Comparative Analysis of Mafriwal (Bos taurus × Bos indicus) and Kedah Kelantan (Bos indicus) Sperm Proteome Identifies Sperm Proteins Potentially Responsible for Higher Fertility in a Tropical Climate
Abstract
:1. Introduction
2. Results and Discussion
2.1. 2DGE, Protein Identification by LC MS/MS Q-TOF and GO Analysis
2.2. Protein Network Analysis
2.3. Proteins with Functions Related to Fertility
2.4. Studies on Sperm Motility
3. Experimental Section
3.1. Semen Sample
3.2. Protein Extraction
3.3. Two-Dimensional Gel Electrophoresis (2DGE)
3.4. LC-MS/MS Q-TOF Protein Identification
3.4.1. In-Gel Tryptic Digestion
3.4.2. Mass Spectrometry
3.4.3. Database Searching
3.4.4. Bioinformatics Analysis
3.4.5. Sperm Motility Test by Computer Assisted Semen Analysis (CASA) System
4. Conclusions
Acknowledgments
Conflict of Interest
References
- MIDA Food Industry Malaysian Industrial Development Authority (MIDA). Available online: http://www.mida.gov.my (accessed on 5 January 2013).
- Lehr, H.; Chang, K.W. Malaysian Milk Production. Available online: http://www.scribd.com/doc/43716997/Milk-Production-in-Malaysia (accessed on 15 December 2012).
- Panandam, J.M.; Raymond, A.K. Development of the Mafriwal dairy cattle of Malaysia. Available online: http://agtr.ilri.cgiar.org/casestudy/jothi/Jothi.htm (accessed on 6 February 2013).
- Perea-Ganchou, F.; Belloso, E.; Stagnaro, C.; Castillo, G.; Fonseca, H. Factors affecting fertility according to the postpartum period in crossbred dual-purpose suckling cows in the tropics. Trop. Anim. Health Prod 2005, 37, 559–572. [Google Scholar]
- Dobson, H.; Tebble, J.E.; Smith, R.F.; Ward, W.R. Is stress really all that important? Theriogenology 2001, 55, 65–73. [Google Scholar]
- Nissim, S. Effects of heat stress on the welfare of extensively managed domestic ruminants. Livest. Prod. Sci 2000, 67, 1–18. [Google Scholar]
- Rensis, F.D.; Scaramuzzi, R.J. Heat stress and seasonal effects on reproduction in the dairy cow—A review. Theriogenology 2003, 60, 1139–1151. [Google Scholar]
- Nichi, M.; Bols, P.E.J.; Züge, R.M.; Barnabe, V.H.; Goovaerts, I.G.F.; Barnabe, R.C.; Cortada, C.N.M. Seasonal variation in semen quality in Bos indicus and Bos taurus bulls raised under tropical conditions. Theriogenology 2006, 66, 822–828. [Google Scholar]
- Rahman, M.B.; Vandaele, L.; Rijsselaere, T.; Maes, D.; Hoogewijs, M.; Frijters, A.; Noordman, J.; Granados, A.; Dernelle, E.; Shamsuddin, M.; et al. Scrotal insulation and its relationship to abnormal morphology, chromatin protamination and nuclear shape of spermatozoa in Holstein-Friesian and Belgian Blue bulls. Theriogenology 2011, 76, 1246–1257. [Google Scholar]
- Fernandes, C.E.; Dode, M.A.N.; Pereira, D.; Silva, A.E.D.F. Effects of scrotal insulation in Nellore bulls (Bos taurus indicus) on seminal quality and its relationship with in vitro fertilizing ability. Theriogenology 2008, 70, 1560–1568. [Google Scholar]
- Cammack, K.M.; Mesa, H.; Lamberson, W.R. Genetic variation in fertility of heat-stressed male mice. Theriogenology 2006, 66, 2195–2201. [Google Scholar]
- Hansen, P.J. Physiological and cellular adaptations of zebu cattle to thermal stress. Anim. Reprod. Sci. 2004, 82–83, 349–360. [Google Scholar]
- Brito, L.F.C.; Silva, A.E.D.F.; Barbosa, R.T.; Kastelic, J.P. Testicular thermoregulation in Bos indicus, crossbred and Bos taurus bulls: Relationship with scrotal, testicular vascular cone and testicular morphology, and effects on semen quality and sperm production. Theriogenology 2004, 61, 511–528. [Google Scholar]
- Von Mering, C.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res 2003, 31, 258–261. [Google Scholar]
- HUGO Gene Nomenclature Committee. Available online: http://www.genenames.org (accessed on 5 January 2013).
