; Baban, A.; Gaspar, H.; Mammi, I.; Garavelli, L.; Cerone, R.; et al. A spectrum of LMX1B mutations in nail patella syndrome: New point mutations, deletion, and evidence of mosaicism in unaffected parents. Genet. Med. 2010, 12, 431–439. [Google Scholar]
- Ham, J.H.; Shin, S.J.; Joo, K.R.; Park, S.M.; Sung, H.Y.; Kim, J.S.; Choi, J.S.; Choi, Y.J.; Song, H.C.; Choi, E.J. A synonymous genetic alteration of LMX1B in a family with nail patella syndrome. Korean J. Intern. Med. 2009, 24, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Bongers, E.M.; Huysmans, F.T.; Levtchenko, E.; de Rooy, J.W.; Blickman, J.G.; Admiraal, R.J.; Huygen, P.L.; Cruysberg, J.R.; Toolens, P.A.; Prins, J.B.; et al. Genotype-phenotype studies in nail patella syndrome show that LMX1B mutation location is involved in the risk of developing nephropathy. Eur. J. Hum. Genet. 2005, 13, 935–946. [Google Scholar]
- Bongers, E.M.; de Wijs, I.J.; Marcelis, C.; Hoefsloot, L.H.; Knoers, N.V. Identification of entire LMX1B gene deletions in nail patella syndrome: Evidence for haploinsufficiency as the main pathogenic mechanism underlying dominant inheritance in man. Eur. J. Hum. Genet. 2008, 16, 1240–1244. [Google Scholar] [CrossRef] [PubMed]
- Seri, M.; Melchionda, S.; Dreyer, S.; Marini, M.; Carella, M.; Cusano, R.; Piemontese, M.R.; Caroli, F.; Silengo, M.; Zelante, L.; et al. Identification of LMX1B gene point mutations in Italian patients affected with nail patella syndrome. Int. J. Mol. Med. 1999, 4, 285–290. [Google Scholar]
- Bongers, E.M.; Gubler, M.C.; Knoers, N.V. Nail patella syndrome. Overview on clinical and molecular findings. Pediatr. Nephrol. 2002, 17, 703–712. [Google Scholar]
- Knoers, N.V.; Bongers, E.M.; van Beersum, S.E.; Lommen, E.J.; van Bokhoven, H.; Hol, F.A. Nail patella syndrome: Identification of mutations in the LMX1B gene in Dutch families. J. Am. Soc. Nephrol. 2000, 11, 1762–1766. [Google Scholar] [PubMed]
- Hamlington, J.D.; Jones, C.; McIntosh, I. Twenty-two novel LMX1B mutations identified in nail patella syndrome (NPS) patients. Hum. Mutat. 2001, 18, 458. [Google Scholar] [CrossRef]
- Lin, Y.; Zhao, J.; Chen, S.; Zeng, X.; Du, Q.; Yang, Y.; Lu, F.; Pu, Y.; Yang, Z. A novel mutation in LMX1B gene causes nail patella syndrome in a large Chinese family. Bone 2008, 43, 591–595. [Google Scholar] [CrossRef] [PubMed]
- UCSC Genome Browser on Human Feb. 2009 (GRCh37/hg19) Assembly. Available online: http://genome.ucsc.edu (accessed on 1 April 2014).
- Schlaubitz, S.; Yatsenko, S.A.; Smith, L.D.; Keller, K.L.; Vissers, L.E.; Scott, D.A.; Cai, W.W.; Reardon, W.; Abdul-Rahman, O.A.; Lammer, E.J.; et al. Ovotestes and XY sex reversal in a female with an interstitial 9q33.3–q34.1 deletion encompassing NR5A1 and LMX1B causing features of Genitopatellar syndrome. Am. J. Med. Genet. A 2007, 143, 1071–1081. [Google Scholar]
- Lichter, P.R.; Richards, J.E.; Downs, C.A.; Stringham, H.M.; Boehnke, M.; Farley, F.A. Cosegregation of open-angle glaucoma and the nail patella syndrome. Am. J. Ophthalmol. 1997, 124, 506–515. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Arvizu, C.; Sparrow, E.P.; Strube, M.J.; Slavin, C.; de Oleo, C.; James, J.; Hoover-Fong, J.; McIntosh, I.; Tierney, E. Increased symptoms of attention deficit hyperactivity disorder and major depressive disorder symptoms in nail patella syndrome: Potential association with LMX1B loss-of-function. Am. J. Med. Genet. B 2011, 156B, 59–66. [Google Scholar] [CrossRef]
- Sato, U.; Kitanaka, S.; Sekine, T.; Takahashi, S.; Ashida, A.; Igarashi, T. Functional characterization of LMX1B mutations associated with nail patella syndrome. Pediatr. Res. 2005, 57, 783–788. [Google Scholar] [CrossRef] [PubMed]
- Isojima, T.; Harita, Y.; Furuyama, M.; Sugawara, N.; Ishizuka, K.; Horita, S.; Kajiho, Y.; Miura, K.; Igarashi, T.; Hattori, M.; et al. LMX1B mutation with residual transcriptional activity as a cause of isolated glomerulopathy. Nephrol. Dial. Transplant. 2014, 29, 81–88. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).