Integrated -Omics: A Powerful Approach to Understanding the Heterogeneous Lignification of Fibre Crops
Abstract
:1. Introduction
2. Plant Fibres: Nature’s Treasure Trove
3. Lignin: The Lord of the (Aromatic) Rings
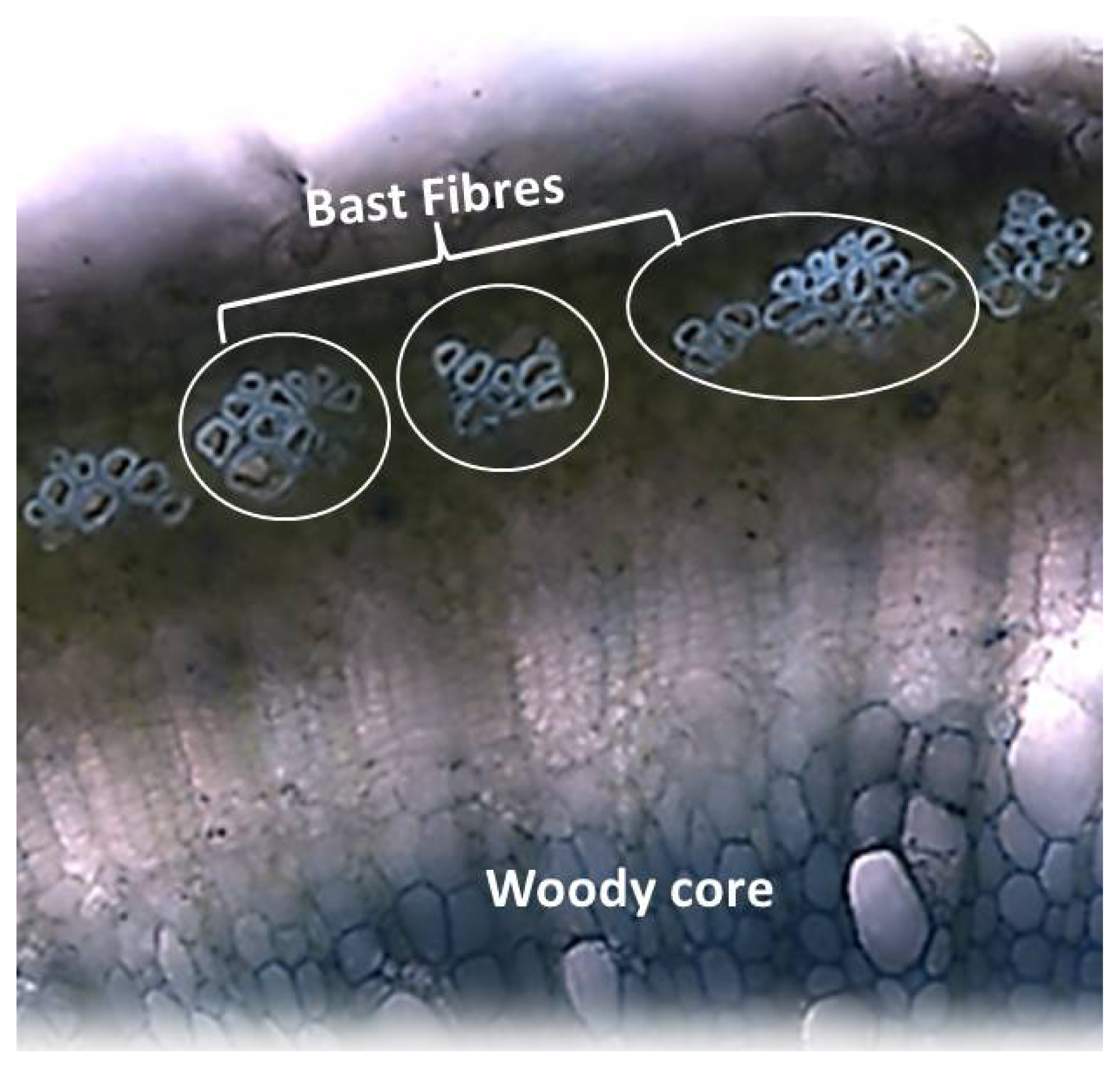
4. The Particular Cell Wall of Bast Fibres
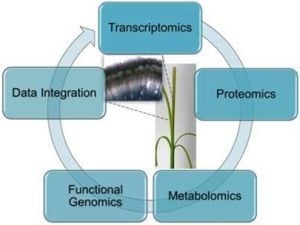
5. Integrated -Omics to Study Differential Lignification Patterns in Fibre Crops
6. Conclusions and Future Perspectives
Conflict of Interest
References
- Marques, G.; Rencoret, J.; Gutiérrez, A.; del Río, J.C. Evaluation of the chemical composition of different non-woody plant fibers used for pulp and paper manufacturing. Open Agric. J 2010, 4, 93–101. [Google Scholar]
- Benning, C.; Pichersky, E. Harnessing plant biomass for biofuels and biomaterials. Plant J 2008, 54, 533–535. [Google Scholar]
- Del Río, J.C.; Rencoret, J.; Gutiérrez, A.; Nieto, L.; Jiménez-Barbero, J.; Martínez, Á.T. Structural characterization of guaiacyl-rich lignins in flax (Linum usitatissimum) fibers and shives. J. Agric. Food Chem. 2011, 59, 11088–11099. [Google Scholar]
- McNutt, J.A.; Haegglom, R.; Raemoe, K. The Global Fiber Resource Picture. In Wood Product Demand and the Environment; Forest Products Research Society: Madison, WI, USA, 1992; pp. 39–53. [Google Scholar]
- Philippou, L.; Karastergiou, S. Lignocellulosic Materials From Annual Plants and Agricultural Residues As Raw Materials For Composite Building Materials. Proceedings of International Conference Forest Research: A Challenge for an Integrated European Approach, Thessaloniki, Greece, 27 August–1 September 2001; II, pp. 817–821.