- Muratori, M.; Luconi, M.; Marchiani, S.; Forti, G.; Baldi, E. Molecular markers of human sperm functions. Int. J. Androl 2009, 32, 25–45. [Google Scholar]
- Turner, R.M.O.; Eriksson, R.L.M.; Gerton, G.L.; Moss, S.B. Relationship between sperm motility and the processing and tyrosine phosphorylation of two human sperm fibrous sheath proteins, pro-hAKAP82 and hAKAP82. Mol. Hum. Reprod 1999, 5, 816–824. [Google Scholar]
- Liao, T.-T.; Xiang, Z.; Zhu, W.-B.; Fan, L.-Q. Proteome analysis of round-headed and normal spermatozoa by 2-D fluorescence difference gel electrophoresis and mass spectrometry. Asian J. Androl 2009, 11, 683–693. [Google Scholar]
- Lefievre, L.; Chen, Y.; Conner, S.J.; Scott, J.L.; Publicover, S.J.; Ford, W.C.L.; Barratt, C.L.R. Human spermatozoa contain multiple targets for protein S-nitrosylation: An alternative mechanism of the modulation of sperm function by nitric oxide? Proteomics 2007, 7, 3066–3084. [Google Scholar]
- Chan, C.C.; Shui, H.A.; Wu, C.H.; Wang, C.Y.; Sun, G.H.; Chen, H.M.; Wu, G.J. Motility and protein phosphorylation in healthy and asthenozoospermic sperm. J. Proteome Res 2009, 8, 5382–5386. [Google Scholar]
- Lee, W.-H.; Choi, J.-S.; Byun, M.-R.; Koo, K.-T.; Shin, S.; Lee, S.-K.; Surh, Y.-J. Functional inactivation of triosephosphate isomerase through phosphorylation during etoposide-induced apoptosis in HeLa cells: Potential role of Cdk2. Toxicology 2010, 278, 224–228. [Google Scholar]
- Siva, A.B.; Kameshwari, D.B.; Singh, V.; Pavani, K.; Sundaram, C.S.; Rangaraj, N.; Deenadayal, M.; Shivaji, S. Proteomics-based study on asthenozoospermia: Differential expression of proteasome alpha complex. Mol. Hum. Reprod 2010, 16, 452–462. [Google Scholar]
- Olshan, A.F.; Luben, T.J.; Hanley, N.M.; Perreault, S.D.; Chan, R.L.; Herring, A.H.; Basta, P.V.; DeMarini, D.M. Preliminary examination of polymorphisms of GSTM1, GSTT1, and GSTZ1 in relation to semen quality. Mutat. Res. Fundam. Mol. Mech. Mutagen 2010, 688, 41–46. [Google Scholar]
