Construction of a Full-Length Enriched cDNA Library and Preliminary Analysis of Expressed Sequence Tags from Bengal Tiger Panthera tigris tigris
Abstract
:1. Introduction
2. Results
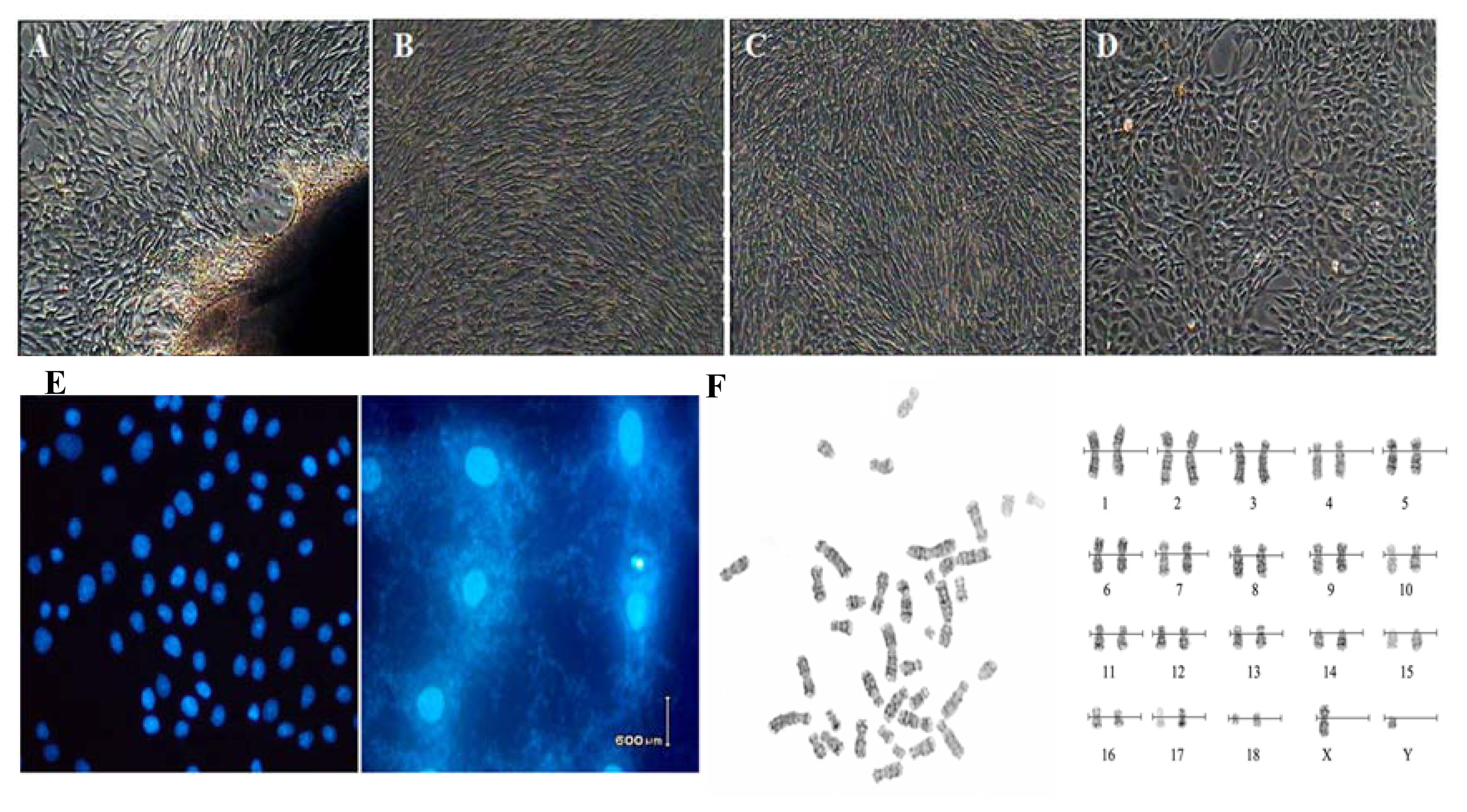
2.1. Cell cultures and Characteristic Tests
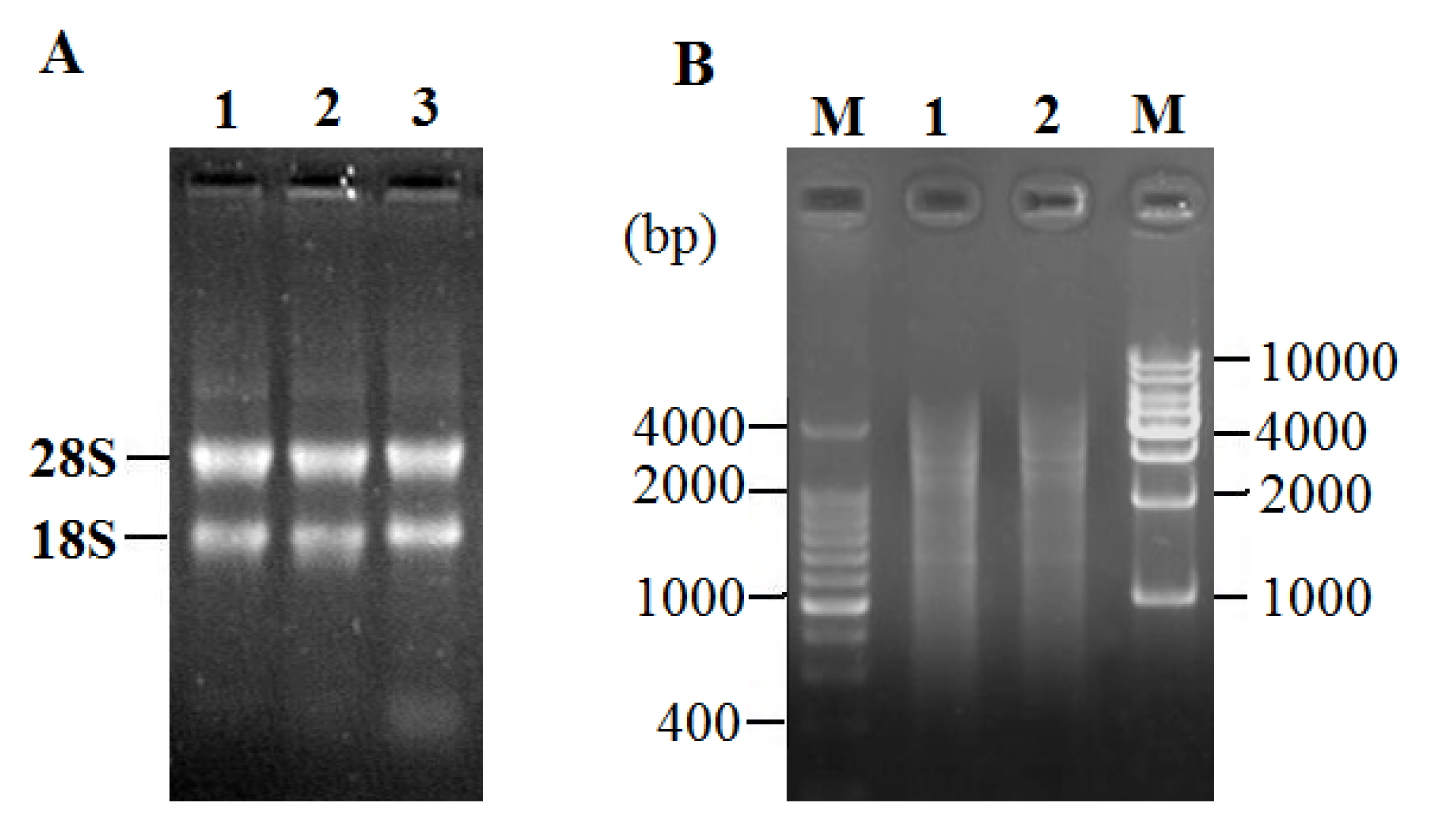
2.2. Total RNA Extraction and LD-PCR
2.3. Characterization of cDNA Library
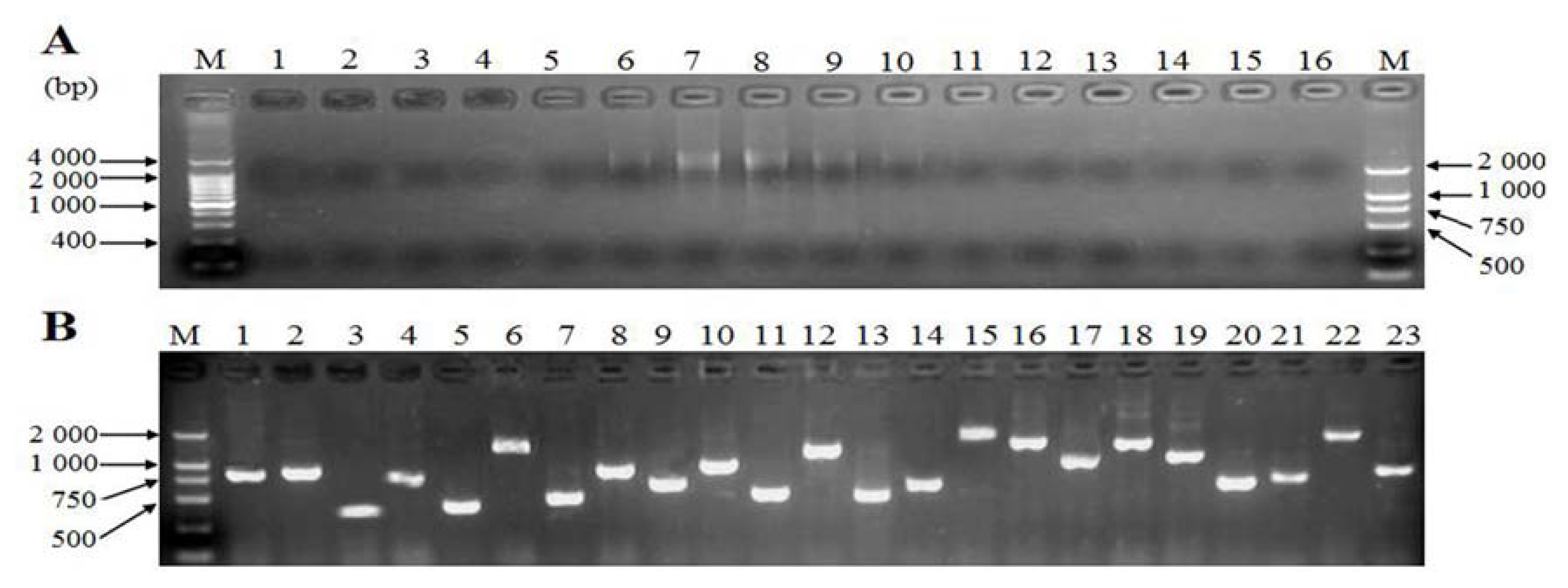
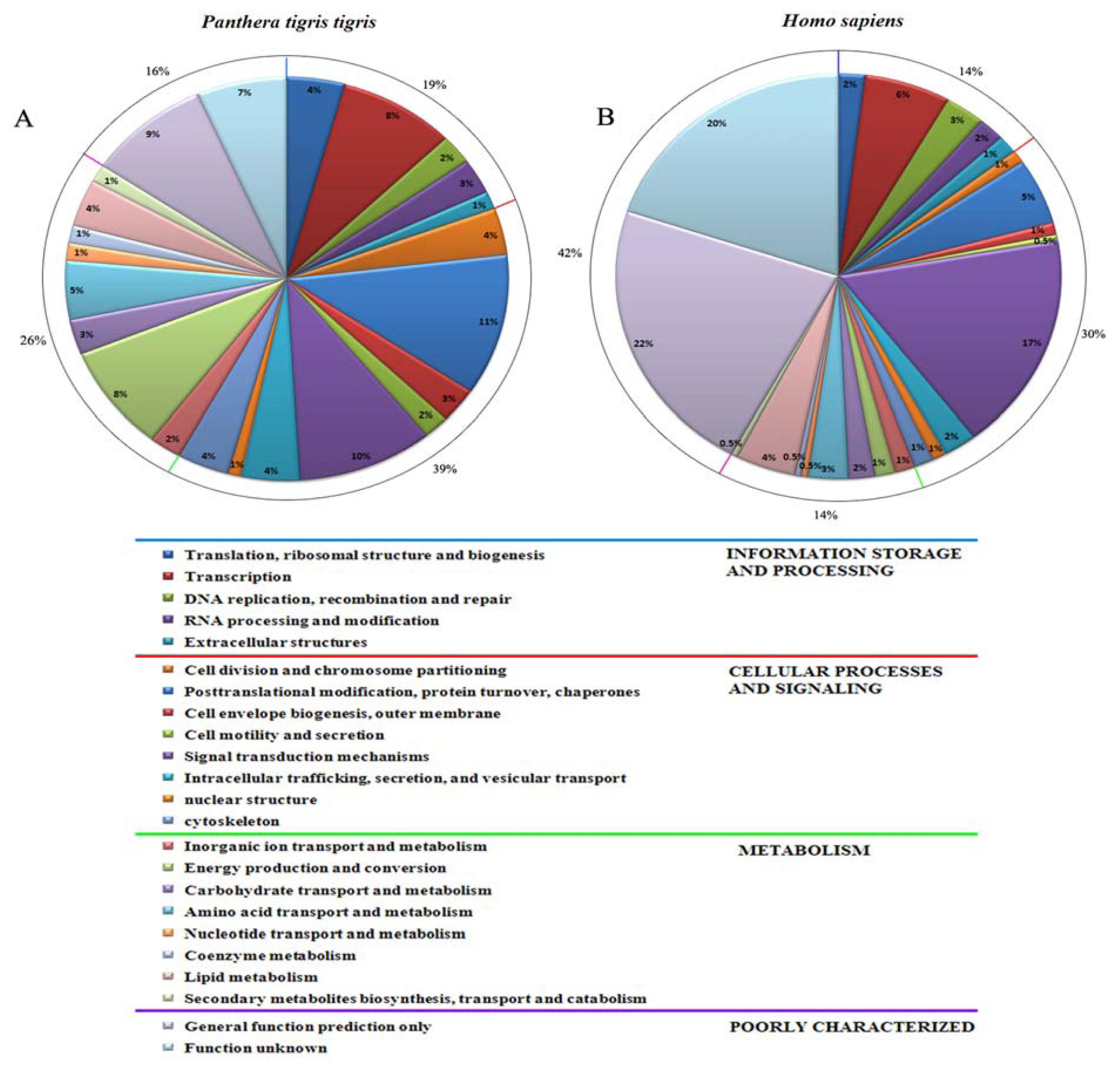
2.4. Generation of Expressed Sequence Tags and Sequence Analysis
2.5. Cloning and Sequence Analysis of Bengal Tigers Ferritin cDNA
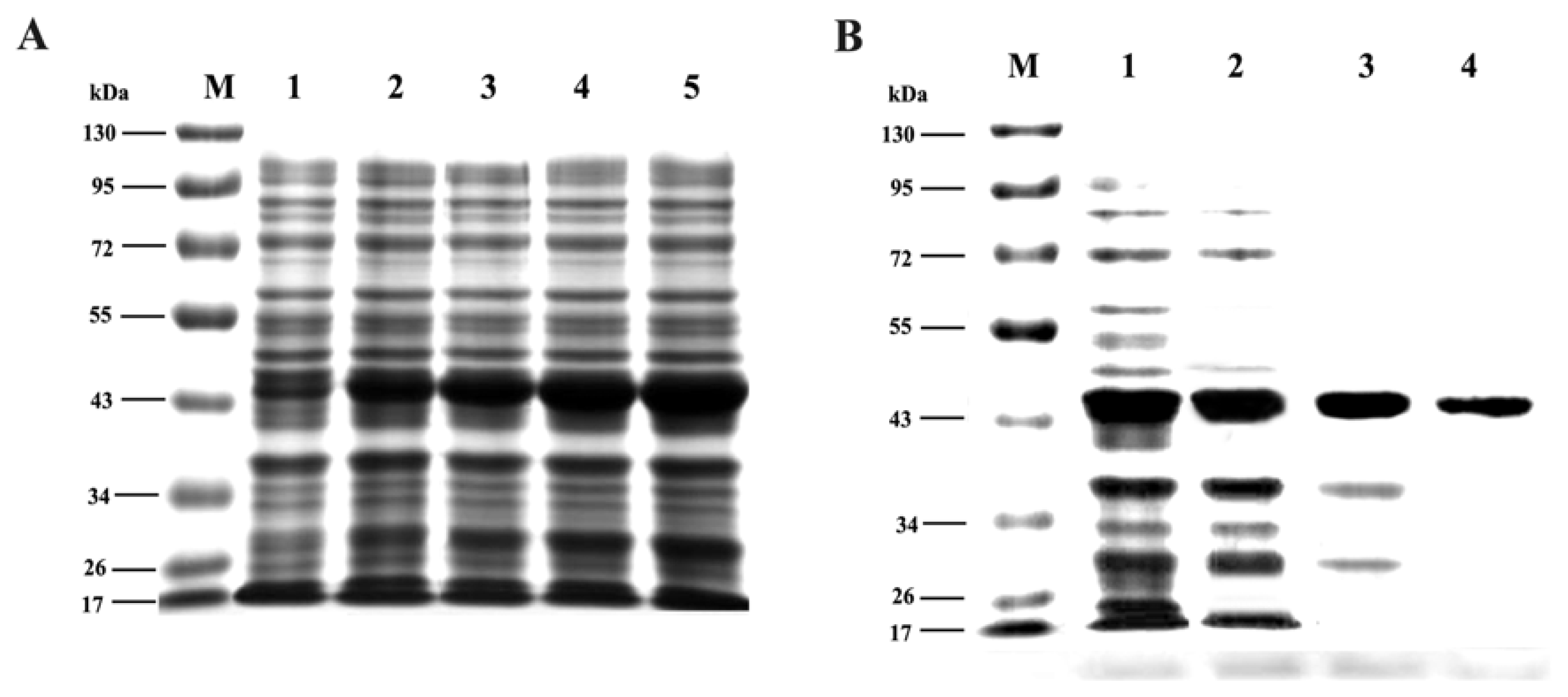
2.6. Expression and Purification of Trx-TAT-Ferritin Fusion Protein
3. Discussions
3.1. Characterization of cDNA Library of Bengal Tiger
3.2. Generation and Analysis of ESTs
3.3. Expression and Purification of Trx-TAT-Ferritin Fusion Protein
4. Materials and Methods
4.1. Material and Reagents
4.2. Cell Cultures and Biological Analysis
4.3. cDNA Library Construction
4.4. Titration of the Primary Library
4.5. Sequence Analysis Method
4.6. Expression and Purification of pET32a-TAT-Ferritin
5. Conclusions
Acknowledgements
Conflict of Interest