- Reddy, N.; Yang, Y. Biofibers from agricultural byproducts for industrial applications. Trends Biotechnol 2005, 23, 22–27. [Google Scholar]
- Davies, G.C.; Bruce, D.M. Effect of environmental relative humidity and damage on the tensile properties of flax and nettle fibers. Textil. Res. J 1998, 68, 623–629. [Google Scholar]
- Fathima, M.; Balasubramanian, A. Effect of plant growth regulators on the quality of bast fibres in Abelmoschus esculentus (Linn.) Moench. Acta Bot. Croat 2006, 65, 101–112. [Google Scholar]
- Gorshkova, T.A.; Gurjanov, O.P.; Mikshina, P.V.; Ibragimova, N.N.; Mokshina, N.E.; Salnikov, V.V.; Ageeva, M.V.; Amenitskii, S.I.; Chernova, T.E.; Chemikosova, S.B. Specific type of secondary cell wall formed by plant fibers. Russ. J. Plant Physiol 2010, 57, 328–341. [Google Scholar]
- Lev-Yadun, S. Plant fibers: Initiation, growth, model plants, and open questions. Russ. J. Plant Physiol 2010, 57, 305–315. [Google Scholar]
- Gorshkova, T.; Brutch, N.; Chabbert, B.; Deyholos, M.; Hayashi, T.; Lev-Yadun, S.; Mellerowicz, E.J.; Morvan, C.; Neutelings, G.; Pilate, G. Plant fiber formation: State of the art, recent and expected progress, and open questions. Crit. Rev. Plant Sci 2012, 31, 201–228. [Google Scholar]
- Ko, J.H.; Yang, S.H.; Park, A.H.; Lerouxel, O.; Han, K.H. ANAC012, a member of the plant–specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. Plant J 2007, 50, 1035–1048. [Google Scholar]
- Mitsuda, N.; Iwase, A.; Yamamoto, H.; Yoshida, M.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 2007, 19, 270–280. [Google Scholar]
- Zhong, R.; Richardson, E.A.; Ye, Z.H. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 2007, 19, 2776–2792. [Google Scholar]
- Ko, J.H.; Kim, W.C.; Han, K.H. Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. Plant J 2009, 60, 649–665. [Google Scholar]
- Bhargava, A.; Mansfield, S.D.; Hall, H.C.; Douglas, C.J.; Ellis, B.E. MYB75 functions in regulation of secondary cell wall formation in the Arabidopsis inflorescence stem. Plant Physiol 2010, 154, 1428–1438. [Google Scholar]
- Demura, T.; Ye, Z.H. Regulation of plant biomass production. Curr. Opin. Plant Biol 2010, 13, 299–304. [Google Scholar]
- Ohashi–Ito, K.; Oda, Y.; Fukuda, H. Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 2010, 22, 3461–3473. [Google Scholar]
- Zhong, R.; Lee, C.; Ye, Z.H. Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol. Plant 2010, 3, 1087–1103. [Google Scholar]
- Yamaguchi, M.; Goué, N.; Igarashi, H.; Ohtani, M.; Nakano, Y.; Mortimer, J.C.; Nishikubo, N.; Kubo, M.; Katayama, Y.; Kakegawa, K.; et al. VASCULAR-RELATED NAC-DOMAIN6 and VASCULAR-RELATED NAC-DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiol 2010, 153, 906–914. [Google Scholar]
- Yamaguchi, M.; Mitsuda, N.; Ohtani, M.; Ohme-Takagi, M.; Kato, K.; Demura, T. VASCULAR-RELATED NAC-DOMAIN7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J 2011, 66, 579–590. [Google Scholar]
- Wang, H.; Zhao, Q.; Chen, F.; Wang, M.; Dixon, R.A. NAC domain function and transcriptional control of a secondary cell wall master switch. Plant J 2011, 68, 1104–1114. [Google Scholar]
- Li, E.; Wang, S.; Liu, Y.; Chen, J.G.; Douglas, C.J. OVATE FAMILY PROTEIN4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana. Plant J 2011, 67, 328–341. [Google Scholar]
- Kim, W.C.; Ko, J.H.; Han, K.H. Identification of a cis-acting regulatory motif recognized by MYB46, a master transcriptional regulator of secondary wall biosynthesis. Plant Mol. Biol 2012, 78, 489–501. [Google Scholar]
- Li, E.; Bhargava, A.; Qiang, W.; Friedmann, M.C.; Forneris, N.; Savidge, R.A.; Johnson, L.A.; Mansfield, S.D.; Ellis, B.E.; Douglas, C.J. The Class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus. New Phytol 2012, 194, 102–115. [Google Scholar]
- Bhargava, A.; Ahad, A.; Wang, S.; Mansfield, S.D.; Haughn, G.W.; Douglas, C.J.; Ellis, B.E. The interacting MYB75 and KNAT7 transcription factors modulate secondary cell wall deposition both in stems and seed coat in Arabidopsis. Planta 2013, 237, 1199–11211. [Google Scholar]
- Williamson, R.E.; Burn, J.E.; Birch, R.; Baskin, T.I.; Arioli, T.; Betzner, A.S.; Cork, A. Morphology of rsw1, a cellulose-deficient mutant of Arabidopsis thaliana. Protoplasma 2001, 215, 116–127. [Google Scholar]
- Burn, J.E.; Hurley, U.A.; Birch, R.J.; Arioli, T.; Cork, A.; Williamson, R.E. The cellulose-deficient Arabidopsis mutant rsw3 is defective in a gene encoding a putative glucosidase II, an enzyme processing N-glycans during ER quality control. Plant J 2002, 32, 949–960. [Google Scholar]
- Zhong, R.; Morrison, W.H., 3rd; Freshour, G.D.; Hahn, M.G.; Ye, Z.H. Expression of a mutant form of cellulose synthase AtCesA7 causes dominant negative effect on cellulose biosynthesis. Plant Physiol 2003, 132, 786–795. [Google Scholar]
- Szyjanowicz, P.M.; McKinnon, I.; Taylor, N.G.; Gardiner, J.; Jarvis, M.C.; Turner, S.R. The irregular xylem 2 mutant is an allele of korrigan that affects the secondary cell wall of Arabidopsis thaliana. Plant J 2004, 37, 730–740. [Google Scholar]