- Shamsi, M.B.; Venkatesh, S.; Tanwar, M.; Talwar, P.; Sharma, R.K.; Dhawan, A.; Kumar, R.; Gupta, N.P.; Malhotra, N.; Singh, N.; et al. DNA integrity and semen quality in men with low seminal antioxidant levels. Mutat. Res. Fundam. Mol. Mech. Mutagen 2009, 665, 29–36. [Google Scholar]
- Agnew, L.L.; Colditz, I.G. Development of a method of measuring cellular stress in cattle and sheep. Vet. Immunol. Immunopathol 2008, 123, 197–204. [Google Scholar]
- Lachance, C.; Fortier, M.; Thimon, V.; Sullivan, R.; Bailey, J.L.; Leclerc, P. Localization of Hsp60 and Grp78 in the human testis, epididymis and mature spermatozoa. Int. J. Androl 2010, 33, 33–44. [Google Scholar]
- Martinez-Heredia, J.; de Mateo, S.; Vidal-Taboada, J.M.; Ballesca, J.L.; Oliva, R. Identification of proteomic differences in asthenozoospermic sperm samples. Hum. Reprod 2008, 23, 783–791. [Google Scholar]
- Nixon, B.; Aitken, R.J.; McLaughlin, E.A. New insights into the molecular mechanisms of sperm-egg interaction. Cell. Mol. Life Sci 2007, 64, 1805–1823. [Google Scholar]
- Inoue, N.; Ikawa, M.; Isotani, A.; Okabe, M. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature 2005, 434, 234–238. [Google Scholar]
- Toshimori, K. Dynamics of the Acrosome. In Dynamics of the Mammalian Sperm Head; Springer: Berlin, Germany, 2009; Volume 204, pp. 31–41. [Google Scholar]
- Cesari, A.; Cacciato, C.S.; de Castro, R.E.; Sánchez, J.J. Partial purification and characterization of a trypsin-like serine protease from bovine sperm. Int. J. Androl 2004, 27, 311–318. [Google Scholar]
- Coates, P.J.; Nenutil, R.; McGregor, A.; Picksley, S.M.; Crouch, D.H.; Hall, P.A.; Wright, E.G. Mammalian prohibitin proteins respond to mitochondrial stress and decrease during cellular senescence. Exp.Cell Res 2001, 265, 262–273. [Google Scholar]
- Ou, J.-X.; Huang, S.-F.; Chen, H. Research progress in the structure and functions of prohibitin. J. Reprod. Contracept 2010, 21, 117–124. [Google Scholar]
- Wang, M.-J.; Ou, J.-X.; Chen, G.-W.; Wu, J.-P.; Shi, H.-J.; O, W.-S.; Martin-DeLeon, P.A.; Chen, H. Does prohibitin expression regulate sperm mitochondrial membrane potential, sperm motility, and male fertility? Antioxid. Redox Signal 2012, 17, 513–519. [Google Scholar]