References
- Song, J.; Hua, S.; Song, K.; Zhang, Y. Culture, characteristics and chromosome complement of Siberian tiger fibroblasts for nuclear transfer. In Vitro Cell. Dev. Biol. Anim 2007, 43, 203–209. [Google Scholar]
- Taro, S.; Junco, N.; Vladimir, V.; Aramilev, A.B.; Seigo, H.; Dale, R.M. Species and sex identification from faecal samples of sympatric carnivores, Amur leopard and Siberian tiger, in the Russian Far East. Conserv. Genet 2006, 7, 799–802. [Google Scholar]
- Wei, K.; Zhang, Z.H.; Zhang, W.P.; Xu, X.; Liang, X. PCR-CTPP: A rapid and reliable genotyping technique based on ZFX/ZFY alleles for sex identification of tiger (Panthera tigris) and four other endangered felids. Conserv. Genet 2008, 9, 225–228. [Google Scholar]
- Li, J.Y.; Wang, H.Y.; Liu, J.; Liu, Q.; Zhang, J.S.; Wan, F.C.; Liu, F.J.; Jin, S.H.; Zhang, Y.L. Transcriptome analysis of a cDNA library from adult human epididymis. DNA Res 2008, 15, 115–122. [Google Scholar]
- Liu, C.Q.; Lu, T.F.; Feng, B.G.; Liu, D.; Guan, W.J.; Ma, Y.H. Construction of cDNA library and preliminary analysis of expressed sequence tags from Siberian tiger. Int. J. Biol. Sci 2010, 6, 584–589. [Google Scholar]