- Chen, Z.; Hong, X.; Zhang, H.; Wang, Y.; Li, X.; Zhu, J.K.; Gong, Z. Disruption of the cellulose synthase gene, AtCesA8/IRX1, enhances drought and osmotic stress tolerance in Arabidopsis. Plant J 2005, 43, 273–283. [Google Scholar]
- MacKinnon, I.M.; Sturcová, A.; Sugimoto-Shirasu, K.; His, I; McCann, M.C.; Jarvis, M.C. Cell-wall structure and anisotropy in procuste, a cellulose synthase mutant of Arabidopsis thaliana. Planta 2006, 224, 438–448. [Google Scholar]
- Chu, Z.; Chen, H.; Zhang, Y.; Zhang, Z.; Zheng, N.; Yin, B.; Yan, H.; Zhu, L.; Zhao, X.; Yuan, M.; et al. Knockout of the AtCESA2 gene affects microtubule orientation and causes abnormal cell expansion in Arabidopsis. Plant Physiol 2007, 143, 213–224. [Google Scholar]
- Persson, S.; Caffall, K.H.; Freshour, G.; Hilley, M.T.; Bauer, S.; Poindexter, P.; Hahn, M.G.; Mohnen, D.; Somerville, C. The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. Plant Cell 2007, 19, 237–255. [Google Scholar]
- Bringmann, M.; Li, E.; Sampathkumar, A.; Kocabek, T.; Hauser, M.T.; Persson, S. POM-POM2/cellulose synthase interacting1 is essential for the functional association of cellulose synthase and microtubules in Arabidopsis. Plant Cell 2012, 24, 163–177. [Google Scholar]
- Sánchez-Rodríguez, C.; Bauer, S.; Hématy, K.; Saxe, F.; Ibáñez, A.B.; Vodermaier, V.; Konlechner, C.; Sampathkumar, A.; Rüggeberg, M.; Aichinger, E.; et al. Chitinase-like1/pom-pom1 and its homolog CTL2 are glucan-interacting proteins important for cellulose biosynthesis in Arabidopsis. Plant Cell 2012, 24, 589–607. [Google Scholar]
- Rubio-Díaz, S.; Pérez-Pérez, J.M.; González-Bayón, R.; Muñoz-Viana, R.; Borrega, N.; Mouille, G.; Hernández-Romero, D.; Robles, P.; Höfte, H.; Ponce, M.R.; et al. Cell expansion-mediated organ growth is affected by mutations in three EXIGUA genes. PLoS One 2012, 7, e36500. [Google Scholar]
- Wyatt, S.E.; Sederoff, R.; Flaishman, M.A.; Lev-Yadun, S. Arabidopsis thaliana as a model for gelatinous fiber formation. Russ. J. Plant Physiol 2010, 57, 363–367. [Google Scholar]
- Rajangam, A.S.; Kumar, M.; Aspeborg, H.; Guerriero, G.; Arvestad, L.; Pansri, P.; Brown, C.J.; Hober, S.; Blomqvist, K.; Divne, C.; et al. MAP20, a microtubule-associated protein in the secondary cell walls of hybrid aspen, is a target of the cellulose synthesis inhibitor 2,6-dichlorobenzonitrile. Plant Physiol 2008, 148, 1283–1294. [Google Scholar]
- Winzell, A.; Aspeborg, H.; Wang, Y.; Ezcurra, I. Conserved CA-rich motifs in gene promoters of Pt x tMYB021-responsive secondary cell wall carbohydrate-active enzymes in Populus. Biochem. Biophys. Res. Commun 2010, 394, 848–853. [Google Scholar]
- Grant, E.H.; Fujino, T.; Beers, E.P.; Brunner, A.M. Characterization of NAC domain transcription factors implicated in control of vascular cell differentiation in Arabidopsis and Populus. Planta 2010, 232, 337–352. [Google Scholar]
- Zhong, R.; McCarthy, R.L.; Lee, C.; Ye, Z.H. Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiol 2011, 157, 1452–1468. [Google Scholar]
- Yang, X.; Ye, C.Y.; Bisaria, A.; Tuskan, G.A.; Kalluri, U.C. Identification of candidate genes in Arabidopsis and Populus cell wall biosynthesis using text-mining, co-expression network analysis and comparative genomics. Plant Sci 2011, 181, 675–687. [Google Scholar]
- Li, Q.; Lin, Y.C.; Sun, Y.H.; Song, J.; Chen, H.; Zhang, X.H.; Sederoff, R.R.; Chiang, V.L. Splice variant of the SND1 transcription factor is a dominant negative of SND1 members and their regulation in Populus trichocarpa. Proc. Natl. Acad. Sci. USA 2012, 109, 14699–14704. [Google Scholar]
- Tian, X.; Xie, J.; Zhao, Y.; Lu, H.; Liu, S.; Qu, L.; Li, J.; Gai, Y.; Jiang, X. Sense-, antisense- and RNAi-4CL1 regulate soluble phenolic acids, cell wall components and growth in transgenic Populus tomentosa Carr. Plant Physiol. Biochem 2013, 65, 111–119. [Google Scholar]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar]
- Ralph, J.; Lapierre, C.; Marita, J.M.; Kim, H.; Lu, F.; Hatfield, R.D.; Ralph, S.; Chapple, C.; Franke, R.; Hemm, M.R.; et al. Elucidation of new structures in lignins of CAD- and COMT-deficient plants by NMR. Phytochemistry 2001, 57, 993–1003. [Google Scholar]
- Tesfaye, M.; Yang, S.S.; Lamb, J.F.S.; Jung, H-J.G.; Samac, D.A.; Vance, C.P.; Gronwald, J.W.; VandenBosch, K.A. Medicago truncatula as a model for dicot cell wall development. Bioenergy Res. 2009, 2, 59–76. [Google Scholar]
- Zhao, Q.; Gallego-Giraldo, L.; Wang, H.; Zeng, Y.; Ding, S.Y.; Chen, F.; Dixon, R.A. An NAC transcription factor orchestrates multiple features of cell wall development in Medicago truncatula. Plant J 2010, 63, 100–114. [Google Scholar]
- Yang, S.S.; Tu, Z.J.; Cheung, F.; Xu, W.W.; Lamb, J.F.; Jung, H.J.; Vance, C.P.; Gronwald, J.W. Using RNA-Seq for gene identification, polymorphism detection and transcript profiling in two alfalfa genotypes with divergent cell wall composition in stems. BMC Genomics 2011, 12, 199. [Google Scholar]
- Lee, Y.; Chen, F.; Gallego-Giraldo, L.; Dixon, R.A.; Voit, E.O. Integrative analysis of transgenic alfalfa (Medicago sativa L.) suggests new metabolic control mechanisms for monolignol biosynthesis. PLoS Comput. Biol 2011, 7, e1002047. [Google Scholar]