- Berkelman, T. Quantitation of Protein in Samples Prepared for 2-D Electrophoresis. In 2D PAGE: Sample Preparation and Fractionation; Posch, A., Ed.; Humana Press: Totowa, NJ, USA, 2008; Volume 1, pp. 43–49. [Google Scholar]
- Yan, J.X.; Wait, R.; Berkelman, T.; Harry, R.A.; Westbrook, J.A.; Wheeler, C.H.; Dunn, M.J. A modified silver staining protocol for visualization of proteins compatible with matrix-assisted laser desorption/ionization and electrospray ionization-mass spectrometry. Electrophoresis 2000, 21, 3666–3672. [Google Scholar]
- Dahlan, H.M.; Karsani, S.A.; Rahman, M.A.; Hamid, N.A.A.; Top, A.G.M.; Ngah, W.Z.W. Proteomic analysis reveals that treatment with tocotrienols reverses the effect of H2O2 exposure on peroxiredoxin expression in human lymphocytes from young and old individuals. J. Nutr. Biochem 2012, 23, 741–751. [Google Scholar]
- Tan, E.C.; Karsani, S.A.; Foo, G.T.; Wong, S.M.; Rahman, N.A.; Khalid, N.; Othman, S.; Yusof, R. Proteomic analysis of cell suspension cultures of Boesenbergia rotunda induced by phenylalanine: Identification of proteins involved in flavonoid and phenylpropanoid biosynthesis pathways. Plant Cell Tissue Organ 2012, 111, 219–229. [Google Scholar]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar]
- Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database v9.05. Available online: http://www.string-db.org (accessed on 1 February 2013).

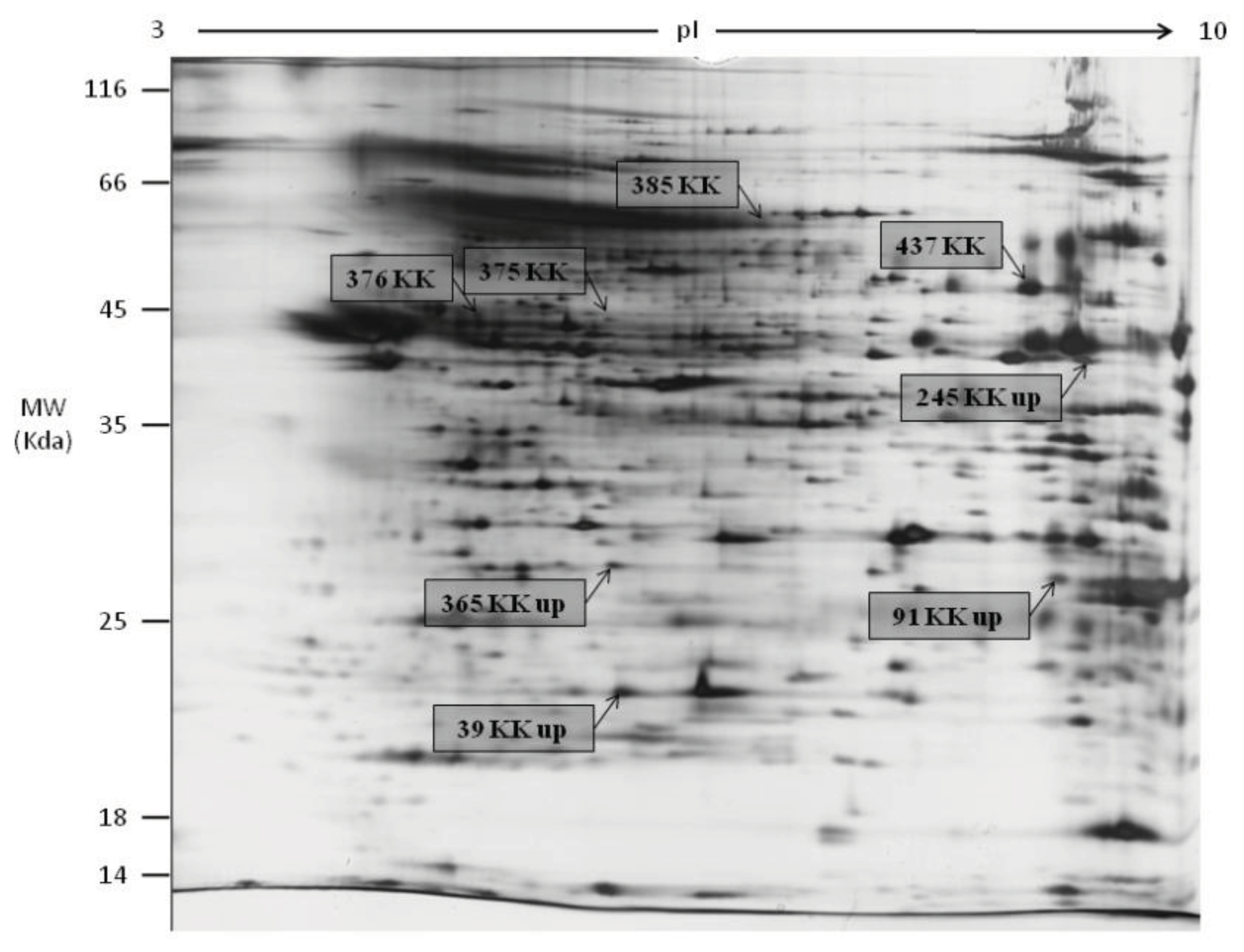
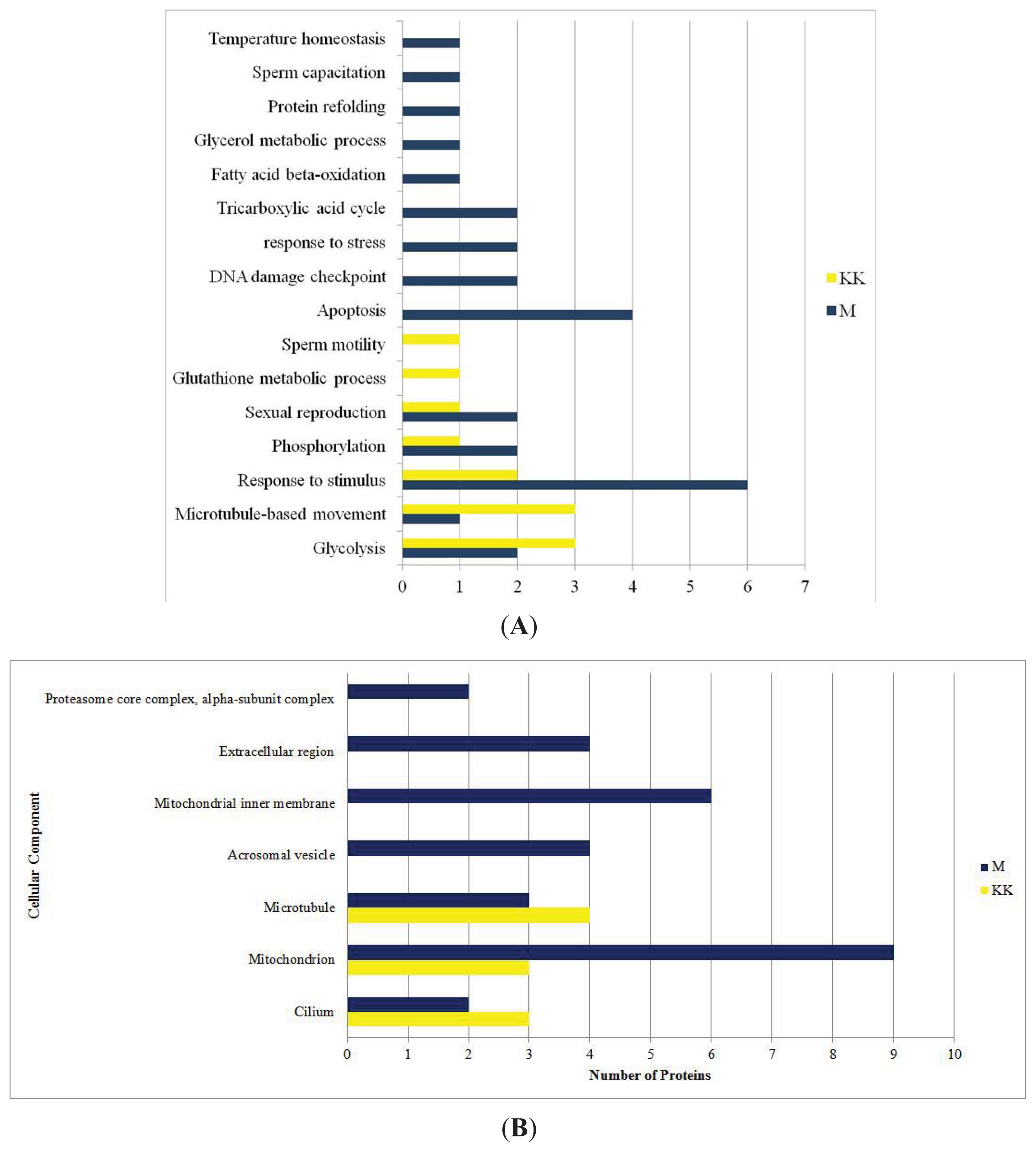
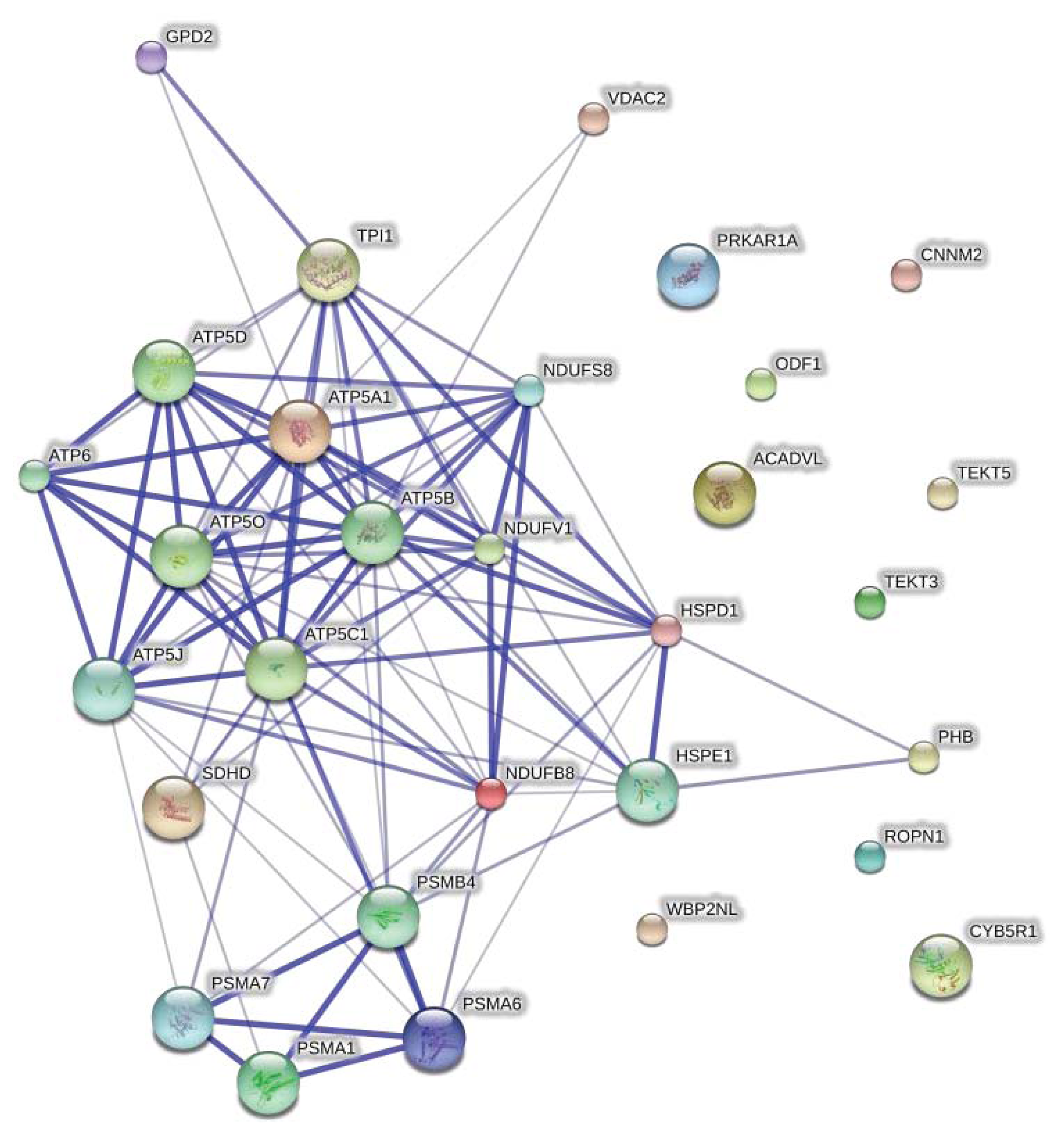
| Ascension number | Full name | Species | Score | Coverage (%) (number of peptides) | Remarkes | Fold change | Spot ID |
|---|---|---|---|---|---|---|---|
| Q2TA16 | Coiled-coil domain-containing protein 65 | Bos taurus | 73.7 | 21 (13) | KK specific | - | 375 |
| O35774 | A-kinase anchor protein 4 | Rattus norvegicus | 110.8 | 17 (16) | KK specific isoform | - | 385 |
| Q2KJE5 | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific | Bos taurus | 76.8 | 14 (5) | KK specific isoform | - | 437 |
| P81947 | Tubulin alpha-1B chain | Bos taurus | 17.8 | 6 (2) | KK specific isoform | - | 376 |