- Hempstead, P.D.; Yewdall, S.J.; Fernie, A.R.; Lawson, D.M.; Artymiuk, P.J.; Rice, D.W.; Ford, G.C.; Harrison, P.M. Comparison of the three-dimensional structures of recombinant human H and horse L ferritins at high resolution. J. Mol. Biol 1997, 268, 424–448. [Google Scholar]
- Ishiwata, H.; Katsuma, S.; Kizaki, K.; Patel, O.V.; Nakano, H.; Takahashi, T.; Imai, K.; Hirasawa, A.; Shiojima, S.; Ikawa, H.; et al. Characterization of gene expression profiles in early bovine pregnancy using a custom cDNA microarray. Mol. Reprod 2003, 65, 9–18. [Google Scholar]
- Orino, K.; Miura, T.; Muto, S.; Watanabe, K. Sequence analysis of canine and equine ferritin H and L subunit cDNAs. DNA Seq 2005, 16, 58–64. [Google Scholar]
- Collawn, J.F.; Gowan, L.K.; Crow, H.; Schwabe, C.; Fish, W.W. Isolation and partial amino acid sequence of three subunit species of porcine spleen ferritin: Evidence of multiple H subunits. Arch. Biochem. Biophys 1987, 259, 105–113. [Google Scholar]
- Shao, Z.T.; Cong, X.; Yuan, J.D.; Yang, G.W.; Chen, Y.; Pan, J.; An, L.G. Construction and characterization of a cDNA library from head kidney of Japanesesea bass (Lateolabrax japonicus). Mol. Biol. Rep 2009, 36, 2031–2037. [Google Scholar]