- Van Bakel, H.; Stout, J.M.; Cote, A.G.; Tallon, C.M.; Sharpe, A.G.; Hughes, T.R.; Page, J.E. The draft genome and transcriptome of Cannabis sativa. Genome Biol 2011, 12, R102. [Google Scholar]
- Wang, Z.; Hobson, N.; Galindo, L.; Zhu, S.; Shi, D.; McDill, J.; Yang, L.; Hawkins, S.; Neutelings, G.; Datla, R.; et al. The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. Plant J 2012, 72, 461–473. [Google Scholar]
- Bothwell, J.H. The long past of systems biology. New Phytol 2006, 170, 6–10. [Google Scholar]
- Keurentjes, J.J.; Angenent, G.C.; Dicke, M.; Dos Santos, V.A.; Molenaar, J.; van der Putten, W.H.; de Ruiter, P.C.; Struik, P.C.; Thomma, B.P. Redefining plant systems biology: From cell to ecosystem. Trends Plant Sci 2011, 16, 183–190. [Google Scholar]
- Fraser, C.M.; Chapple, C. The phenylpropanoid pathway in Arabidopsis. Arabidopsis Book 2011, 9, e0152. [Google Scholar]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 2001, 126, 485–493. [Google Scholar]
- Millar, D.J.; Long, M.; Donovan, G.; Fraser, P.D.; Boudet, A.M.; Danoun, S.; Bramley, P.M.; Bolwell, G.P. Introduction of sense constructs of cinnamate 4-hydroxylase (CYP73A24) in transgenic tomato plants shows opposite effects on flux into stem lignin and fruit flavonoids. Phytochemistry 2007, 68, 1497–1509. [Google Scholar]
- Bonawitz, N.D.; Chapple, C. The genetics of lignin biosynthesis: Connecting genotype to phenotype. Annu. Rev. Genet 2010, 44, 337–363. [Google Scholar]
- Vanholme, R.; Morreel, K.; Darrah, C.; Oyarce, P.; Grabber, J.H.; Ralph, J.; Boerjan, W. Metabolic engineering of novel lignin in biomass crops. New Phytol 2012, 196, 978–1000. [Google Scholar]
- Song, L.; Siguier, B.; Dumon, C.; Bozonnet, S.; O’Donohue, M.J. Engineering better biomass-degrading ability into a GH11 xylanase using a directed evolution strategy. Biotechnol. Biofuels 2012, 5, 3. [Google Scholar]
- Petersen, P.D.; Lau, J.; Ebert, B.; Yang, F.; Verhertbruggen, Y.; Kim, J.S.; Varanasi, P.; Suttangkakul, A.; Auer, M.; Loqué, D.; et al. Engineering of plants with improved properties as biofuels feedstocks by vessel-specific complementation of xylan biosynthesis mutants. Biotechnol. Biofuels 2012, 5, 84. [Google Scholar]
- Bonawitz, N.D.; Chapple, C. Can genetic engineering of lignin deposition be accomplished without an unacceptable yield penalty? Curr. Opin. Biotechnol 2013, 24, 336–343. [Google Scholar]
- Chen, F.; Srinivasa Reddy, M.S.; Temple, S.; Jackson, L.; Shadle, G.; Dixon, R.A. Multi-site genetic modulation of monolignol biosynthesis suggests new routes for formation of syringyl lignin and wall-bound ferulic acid in alfalfa (Medicago sativa L.). Plant J 2006, 48, 113–124. [Google Scholar]
- Shadle, G.; Chen, F.; Srinivasa Reddy, M.S.; Jackson, L.; Nakashima, J.; Dixon, R.A. Down-regulation of hydroxycinnamoyl CoA: Shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry 2007, 68, 1521–1529. [Google Scholar]
- Nakashima, J.; Chen, F.; Jackson, L.; Shadle, G.; Dixon, R.A. Multi-site genetic modification of monolignol biosynthesis in alfalfa (Medicago sativa): Effects on lignin composition in specific cell types. New Phytol 2008, 179, 738–750. [Google Scholar]
- Xu, B.; Escamilla-Treviño, L.L.; Sathitsuksanoh, N.; Shen, Z.; Shen, H.; Zhang, Y.H.; Dixon, R.A.; Zhao, B. Silencing of 4-coumarate:coenzyme A ligase in switchgrass leads to reduced lignin content and improved fermentable sugar yields for biofuel production. New Phytol 2011, 192, 611–625. [Google Scholar]
- Dien, B.S.; Miller, D.J.; Hector, R.E.; Dixon, R.A.; Chen, F.; McCaslin, M.; Reisen, P.; Sarath, G.; Cotta, M.A. Enhancing alfalfa conversion efficiencies for sugar recovery and ethanol production by altering lignin composition. Bioresour. Technol 2011, 102, 6479–6486. [Google Scholar]
- Ambavaram, M.M.; Krishnan, A.; Trijatmiko, K.R.; Pereira, A. Coordinated activation of cellulose and repression of lignin biosynthesis pathways in rice. Plant Physiol 2011, 155, 916–931. [Google Scholar]
- Fursova, O.; Pogorelko, G.; Zabotina, O.A. An efficient method for transient gene expression in monocots applied to modify the Brachypodium distachyon cell wall. Ann Bot 2012, 110, 47–56. [Google Scholar]
- Jung, J.H.; Vermerris, W.; Gallo, M.; Fedenko, J.R.; Erickson, J.E.; Altpeter, F. RNA interference suppression of lignin biosynthesis increases fermentable sugar yields for biofuel production from field-grown sugarcane. Plant Biotechnol. J. 2013. [Google Scholar] [CrossRef]
- Grabber, J.H.; Schatz, P.F.; Kim, H.; Lu, F.; Ralph, J. Identifying new lignin bioengineering targets: 1. Monolignol-substitute impacts on lignin formation and cell wall fermentability. BMC Plant Biol 2010, 10, 114. [Google Scholar]
- Zhang, K.; Bhuiya, M.W.; Pazo, J.R.; Miao, Y.; Kim, H.; Ralph, J.; Liu, C.J. An engineered monolignol 4-O-methyltransferase depresses lignin biosynthesis and confers novel metabolic capability in Arabidopsis. Plant Cell 2012, 24, 3135–3152. [Google Scholar]
- Elumalai, S.; Tobimatsu, Y.; Grabber, J.H.; Pan, X.; Ralph, J. Epigallocatechin gallate incorporation into lignin enhances the alkaline delignification and enzymatic saccharification of cell walls. Biotechnol. Biofuels 2012, 5, 59. [Google Scholar]