| Q2HJ86 | Tubulin alpha-1D chain | Bos taurus | 17.8 | 6 (2) | KK specific isoform | - | 376 |
| Q32KN8 | Tubulin alpha-3 chain | Bos taurus | 73.3 | 10 (5) | KK specific isoform | - | 375 |
| Q32KN8 | Tubulin alpha-3 chain | Bos taurus | 74.0 | 15 (6) | KK specific isoform | - | 376 |
| P11979 | Pyruvate kinase isozymes M1/M2 | Felis catus | 84.2 | 11 (6) | KK specific isoform | - | 437 |
| Q9MZ13 | Voltage-dependent anion-selective channel protein 3 | Bos taurus | 87.5 | 16 (5) | KK up regulated | 15.9 | 91 |
| Q9Z1B2 | Glutathione S-transferase Mu 5 | Rattus norvegicus | 139.6 | 61 (17) | KK up regulated | 2.0 | 39 |
| Q3MHW9 | NADH-cytochrome b5 reductase 1 | Bos taurus | 45.2 | 9 (3) | KK up regulated | 15.9 | 91 |
| Q29438 | Outer dense fiber Protein 1 | Bos taurus | 168.2 | 44 (20) | KK up regulated | 15.9 | 91 |
| Q29438 | Outer dense fiber protein 1 | Bos taurus | 93.0 | 22 (6) | KK up regulated | 2.3 | 365 |
| Q5E956 | Triose phosphate isomerase | Bos taurus | 119.6 | 71 (14) | KK up regulated | 2.3 | 365 |
| P53353 | Sperm acrosomal protein FSA-ACR.1 (Fragment) | Vulpes vulpes | 54.2 | 4 (2) | M specific | - | 472 |
| P53353 | Sperm acrosomal protein FSA-ACR.1 (Fragment) | Vulpes vulpes | 48.3 | 9 (3) | M specific | - | 476 |
| Q3MHM5 | Tubulin beta-4B chain | Bos taurus | 45.3 | 5 (2) | M specific | - | 537 |
| P48818 | Very long-chain specific acyl-CoA dehydrogenase, mitochondrial | Bos taurus | 66.1 | 6 (4) | M specific | - | 551 |
| P26436 | Acrosomal protein SP-10 | Homo sapiens | 54.2 | 6 (2) | M specific | - | 472 |
| A6QLU1 | Glycerol-3-phosphate dehydrogenase, mitochondrial | Bos taurus | 103.0 | 24 (19) | M specific | - | 559 |
| A3KFF6 | Postacrosomal sheath WW domain-binding protein | Bos taurus | 94.3 | 25 (7) | M specific | - | 509 |
| Q3T0X5 | Proteasome subunit alpha type-1 | Bos taurus | 30.3 | 7 (2) | M specific | - | 494 |
| Q2YDE4 | Proteasome subunit alpha type-6 | Bos taurus | 49.7 | 20 (5) | M specific | - | 491 |
| A6H782 | Tektin-3 | Bos taurus | 120.7 | 31 (16) | M specific | - | 542 |
| Q1ZYL8 | Izumo sperm-egg fusion protein 4 | Homo sapiens | 40.5 | 6 (2) | M specific isoform | - | 491 |
