Enzymatic Properties of Populus α- and β-NAD-ME Recombinant Proteins
Abstract
:1. Introduction
2. Results and Discussion
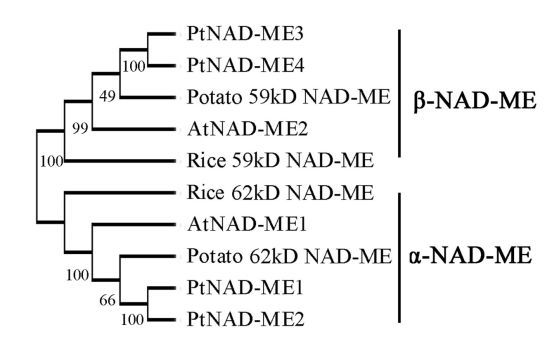
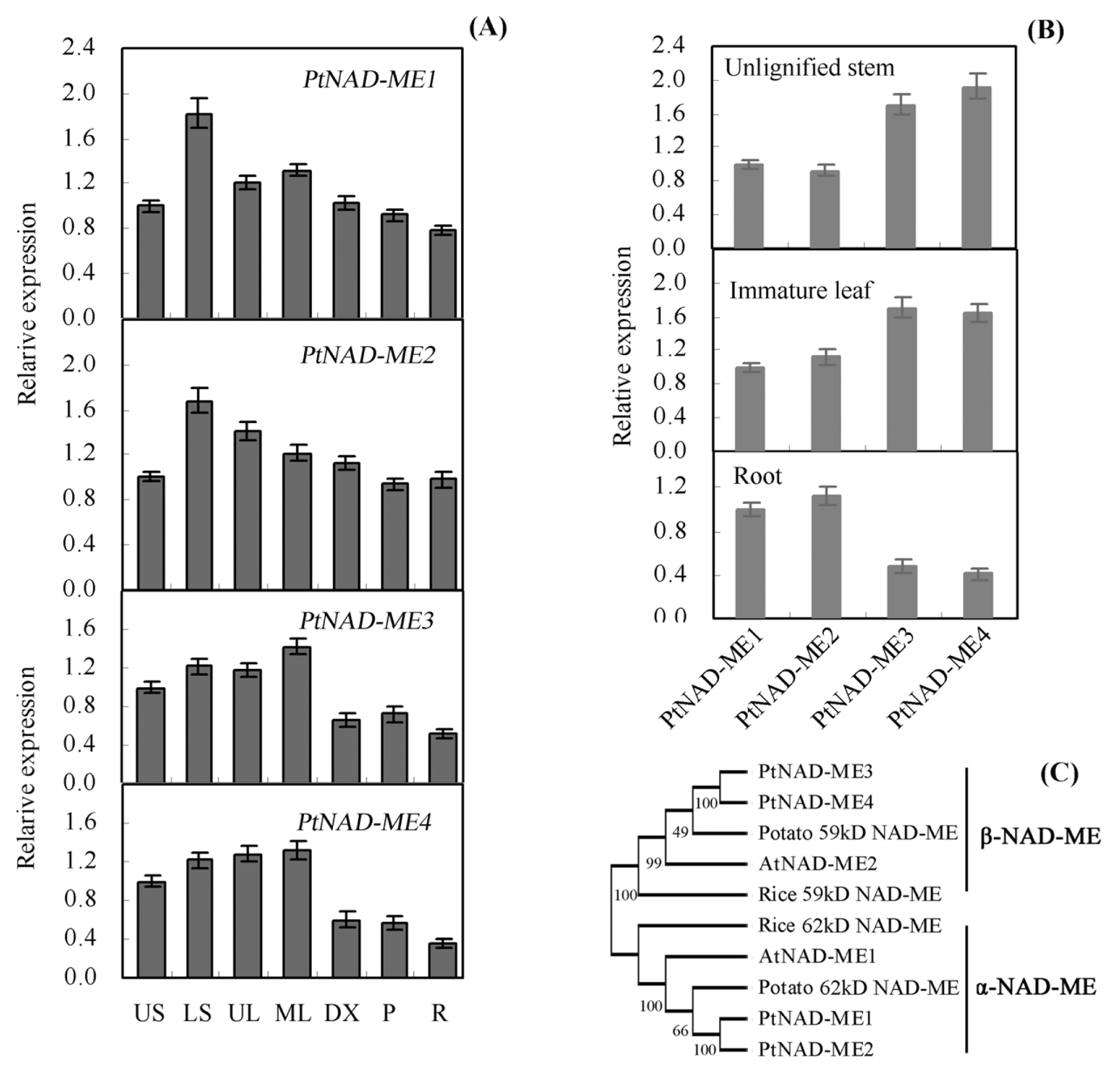
2.1. Characterization and Expression Pattern of the PtNAD-ME Gene Family from Populus
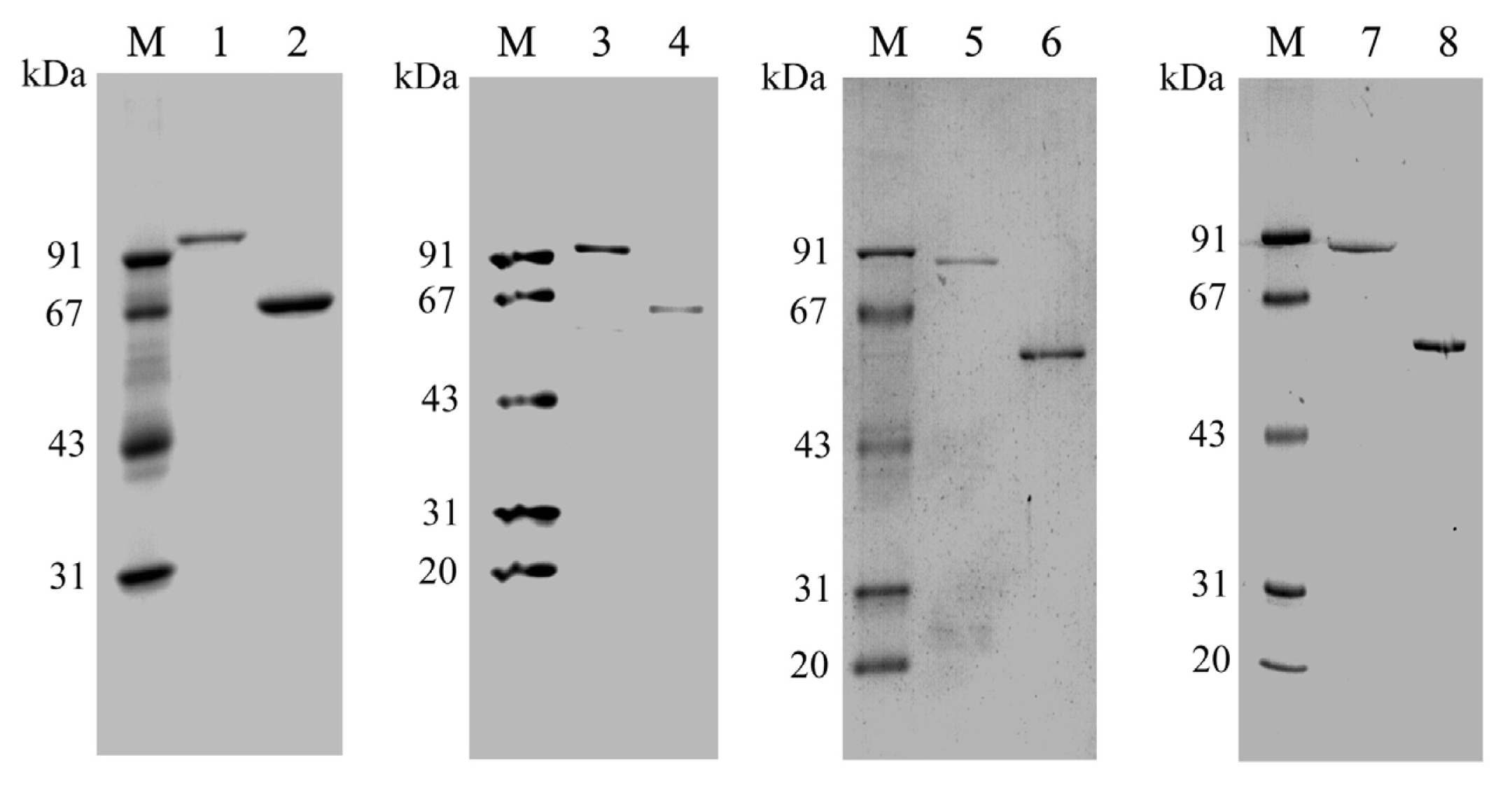
2.2. Expression and Purification of PtNAD-ME Proteins in E. coli
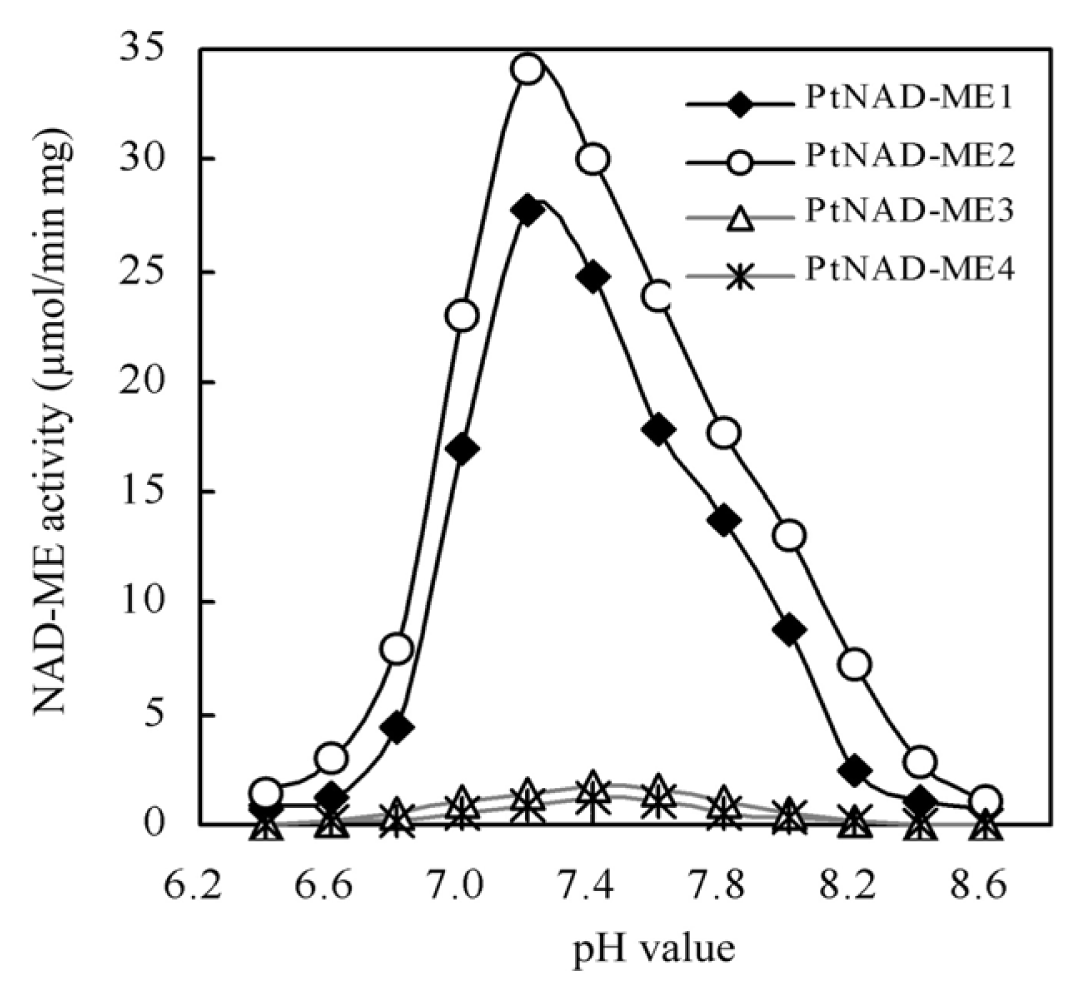
2.3. Optimum pH of Recombinant PtNAD-ME Proteins
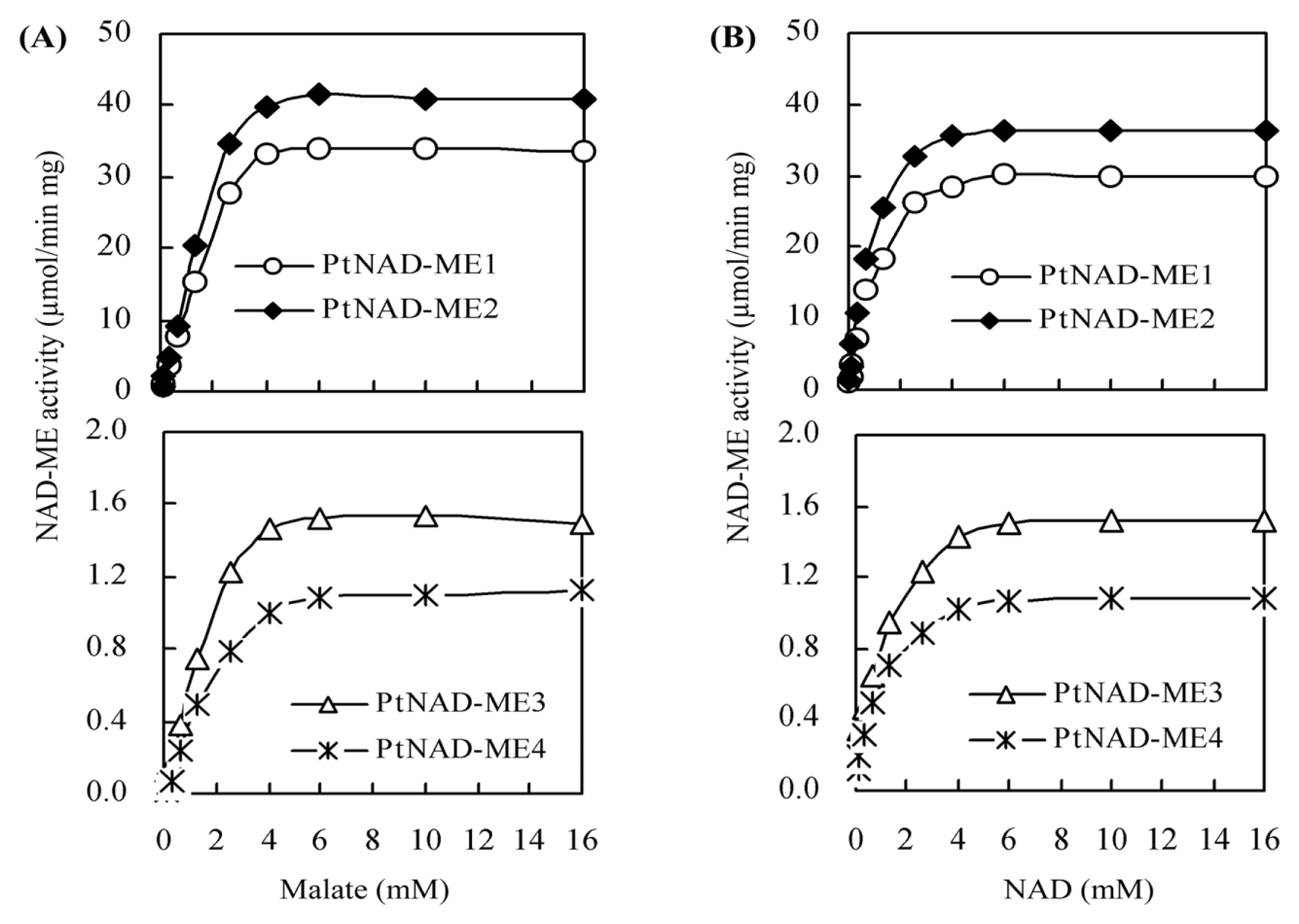
2.4. Kinetic Parameters for Recombinant PtNAD-ME Proteins
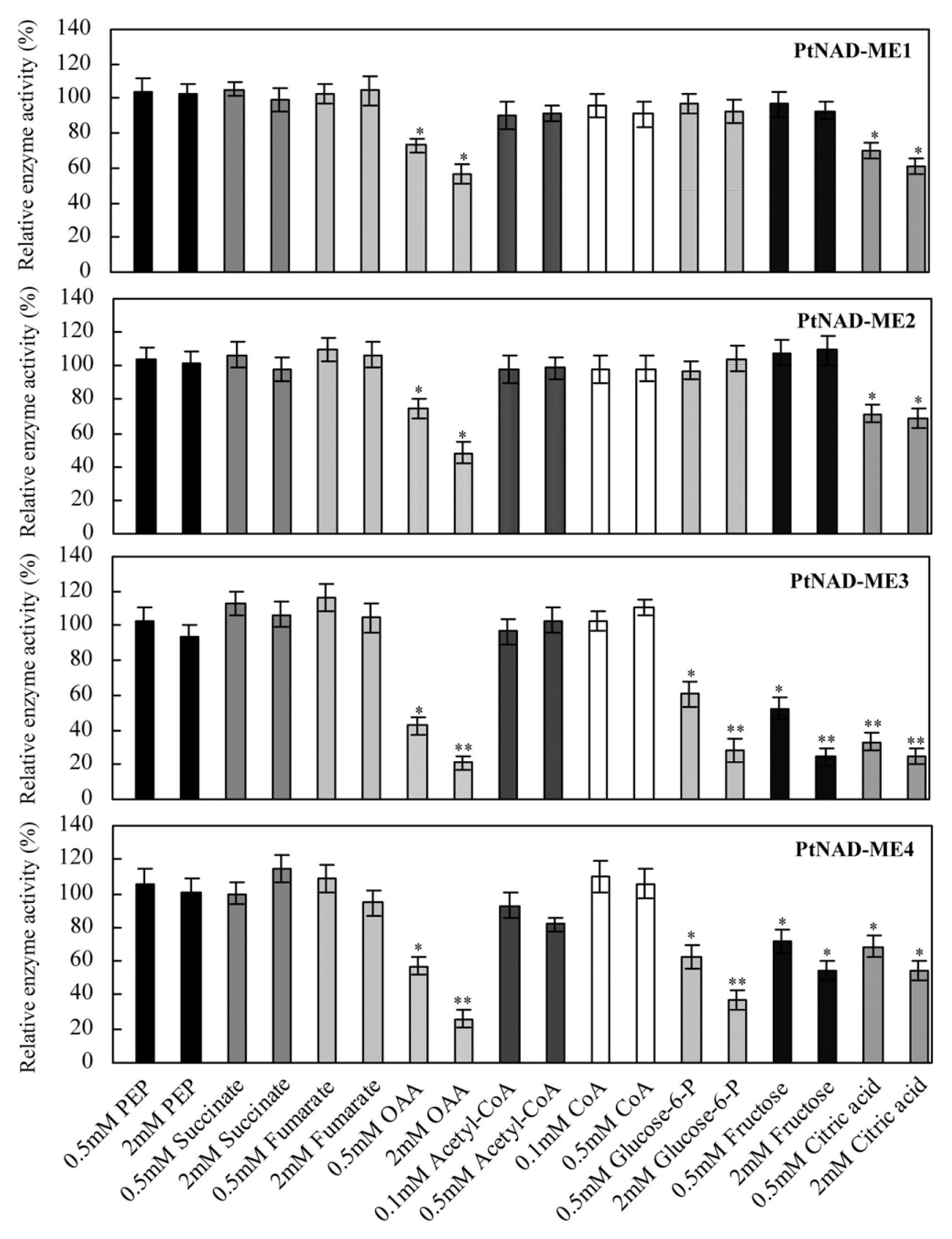
2.5. Effect of Catabolites on the Activities of Recombinant PtNAD-MEs
3. Experimental Section
3.1. Plant Growth
3.2. Isolation of PtNAD-ME cDNAs and Real-Time PCR Analysis
3.3. Construction of Expression Plasmids
3.4. Expression and Purification of Populus PtNAD-ME Proteins in E. coli
3.5. Cleavage of the GST Tag
3.6. SDS-PAGE
3.7. Assay of NAD-ME Activity
3.8. Kinetic Studies on the Recombinant PtNAD-ME Proteins