- Gorshkova, T.A.; Salnikov, V.V.; Pogodina, N.M.; Chemikosova, S.B.; Yablokova, E.V.; Ulanov, A.V.; Ageeva, M.V.; van Dam, J.E.G.; Lozovaya, V.V. Composition and distribution of cell wall phenolic compounds in flax (Linum usitatissimum L.) stem tissues. Ann. Bot 2000, 85, 477–486. [Google Scholar]
- Day, A.; Ruel, K.; Neutelings, G.; Crônier, D.; David, H.; Hawkins, S.; Chabbert, B. Lignification in the flax stem: Evidence for an unusual lignin in bast fibers. Planta 2005, 222, 234–245. [Google Scholar]
- Day, A.; Addi, M.; Kim, W.; David, H.; Bert, F.; Mesnage, P.; Rolando, C.; Chabbert, B.; Neutelings, G.; Hawkins, S. ESTs from the fibre-bearing stem tissues of flax (Linum usitatissimum L.): Expression analyses of sequences related to cell wall development. Plant Biol 2005, 7, 23–32. [Google Scholar]
- Crônier, D.; Monties, B.; Chabbert, B. Structure and chemical composition of bast fibers isolated from developing hemp stem. J. Agric. Food Chem 2005, 53, 8279–8289. [Google Scholar]
- De Pauw, M.A.; Vidmar, J.J.; Collins, J.; Bennett, R.A.; Deyholos, M.K. Microarray analysis of bast fibre producing tissue of Cannabis sativa identifies transcripts associated with conserved and specialised processes of secondary wall development. Func. Plant Biol 2007, 34, 737–749. [Google Scholar]
- Roach, M.J.; Deyholos, M.K. Microarray analysis of flax (Linum usitatissimum L.) stems identifies transcripts enriched in fibre-bearing phloem tissues. Mol. Genet. Genomics 2007, 278, 149–165. [Google Scholar]
- Van den Broeck, H.C.; Maliepaard, C.; Ebskam, M.J.M.; Toonen, M.A.J.; Koops, A.J. Differential expression of genes involved in C1 metabolism and lignin biosynthesis in wooden core and bast tissues of fibre hemp (Cannabis sativa L.). Plant Sci 2008, 2, 205–220. [Google Scholar]
- Hotte, N.S.; Deyholos, M.K. A flax fibre proteome: Identification of proteins enriched in bast fibres. BMC Plant Biol 2008, 8, 52. [Google Scholar]
- Roach, M.J.; Deyholos, M.K. Microarray analysis of developing flax hypocotyls identifies novel transcripts correlated with specific stages of phloem fibre differentiation. Ann. Bot 2008, 102, 317–330. [Google Scholar]
- Fénart, S.; Ndong, Y.P.; Duarte, J.; Rivière, N.; Wilmer, J.; van Wuytswinkel, O.; Lucau, A.; Cariou, E.; Neutelings, G.; Gutierrez, L.; et al. Development and validation of a flax (Linum usitatissimum L.) gene expression oligo microarray. BMC Genomics 2010, 11, 592. [Google Scholar]
- Long, S.H.; Deng, X.; Wang, Y.F.; Li, X.; Qiao, R.Q.; Qiu, C.S.; Guo, Y.; Hao, D.M.; Jia, W.Q.; Chen, X.B. Analysis of 2.297 expressed sequence tags (ESTs) from a cDNA library of flax (Linum ustitatissimum L.) bark tissue. Mol. Biol. Rep 2012, 39, 6289–6296. [Google Scholar]
- Huis, R.; Morreel, K.; Fliniaux, O.; Lucau-Danila, A.; Fénart, S.; Grec, S.; Neutelings, G.; Chabbert, B.; Mesnard, F.; Boerjan, W.; et al. Natural hypolignification is associated with extensive oligolignol accumulation in flax stems. Plant Physiol 2012, 158, 1893–1915. [Google Scholar]
- Neutelings, G.; Fénart, S.; Lucau-Danila, A.; Hawkins, S. Identification and characterization of miRNAs and their potential targets in flax. J. Plant Physiol 2012, 169, 1754–1766. [Google Scholar]
- Ibragimova, N.N.; Mokshina, N.E.; Gorshkova, T.A. Cell wall proteins of flax phloem fibers. Russ. J. Bioorg. Chem 2012, 38, 117–125. [Google Scholar]
- Day, A.; Fénart, S.; Neutelings, G.; Hawkins, S.; Rolando, C.; Tokarski, C. Identification of cell wall proteins in the flax (Linum usitatissimum) stem. Proteomics 2013, 13, 812–825. [Google Scholar]
- Liu, T.; Zhu, S.; Tang, Q.; Chen, P.; Yu, Y.; Tang, S. De novo assembly and characterization of transcriptome using Illumina paired-end sequencing and identification of CesA gene in ramie (Boehmeria nivea L. Gaud). BMC Genomics 2013, 14, 125. [Google Scholar]
- Achyuthan, K.E.; Achyuthan, A.M.; Adams, P.D.; Dirk, S.M.; Harper, J.C.; Simmons, B.A.; Singh, A.K. Supramolecular self-assembled chaos: Polyphenolic lignin’s barrier to cost-effective lignocellulosic biofuels. Molecules 2010, 15, 8641–8488. [Google Scholar]
- Gandolfi, S.; Ottolina, G.; Riva, S.; Predrocchi Fantoni, G.; Patel, I. Complete chemical analysis of carmagnola hemp hurds and structural features of its components. BioResources 2013, 8, 2641–2656. [Google Scholar]
- Del Río, J.C.; Rencoret, J.; Prinsen, P.; Martinez, A.T.; Ralph, J.; Gutierrez, A. Structural characterization of wheat straw lignin as revealed by analytical pyrolysis, 2D-NMR, and reductive cleavage methods. J. Agric. Food Chem 2012, 60, 5922–5935. [Google Scholar]
- Amthor, J.S. Efficiency of lignin biosynthesis: A quantitative analysis. Ann. Bot 2003, 91, 673–695. [Google Scholar]
- Casler, M.D.; Buxton, D.R.; Vogel, K.P. Genetic modification of lignin concentration affects fitness of perennial herbaceous plants. Theor. Appl. Genet 2002, 104, 127–131. [Google Scholar]
- Moura, J.C.; Bonine, C.A.; de Oliveira, F.V.J.; Dornelas, M.C.; Mazzafera, P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol 2010, 52, 360–376. [Google Scholar]