- Du, L.X.; Liu, S.F.; Zhu, J.; Li, H.B.; Li, S.G.; Song, X.M.; Wang, A.H. Construction of SMART cDNA library of sheep ovary and identification of candidate gene by homologous cloning. Agric. Sci. China 2007, 6, 1390–1395. [Google Scholar]
- Wellenreuther, R.; Schupp, I.; Poustka, A.; Wiemann, S. German c DNA Consortium. SMART amplification combined with cDNA size fractionation in order to obtain large full-length clones. BMC Genomics 2004, 5. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.H.; Chen, Z.; Yao, H.P.; Chen, F.; Zhu, H.H.; Zhou, H.J. Construction and characterization of a cDNA library from human liver tissue with chronic hepatitis B. J. Zhejiang Univ. Sci 2005, 6B, 288–294. [Google Scholar]
- Li, Y.P.; Xia, R.X.; Wang, H.; Li, X.S.; Liu, Y.Q.; Wei, Z.J.; Lu, C.; Xiang, Z.H. Construction of a full-length cDNA Library from Chinese oak silkworm pupa and identification of a KK-42-binding protein gene in relation to pupa-diapause termination. Int. J. Biol. Sci 2009, 5, 451–457. [Google Scholar]
- Wu, H.L.; Wan, Q.H.; Fang, S.G.; Zhang, L.Y.; Xia, J.S.; Zhong, Z.Y. Construction of a cDNA library of blood of Elaphurus davidianus. Acta Theriol. Sin 2007, 27, 380–384. [Google Scholar]