4. Conclusions
Supplementary Information
ijms-14-12994-s001.pdfAcknowledgments
Conflict of Interest
References
- Maier, A.; Zell, M.B.; Maurino, V.G. Malate decarboxylases: Evolution and roles of NAD(P)-ME isoforms in species performing C(4) and C(3) photosynthesis. J. Exp. Bot 2011, 62, 3061–3069. [Google Scholar]
- Winning, B.M.; Bourguignon, J.; Leaver, C.J. Plant mitochondrial NAD-dependent malic enzyme, cDNA cloning, deduced primary structure of the 59- and 62-kDa subunit, import, gene complexity and expression analysis. J. Biol. Chem 1994, 269, 4780–4786. [Google Scholar]
- Hatch, M.D.; Kagawa, T. Activity, location and role of NAD malic enzyme in the leaves of C4 photosynthesis. Aust. J. Plant Physiol 1974, 1, 357–369. [Google Scholar]
- Artus, N.N.; Edwards, G.E. Properties of leaf NAD-malic enzyme from the inducible crassulacean acid metabolism species Mesembryanthemum crystallinum. Plant Cell Physiol 1985, 26, 341–350. [Google Scholar]
- Artus, N.N.; Edwards, G.E. NAD-malic enzyme from plants. FEBS Lett 1985, 182, 225–233. [Google Scholar]