- Hamberger, B.; Hahlbrock, K. The 4-coumarate:CoA ligase gene family in Arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. Proc. Natl. Acad. Sci. USA 2004, 101, 2209–2214. [Google Scholar]
- Costa, M.A.; Bedgar, D.L.; Moinuddin, S.G.; Kim, K.W.; Cardenas, C.L.; Cochrane, F.C.; Shockey, J.M.; Helms, G.L.; Amakura, Y.; Takahashi, H.; et al. Characterization in vitro and in vivo of the putative multigene 4-coumarate:CoA ligase network in Arabidopsis: Syringyl lignin and sinapate/sinapyl alcohol derivative formation. Phytochemistry 2005, 66, 2072–2091. [Google Scholar]
- Lu, S.; Zhou, Y.; Li, L.; Chiang, V.L. Distinct roles of cinnamate 4-hydroxylase genes in Populus. Plant Cell Physiol 2006, 47, 905–914. [Google Scholar]
- Soltani, B.M.; Ehlting, J.; Hamberger, B.; Douglas, C.J. Multiple cis-regulatory elements regulate distinct and complex patterns of developmental and wound-induced expression of Arabidopsis thaliana 4CL gene family members. Planta 2006, 224, 1226–1238. [Google Scholar]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Chen, Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol 2010, 153, 1526–1538. [Google Scholar]
- Rasmussen, S.; Dixon, R.A. Transgene-mediated and elicitor-induced perturbation of metabolic channeling at the entry point into the phenylpropanoid pathway. Plant Cell 1999, 11, 1537–1552. [Google Scholar]
- Achnine, L.; Blancaflor, E.B.; Rasmussen, S.; Dixon, R.A. Colocalization of L-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell 2004, 16, 3098–3109. [Google Scholar]
- Bassard, J.E.; Richert, L.; Geerinck, J.; Renault, H.; Duval, F.; Ullmann, P.; Schmitt, M.; Meyer, E.; Mutterer, J.; Boerjan, W.; et al. Protein-protein and protein-membrane associations in the lignin pathway. Plant Cell 2012, 24, 4465–4482. [Google Scholar]
- Jones, L.; Ennos, A.R.; Turner, S.R. Cloning and characterization of irregular xylem4 (irx4): A severely lignin-deficient mutant of Arabidopsis. Plant J 2001, 26, 205–216. [Google Scholar]
- Leplé, J.C.; Dauwe, R.; Morreel, K.; Storme, V.; Lapierre, C.; Pollet, B.; Naumann, A.; Kang, K.Y.; Kim, H.; Ruel, K.; et al. Downregulation of cinnamoyl-coenzyme A reductase in poplar: Multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell 2007, 19, 3669–3691. [Google Scholar]
- Zhou, R; Jackson, L; Shadle, G; Nakashima, J; Temple, S; Chen, F; Dixon, R.A. Distinct cinnamoyl CoA reductases involved in parallel routes to lignin in Medicago truncatula. Proc. Natl. Acad. Sci. USA 2010, 107, 17803–17808. [Google Scholar]
- Kaneda, M.; Schuetz, M.; Lin, B.S.; Chanis, C.; Hamberger, B.; Western, T.L.; Ehlting, J.; Samuels, A.L. ABC transporters coordinately expressed during lignification of Arabidopsis stems include a set of ABCBs associated with auxin transport. J. Exp. Bot 2011, 62, 2063–2077. [Google Scholar]
- Alejandro, S.; Lee, Y.; Tohge, T.; Sudre, D.; Osorio, S.; Park, J.; Bovet, L.; Lee, Y.; Geldner, N.; Fernie, A.R.; et al. AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr. Biol 2012, 22, 1207–1212. [Google Scholar]
- Liu, C.J. Deciphering the enigma of lignification: Precursor transport, oxidation, and the topochemistry of lignin assembly. Mol. Plant 2012, 5, 304–317. [Google Scholar]
- Neutelings, G. Lignin variability in plant cell walls: Contribution of new models. Plant Sci 2011, 181, 379–386. [Google Scholar]
- Cabané, M.; Pireaux, J.C.; Léger, E.; Weber, E.; Dizengremel, P.; Pollet, B.; Lapierre, C. Condensed lignins are synthesized in poplar leaves exposed to ozone. Plant Physiol 2004, 134, 586–594. [Google Scholar]
- Pejic, B.M.; Kostic, M.M.; Skundric, P.D.; Praskalo, J.Z. The effects of hemicelluloses and lignin removal on water uptake behavior of hemp fibers. Bioresour. Technol 2008, 99, 7152–7179. [Google Scholar]
- Mikshina, P.V.; Chemikosova, S.B.; Mokshina, N.E.; Ibragimova, N.N.; Gorshkova, T.A. Free galactose and galactosidase activity in the course of flax fiber development. Russ. J. Plant Physiol 2009, 56, 58–67. [Google Scholar]
- Hobson, N.; Roach, M.J.; Deyholos, M.K. Gene expression in tension wood and bast fibres. Russ. J. Plant Physiol. 2010, 57, 321–327. [Google Scholar]
- Thygesen, A. Properties of Hemp Fibre Polymer Composites—An Optimisation of Fibre Properties Using Novel Defibration Methods and Fibre Characterization. Ph.D. Thesis, The Royal Agricultural and Veterinary University, Frederiksberg, Denmark, May 2005. [Google Scholar]
- Clair, B.; Alméras, T.; Yamamoto, H.; Okuyama, T.; Sugiyama, J. Mechanical behavior of cellulose microfibrils in tension wood, in relation with maturation stress generation. Biophys. J 2006, 91, 1128–1135. [Google Scholar]
- Mellerowicz, E.J.; Immerzeel, P.; Hayashi, T. Xyloglucan: The molecular muscle of trees. Ann. Bot 2008, 102, 659–665. [Google Scholar]
- Gorshkova, T.A.; Chemikosova, S.B.; Salnikov, V.V.; Pavlencheva, N.V.; Gurjanov, O.P.; Stolle-Smits, T.; van Dam, J.E.G. Occurrence of cell-specific galactan is coinciding with bast fiber developmental transition in flax. Ind. Crops Prod 2004, 19, 217–224. [Google Scholar]
- Mellerowicz, E.J.; Gorshkova, T.A. Tensional stress generation in gelatinous fibres: A review and possible mechanism based on cell-wall structure and composition. J. Exp. Bot 2012, 63, 551–565. [Google Scholar]