- Al-Taweel, K.; Dilantha Fernando, W.G.; Brûlé-Babel, A.L. Construction and Characterization of a cDNA library from wheat infected with Fusarium graminearum Fg 2. Int. J. Mol. Sci 2011, 12, 613–626. [Google Scholar]
- Ling, P.; Wang, M.; Chen, X.; Campbell, K.G. Construction and characterization of a full-length cDNA library for the wheat stripe rust pathogen (Puccinia striiformis f. sp. tritici). BMC Genomics 2007, 8, 145. [Google Scholar]
- Wiemann, S.; Mehrle, A.; Bechtel, S.; Wellenreuther, R.; Pepperkok, R.; Poustka, A. cDNAs for functional genomics and proteomics: The German sonsortium. C. R. Biol. 2003, 326, 1003–1009. [Google Scholar]
- Blair, M.W.; Fernandez, A.C.; Ishitani, M.; Moreta, D.; Seki, M.; Ayling, S.; Shinozaki, K. Construction and EST sequencing of full-length, drought stress cDNA libraries for common beans (Phaseolus vulgaris L.). BMC Plant Biol 2011, 11, 171. [Google Scholar]
- Liu, C.Q.; Guo, Y.; Liu, D.; Guan, W.J.; Ma, Y.H. Establishment and characterization of fibroblast cell line derived from Siberian tiger (Panthera tigris altaica). Biopreserv. Biobank 2010, 8, 99–105. [Google Scholar]