- Tronconi, M.A.; Fahnenstich, H.; Gerrard Wheeler, M.C.; Andreo, C.S.; Flugge, U.I.; Drincovich, M.F.; Maurino, V.G. Arabidopsis NAD-malic enzyme functions as a homodimer and heterodimer and has a major impact during nocturnal metabolism. Plant Physiol 2008, 146, 1540–1552. [Google Scholar]
- Grover, S.D.; Wedding, R.T. Kinetic ramifications of the association-dissociation behavior of NAD-malic enzyme. Plant Physiol 1982, 70, 1169–1172. [Google Scholar]
- Burnell, J.N. Photosynthesis in phosphoenolpyruvate carboxykinase type C4 species: Properties of NAD-malic enzyme from Urochloa panicoides. Aust. J. Plant Physiol 1987, 14, 517–525. [Google Scholar]
- Willeford, K.O.; Wedding, R.T. Evidence for a multiple subunit composition of plant NAD malic enzyme. J. Biol. Chem 1987, 262, 8423–8429. [Google Scholar]
- Long, J.L.; Wang, J.L.; Berry, J.O. Cloning and analysis of the C4 photosynthetic NAD-dependent malic enzyme of Amaranth mitochondria. J. Biol. Chem 1994, 269, 2817–1833. [Google Scholar]