- Snegireva, A.V.; Ageeva, M.V.; Amenitskii, S.I.; Chernova, T.E.; Ebskamp, M.; Gorshkova, T.A. Intrusive growth of sclerenchyma fibers. Russ. J. Plant Physiol 2010, 57, 342–355. [Google Scholar]
- Gorshkova, T.A.; Sal’nikov, V.V.; Chemikosova, S.B.; Ageeva, M.V.; Pavlencheva, N.V. The snap point: A transition point in Linum usitatissimum bast fiber development. Ind. Crops Prod 2003, 18, 213–221. [Google Scholar]
- Ageeva, M.V.; Petrovská, B.; Kieft, H.; Sal’nikov, V.V.; Snegireva, A.V.; van Dam, J.E.; van Veenendaal, W.L.; Emons, A.M.; Gorshkova, T.A.; van Lammeren, A.A. Intrusive growth of flax phloem fibers is of intercalary type. Planta 2005, 222, 565–574. [Google Scholar]
- Gorshkova, T.; Morvan, C. Secondary cell-wall assembly in flax phloem fibres: Role of galactans. Planta 2006, 223, 149–158. [Google Scholar]
- Salnikov, V.V.; Ageeva, M.V.; Gorshkova, T.A. Homofusion of Golgi secretory vesicles in flax phloem fibers during formation of the gelatinous secondary cell wall. Protoplasma 2008, 233, 269–273. [Google Scholar]
- Fukushima, A.; Kusano, M.; Redestig, H.; Arita, M.; Saito, K. Integrated omics approaches in plant systems biology. Curr. Opin. Chem. Biol 2009, 13, 532–538. [Google Scholar]
- Weckwerth, W. Green systems biology—From single genomes, proteomes and metabolomes to ecosystems research and biotechnology. J. Proteomics 2011, 75, 284–305. [Google Scholar]
- García-Alcalde, F.; García-López, F.; Dopazo, J.; Conesa, A. Paintomics: A web based tool for the joint visualization of transcriptomics and metabolomics data. Bioinformatics 2011, 27, 137–139. [Google Scholar]
- Lin, K.; Kools, H.; de Groot, P.J.; Gavai, A.K.; Basnet, R.K.; Cheng, F.; Wu, J.; Wang, X.; Lommen, A.; Hooiveld, G.J.; et al. MADMAX—Management and analysis database for multiple ~omics experiments. J. Integr. Bioinform 2011, 8, 160. [Google Scholar]
- Syed, M.H.; Karpinets, T.V.; Parang, M.; Leuze, M.R.; Park, B.H.; Hyatt, D.; Brown, S.D.; Moulton, S.; Galloway, M.D.; Uberbacher, E.C. BESC knowledgebase public portal. Bioinformatics 2012, 28, 750–751. [Google Scholar]
- Sun, X.; Weckwerth, W. COVAIN: A toolbox for uni- and multivariate statistics, time-series and correlation network analysis and inverse estimation of the differential Jacobian from metabolomics covariance data. Metabolomics 2012, 8, 81–93. [Google Scholar]
- Fukushima, A.; Kusano, M. Recent progress in the development of metabolome databases for plant systems biology. Front. Plant Sci 2013, 4, 73. [Google Scholar]
- Liepe, J.; Filippi, S.; Komorowski, M.; Stumpf, M.P. Maximizing the information content of experiments in systems biology. PLoS Comput. Biol 2013, 9, e1002888. [Google Scholar]
- Chiaiese, P.; Ruotolo, G.; di Matteo, A.; de Santo Virzo, A.; de Marco, A.; Filippone, E. Cloning and expression analysis of kenaf (Hibiscus cannabinus L.) major lignin and cellulose biosynthesis gene sequences and polymer quantification during plant development. Ind. Crops Prod 2011, 34, 1072–1078. [Google Scholar]
- Jeong, M.-J.; Choi, B.S.; Bae, D.W.; Shin, S.C.; Park, S.U.; Lim, H.-S.; Kim, J.; Kim, J.B.; Cho, B.-K.; Bae, H. Differential expression of kenaf phenylalanine ammonia-lyase (PAL) ortholog during developmental stages and in response to abiotic stresses. Plant Omics 2012, 4, 392–399. [Google Scholar]
- Chowdhury, E.M.D.; Choi, B.S.; Park, S.U.; Lim, H.-S.; Bae, H. Transcriptional analysis of hydroxycinnamoyl transferase (HCT) in various tissues of Hibiscus cannabinus in response to abiotic stress conditions. Plant Omics 2012, 5, 305–313. [Google Scholar]
- Kumar, S.; You, F.M.; Cloutier, S. Genome wide SNP discovery in flax through next generation sequencing of reduced representation libraries. BMC Genomics 2012, 13, 684. [Google Scholar]
- Martin, L.B.; Fei, Z.; Giovannoni, J.J.; Rose, J.K. Catalyzing plant science research with RNA-seq. Front. Plant Sci 2013, 4, 66. [Google Scholar]
- Puzey, J.R.; Karger, A.; Axtell, M.; Kramer, E.M. Deep annotation of Populus trichocarpa microRNAs from diverse tissue sets. PLoS One 2012, 7, e33034. [Google Scholar]
- Ong, S.S.; Wickneswari, R. Characterization of microRNAs expressed during secondary wall biosynthesis in Acacia mangium. PLoS One 2012, 7, e49662. [Google Scholar]
- Lu, S.; Sun, Y.H.; Shi, R.; Clark, C.; Li, L.; Chiang, V.L. Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 2005, 17, 2186–2203. [Google Scholar]
- Fagerstedt, K.V.; Kukkola, E.M.; Koistinen, V.V.; Takahashi, J.; Marjamaa, K. Cell wall lignin is polymerised by class III secretable plant peroxidases in Norway spruce. J. Integr. Plant Biol 2010, 52, 186–194. [Google Scholar]
- Marjamaa, K.; Kukkola, E.M.; Fagerstedt, K.V. The role of xylem class III peroxidases in lignification. J. Exp. Bot 2009, 60, 367–376. [Google Scholar]
- Li, X.; Weng, J.K.; Chapple, C. Improvement of biomass through lignin modification. Plant J 2008, 54, 569–581. [Google Scholar]
- Raes, J.; Rohde, A.; Christensen, J.H.; Van de Peer, Y.; Boerjan, W. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 2003, 133, 1051–1071. [Google Scholar]