- Ye, J.; McGinnis, S.; Madden, T.L. BLAST: Improvements for better sequence analysis. Nucleic Acids Res 2006, 34, W6–W9. [Google Scholar]
- Wheeler, D.L.; Barrett, T.; Benson, D.A.; Bryant, S.H.; Canese, K.; Chetvernin, V.; Church, D.M.; DiCuccio, M.; Edgar, R.; Federhen, S.; et al. Database resources of the National Center for Biotechnology Information. Nucleic. Acids Res 2008, 36, D13–D21. [Google Scholar]
- Thanh, T.; Chi, V.T.; Abdullah, M.P.; Omar, H.; Noroozi, M.; Ky, H.; Napis, S. Construction of cDNA library and preliminary analysis of expressed sequence tags from green microalga Ankistrodesmus convolutus Corda. Mol. Biol. Rep 2011, 38, 177–182. [Google Scholar]
- Thompson, J.D.; Higgin, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitive of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weigh matrix choice. Nucleic Acids Res 1994, 22, 4673–4680. [Google Scholar]

© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Liu, C.; Liu, D.; Guo, Y.; Lu, T.; Li, X.; Zhang, M.; Ma, J.; Ma, Y.; Guan, W. Construction of a Full-Length Enriched cDNA Library and Preliminary Analysis of Expressed Sequence Tags from Bengal Tiger Panthera tigris tigris. Int. J. Mol. Sci. 2013, 14, 11072-11083. https://doi.org/10.3390/ijms140611072