- Grover, S.D.; Wedding, R.T. Modulation of the activity of NAD malic enzyme from Solanum tuberosum by changes in oligomeric state. Arch. Biochem. Biophys 1984, 234, 418–425. [Google Scholar]
- Tronconi, M.A.; Gerrard Wheeler, M.C.; Maurino, V.G.; Drincovich, M.F.; Andreo, C.S. NAD-malic enzymes of Arabidopsis thaliana display distinct kinetic mechanisms that support differences in physiological control. Biochem J 2010, 430, 295–303. [Google Scholar]
- Jenner, H.L.; Winning, B.M.; Millar, A.H.; Tomlinson, K.L.; Leaver, C.J.; Hill, S.A. NAD malic enzyme and the control of carbohydrate metabolism in potato tubers. Plant Physiol 2001, 126, 1139–1149. [Google Scholar]
- Tronconi, M.A.; Maurino, V.G.; Andreo, C.S.; Drincovich, M.F. Three different and tissue-specific NAD-malic enzymes generated by alternative subunit association in Arabidopsis thaliana. J. Biol. Chem 2010, 285, 11870–11879. [Google Scholar]
- Day, D.A.; Neuberger, M.; Douce, R. Activation of NAD-linked malic enzyme in intact mitochondria by exogenous CoA. Arch Biochem. Biophys 1984, 231, 233–234. [Google Scholar]