| P31081 | 60 kDa heat shock protein, mitochondrial | Bos taurus | 104.8 | 19 (12) | M specific isoform | - | 548 |
| P00514 | cAMP-dependent protein kinase type I-alpha regulatory subunit | Bos taurus | 72.3 | 12 (5) | M specific isoform | - | 524 |
| P10173 | Fumarate hydratase, mitochondrial | Sus scrofa | 88.0 | 21 (10) | M specific isoform | - | 523 |
| P11979 | Pyruvate kinase isozymes M1/M2 | Felis catus | 150.2 | 32 (23) | M specific isoform | - | 556 |
| Q3T064 | Ropporin-1 | Bos taurus | 49.4 | 8 (2) | M specific isoform | - | 476 |
| P31039 | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | Bos taurus | 93.4 | 14 (10) | M specific isoform | - | 559 |
| Q5E956 | Triose phosphate isomerase | Bos taurus | 80.0 | 24 (6) | M specific isoform | - | 498 |
| P68002 | Voltage-dependent anion-selective channel protein 2 | Bos taurus | 120.7 | 21 (5) | M specific isoform | - | 500 |
| Q1ZYL8 | Izumo sperm-egg fusion protein 4 | Homo sapiens | 48.3 | 6 (2) | M up regulated | 2.0 | 72 |
| P53353 | Sperm acrosomal protein FSA-ACR.1 (Fragment) | Vulpes vulpes | 82.6 | 11 (5) | M up regulated | 3.2 | 78 |
| Q8NEB7 | Acrosin-binding protein | Homo sapiens | 41.6 | 4 (2) | M up regulated | 6.5 | 50 |
| Q29016 | Acrosin-binding protein (Fragment) | Sus scrofa | 81.2 | 6 (4) | M up regulated | 3.8 | 69 |
| P19483 | ATP synthase subunit alpha, mitochondrial | Bos taurus | 169.9 | 36 (23) | M up regulated | 3.4 | 246 |
| Q3T165 | Prohibitin | Bos taurus | 135.4 | 40 (12) | M up regulated | 1.5 | 67 |
| Q3T165 | Prohibitin | Bos taurus | 98.1 | 31 (9) | M up regulated | 3.8 | 69 |
| Q3T064 | Ropporin-1 | Bos taurus | 48.9 | 8 (2) | M up regulated | 4.3 | 22 |
| Q2YDI7 | Tektin-5 | Bos taurus | 168.0 | 47 (27) | M up regulated | 3.4 | 246 |
| Q5E956 | Triosephosphate isomerase | Bos taurus | 136.4 | 54 (14) | M up regulated | 1.9 | 86 |
| Abbreviation | Protein name |
|---|---|
| Proteins Subjected to STRING Analysis: | |
| NDUFB8 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8 |
| ATP5A1 | ATP synthase subunit alpha |
| ACADVL | Very long-chain specific acyl-CoA dehydrogenase |