| Species | Type of study | Tissue(s) | Relevant results | Reference |
|---|---|---|---|---|
| L. usitatissimum | histology, GC-analysis | inner and outer stem tissues | different pattern of laccase-gold particles detected in stem tissues which relates to phenolic compounds | [75] |
| L. usitatissimum | histochemistry/chemical analysis | bast fibres at 2 developmental stages | xylem and bast fibres rich in G units, H units present in bast fibres | [76] |
| L. usitatissimum | ESTs from cDNA library/RT-PCR | outer stem tissue | highly expressed cell wall-related ESTs found: CesAs, XTH, peroxidase, β-xylosidase and β-galactosidase | [77] |
| C. sativa | microscopy/chemical analysis | bast fibres from apical and basal regions of the stem | condensed lignin in stem, p-hydroxyphenyl units decline with maturity | [78] |
| C. sativa | cDNA microarray/qPCR | outer stem tissue top, middle, bottom | GT transcripts enriched in the middle and bottom regions, GH transcripts enriched in the top region; top region rich in PRPs; middle and bottom regions rich in AGPs | [79] |
| L. usitatissimum | cDNA library | fibre-bearing phloem tissues from stem/3 segments along stem axis | AGP, LTP enriched in elongation and cell wall-thickening regions; chitinases, β-galactosidases enriched also in specific stages of stem development | [80] |
| C. sativa | cDNA microarray/lignin histochemistry/chemical analysis/Northern blot | wooden core vs. bast fibres in the top, middle, bottom regions | upregulation of PP and shikimate pathways, AAA and lignin biosynthesis, C1 metabolism in the core tissue; AGPs, genes in lipid/wax metabolism and photosynthesis upregulated in bast tissues | [81] |
| L. usitatissimum | histology, 2D-DIGE | isolated fibres, cortical tissue | enrichment of β-galactosidase, rhamnose biosynthetic enzyme | [82] |
| L. usitatissimum | cDNA microarray/qPCR/activity staining | hypocotyls at 7, 9 and 15 days | genes involved in primary wall deposition upregulated in the early stage, AGPS and β-galactosidase expressed in the middle stage, secondary metabolism and GH transcripts upregulated in the last stage | [83] |
| L. usitatissimum | 454 sequencing of high-density oligo-microarray/qPCR | different tissues and cultivars (Drakkar vs. Belinka) | secondary metabolism-associated genes highly represented in stems; 3 FLAs, laccases overexpressed in core tissue; 3 FLAs overexpressed in outer stem tissue; 6 LTPs, lipid/wax metabolism and photosynthesis-associated genes overexpressed in outer tissue; cultivars differ in cell wall- and biotic stress response-related genes | [84] |
| L. usitatissimum | cDNA library | bark | 21 cell wall-related ESTs and C2H2 transcription factors identified | [85] |
| L. usitatissimum | lignomics/microarray | inner and outer stem tissues | hypolignification associated with low abundance of monolignol biosynthetic genes, 81 phenolic compounds found, 65 identified for the first time, lignan-associated genes abundant in inner tissues | [86] |
| L. usitatissimum | miRNA | bottom leaves, stem, leafless apex | 20 miRNA identified, which could regulate cell wall metabolism | [87] |
| L. usitatissimum | Cell wall proteome, 1D-PAGE, MALDI-TOF | isolated fibres | List of potential cell wall proteins of flax fibres | [88] |
| L. usitatissimum | Cell wall proteome, LC-MS/MS | Cortical tissue, cell wall protein enrichment | Identification of > 150 predicted cell wall proteins, major group acting on sugars and glycoproteins | [89] |
| B. nivea | Illumina paired-end sequencing | xylem, shoot, leaves, bark from seedlings, 30- and 60-days old plants | 51 genes of the CesA superfamily identified, 36 showing high expression in bark | [90] |
| Plant | Fibre type | Length (mm) | Diameter (μm) | MFA |
|---|---|---|---|---|
| Hemp | Bast | 5–60 | 20–40 | 4° |
| Hemp | Hurd | 0.2–0.6 | 10–30 | 0°–10° in S2 layer; 70°–90° in S1 layer |
| Flax | Bast | 2–40 | 20–23 | 10° |
| Jute | Bast | 2–3 | 16 | 8° |
| Ramie | Bast | 40–150 | 30 | 8° |
| Sisal | Leaf | 2–4 | 20 | 20° |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Guerriero, G.; Sergeant, K.; Hausman, J.-F. Integrated -Omics: A Powerful Approach to Understanding the Heterogeneous Lignification of Fibre Crops. Int. J. Mol. Sci. 2013, 14, 10958-10978. https://doi.org/10.3390/ijms140610958
Guerriero G, Sergeant K, Hausman J-F. Integrated -Omics: A Powerful Approach to Understanding the Heterogeneous Lignification of Fibre Crops. International Journal of Molecular Sciences. 2013; 14(6):10958-10978. https://doi.org/10.3390/ijms140610958
Chicago/Turabian StyleGuerriero, Gea, Kjell Sergeant, and Jean-François Hausman. 2013. "Integrated -Omics: A Powerful Approach to Understanding the Heterogeneous Lignification of Fibre Crops" International Journal of Molecular Sciences 14, no. 6: 10958-10978. https://doi.org/10.3390/ijms140610958
APA StyleGuerriero, G., Sergeant, K., & Hausman, J.-F. (2013). Integrated -Omics: A Powerful Approach to Understanding the Heterogeneous Lignification of Fibre Crops. International Journal of Molecular Sciences, 14(6), 10958-10978. https://doi.org/10.3390/ijms140610958