- Grissom, C.B.; Canellas, P.F.; Wedding, R.T. Allosteric regulation of the NAD malic enzyme from cauliflower: Activation by fumarate and coenzyme A. Arch. Biochem. Biophys. 1983, 220, 133–144. [Google Scholar]
- Koichiro, T.; Joel, D.; Masatoshi, N.; Sudhir, K. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol 2007, 24, 1596–1599. [Google Scholar]
- Oshugi, R.; Murata, T. Leaf anatomy, post-illumination CO2 burst and NAD-malic enzyme activity in Panicum dichotomiflorum. Plant Cell Physiol 1980, 21, 1329–1333. [Google Scholar]
- Murata, T.; Oshugi, R.; Matsuoka, M.; Nakamoto, M. Purification and characterization of NAD ME from leaves of Eleusine coracana and Panicum dichotomiflorum. Plant Physiol 1989, 89, 316–324. [Google Scholar]
- Rathnam, C.K.M. Studies with isolated bundle sheath mitochondria: Evidence for NAD-malic enzyme-catalyzed decarboxylation of C4 acids in species representing the three C4 metabolic subtypes. FEBS Lett 1978, 96, 367–372. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar]
| Recombinant enzyme | Purification step | Total protein (mg) | Total activity (U) | Specific activity a (U/mg) | Purification fold | Recovery (%) |
|---|---|---|---|---|---|---|
| PtNAD-ME1 | Soluble fraction | 537 | 107.4 | 0.2 | 1 | 100 |
| Cleavage of GST tag | 2.3 | 78.2 | 34.0 | 170 | 72.9 | |
| PtNAD-ME2 | Soluble fraction | 569 | 170.7 | 0.3 | 1 | 100 |
| Cleavage of GST tag | 2.7 | 109.4 | 40.5 | 142 | 64.0 | |
| PtNAD-ME3 | Soluble fraction | 529 | 2.1 | 0.004 | 1 | 100 |
| Cleavage of GST tag | 1.3 | 1.6 | 1.2 | 300 | 74.3 | |
| PtNAD-ME4 | Soluble fraction | 533 | 1.6 | 0.003 | 1 | 100 |
| Cleavage of GST tag | 1.4 | 1.3 | 0.9 | 300 | 78.7 |
| Parameters | Vmax (μmol min−1·mg−1) | Km (mM) | kcat (s−1) | kcat/Km (mM−1·s−1) | ||||
|---|---|---|---|---|---|---|---|---|
| Malate | NAD | Malate | NAD | Malate | NAD | Malate | NAD | |
| PtNAD-ME1 | 31.5 | 30.3 | 2.0 | 0.71 | 35.2 | 33.8 | 17.6 | 47.6 |
| PtNAD-ME2 | 39.1 | 37.8 | 1.8 | 0.62 | 43.0 | 41.6 | 23.8 | 67.0 |
| PtNAD-ME3 | 1.4 | 1.3 | 1.3 | 0.45 | 1.4 | 1.3 | 1.1 | 2.9 |