| ODF1 | Outer dense fiber protein 1 |
| TEKT3 | Tektin-3 |
| PSMA1 | Proteasome subunit alpha type-1 (EC 3.4.25.1) |
| ROPN1 | Ropporin-1 (Rhophilin-associated protein 1) |
| PRKAR1A | cAMP-dependent protein kinase type I-alpha regulatory subunit |
| PSMA6 | Proteasome subunit alpha type-6 (EC 3.4.25.1) |
| GPD2 | Glycerol-3-phosphate dehydrogenase, mitochondrial Precursor (GPDH-M)(GPD-M)(EC 1.1.5.3) |
| HSPD1 | 60 kDa heat shock protein, mitochondrial Precursor (Heat shock protein 60)(HSP-60) |
| CNNM2 | cyclin M2 |
| VDAC2 | Voltage-dependent anion-selective channel protein 2 (VDAC-2) (Outer mitochondrial membrane protein porin 2) |
| WBP2NL | Postacrosomal sheath WW domain-binding protein (WW domain-binding protein 2-like) |
| SDHD | Succinate dehydrogenase [ubiquinone] cytochrome b small subunit |
| TEKT5 | Tektin-5 |
| PHB | Prohibitin |
| TPI1 | Triosephosphate isomerase (TIM)(EC 5.3.1.1) |
| CYB5R1 | NADH-cytochrome b5 reductase 1 (b5R.1)(EC 1.6.2.2) |
| NDUFV1 | NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial Precursor (EC 1.6.5.3) |
| Predicted Functional Partners: | |
| ATP5C1 | ATP synthase subunit gamma |
| ATP5O | ATP synthase subunit O |
| ATP5D | ATP synthase subunit delta |
| ATP5B | ATP synthase subunit beta (EC 3.6.3.14); |
| ATP6 | ATP synthase subunit a |
| PSMB4 | Proteasome subunit beta type-4 Precursor (EC 3.4.25.1) |
| HSPE1 | 10 kDa heat shock protein, mitochondrial (Hsp10) |
| ATP5J | ATP synthase-coupling factor 6 |
| NDUFS8 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 8 (EC 1.6.5.3) |
| PSMA7 | Proteasome subunit alpha type-7 (EC 3.4.25.1) |
| Motile Sperm (%) | |||
|---|---|---|---|
| Breed | Mean | Standard | p value |
| KK | 82 | 1.7321 | 0.018 * |
| Mafriwal | 72.667 | 3.7859 | |
| Progressive (%) | |||
| Breed | Mean | Standard | pvalue |
| KK | 51.333 | 3.7859 | 0.086 |
| Mafriwal | 44.333 | 3.7859 | |
| Rapid Cells (%) | |||
| Breed | Mean | Standard | pvalue |
| KK | 69.667 | 1.1547 | 0.002 ** |
| Mafriwal | 57 | 2.6458 | |
| Path Velocity (μm/s) | |||
| Breed | Mean | Standard | pvalue |
| KK | 130.733 | 3.72 | 0.062 |
| Mafriwal | 120.367 | 5.9 | |