Liu C, Liu D, Guo Y, Lu T, Li X, Zhang M, Ma J, Ma Y, Guan W. Construction of a Full-Length Enriched cDNA Library and Preliminary Analysis of Expressed Sequence Tags from Bengal Tiger Panthera tigris tigris. International Journal of Molecular Sciences. 2013; 14(6):11072-11083. https://doi.org/10.3390/ijms140611072
Chicago/Turabian StyleLiu, Changqing, Dan Liu, Yu Guo, Taofeng Lu, Xiangchen Li, Minghai Zhang, Jianzhang Ma, Yuehui Ma, and Weijun Guan. 2013. "Construction of a Full-Length Enriched cDNA Library and Preliminary Analysis of Expressed Sequence Tags from Bengal Tiger Panthera tigris tigris" International Journal of Molecular Sciences 14, no. 6: 11072-11083. https://doi.org/10.3390/ijms140611072
APA StyleLiu, C., Liu, D., Guo, Y., Lu, T., Li, X., Zhang, M., Ma, J., Ma, Y., & Guan, W. (2013). Construction of a Full-Length Enriched cDNA Library and Preliminary Analysis of Expressed Sequence Tags from Bengal Tiger Panthera tigris tigris. International Journal of Molecular Sciences, 14(6), 11072-11083. https://doi.org/10.3390/ijms140611072