| PtNAD-ME4 | 1.0 | 0.9 | 1.4 | 0.41 | 1.0 | 0.9 | 0.7 | 2.2 |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Liu, J.; Yu, Q.; Elsheery, N.I.; Cheng, Y. Enzymatic Properties of Populus α- and β-NAD-ME Recombinant Proteins. Int. J. Mol. Sci. 2013, 14, 12994-13004. https://doi.org/10.3390/ijms140712994
Liu J, Yu Q, Elsheery NI, Cheng Y. Enzymatic Properties of Populus α- and β-NAD-ME Recombinant Proteins. International Journal of Molecular Sciences. 2013; 14(7):12994-13004. https://doi.org/10.3390/ijms140712994
Chicago/Turabian StyleLiu, Jinwen, Qiguo Yu, Nabil I. Elsheery, and Yuxiang Cheng. 2013. "Enzymatic Properties of Populus α- and β-NAD-ME Recombinant Proteins" International Journal of Molecular Sciences 14, no. 7: 12994-13004. https://doi.org/10.3390/ijms140712994
APA StyleLiu, J., Yu, Q., Elsheery, N. I., & Cheng, Y. (2013). Enzymatic Properties of Populus α- and β-NAD-ME Recombinant Proteins. International Journal of Molecular Sciences, 14(7), 12994-13004. https://doi.org/10.3390/ijms140712994