| Progressive Velocity (μm/s) | |||
| Breed | Mean | Standard | pvalue |
| KK | 111.267 | 4.9743 | 0.272 |
| Mafriwal | 105.767 | 5.6012 | |
| Track Speed (μm/s) | |||
| Breed | Mean | Standard | pvalue |
| KK | 206.767 | 8.5582 | 0.018 * |
| Mafriwal | 178.667 | 9.2511 | |
| Lateral Amplitude (μm) | |||
| Breed | Mean | Standard | pvalue |
| KK | 8.067 | 0.8622 | 0.048 * |
| Mafriwal | 6.6 | 0.2646 | |
| Beat Frequency | |||
| Breed | Mean | Standard | pvalue |
| KK | 26.9 | 2.1703 | 0.279 |
| Mafriwal | 28.733 | 0.5033 | |
| Straightness (%) | |||
| Breed Mean Standardpvalue | |||
| KK | 84.333 | 1.5275 | 0.349 |
| Mafriwal | 85.333 | 0.5774 | |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ashrafzadeh, A.; Nathan, S.; Karsani, S.A. Comparative Analysis of Mafriwal (Bos taurus × Bos indicus) and Kedah Kelantan (Bos indicus) Sperm Proteome Identifies Sperm Proteins Potentially Responsible for Higher Fertility in a Tropical Climate. Int. J. Mol. Sci. 2013, 14, 15860-15877. https://doi.org/10.3390/ijms140815860
Ashrafzadeh A, Nathan S, Karsani SA. Comparative Analysis of Mafriwal (Bos taurus × Bos indicus) and Kedah Kelantan (Bos indicus) Sperm Proteome Identifies Sperm Proteins Potentially Responsible for Higher Fertility in a Tropical Climate. International Journal of Molecular Sciences. 2013; 14(8):15860-15877. https://doi.org/10.3390/ijms140815860
Chicago/Turabian StyleAshrafzadeh, Ali, Sheila Nathan, and Saiful Anuar Karsani. 2013. "Comparative Analysis of Mafriwal (Bos taurus × Bos indicus) and Kedah Kelantan (Bos indicus) Sperm Proteome Identifies Sperm Proteins Potentially Responsible for Higher Fertility in a Tropical Climate" International Journal of Molecular Sciences 14, no. 8: 15860-15877. https://doi.org/10.3390/ijms140815860
APA StyleAshrafzadeh, A., Nathan, S., & Karsani, S. A. (2013). Comparative Analysis of Mafriwal (Bos taurus × Bos indicus) and Kedah Kelantan (Bos indicus) Sperm Proteome Identifies Sperm Proteins Potentially Responsible for Higher Fertility in a Tropical Climate. International Journal of Molecular Sciences, 14(8), 15860-15877. https://doi.org/10.3390/ijms140815860




