Abstract
We explored the contribution of nitrosamine metabolism to lung cancer in a pilot investigation of genetic variation in CYP2B6, a high-affinity enzymatic activator of tobacco-specific nitrosamines with a negligible role in nicotine metabolism. Previously we found that variation in CYP2A6 and CHRNA5-CHRNA3-CHRNB4 combined to increase lung cancer risk in a case-control study in European American ever-smokers (n = 860). However, these genes are involved in the pharmacology of both nicotine, through which they alter smoking behaviours, and carcinogenic nitrosamines. Herein, we separated participants by CYP2B6 genotype into a high- vs. low-risk group (*1/*1 + *1/*6 vs. *6/*6). Odds ratios estimated through logistic regression modeling were 1.25 (95% CI 0.68–2.30), 1.27 (95% CI 0.89–1.79) and 1.56 (95% CI 1.04–2.31) for CYP2B6, CYP2A6 and CHRNA5-CHRNA3-CHRNB4, respectively, with negligible differences when all genes were evaluated concurrently. Modeling the combined impact of high-risk genotypes yielded odds ratios that rose from 2.05 (95% CI 0.39–10.9) to 2.43 (95% CI 0.47–12.7) to 3.94 (95% CI 0.72–21.5) for those with 1, 2 and 3 vs. 0 high-risk genotypes, respectively. Findings from this pilot point to genetic variation in CYP2B6 as a lung cancer risk factor supporting a role for nitrosamine metabolic activation in the molecular mechanism of lung carcinogenesis.
1. Introduction
Variation in genes participating in nicotine and nitrosamine pharmacology has been associated with lung cancer risk among smokers. We have reported that variation in the nicotine and nitrosamine metabolizing gene, CYP2A6, and the nicotinic receptor subunit gene cluster, CHRNA5-CHRNA3-CHRNB4 (CHRNA5-A3-B4), combined to increase cigarette consumption and lung cancer risk among European American cigarette smokers [1]. Owing to the contribution of these genes to nicotine pharmacology, CYP2A6 and CHRNA5-A3-B4 increase lung cancer risk indirectly through increased cigarette consumption and thus increased carcinogen exposure. Additionally, these genes could be contributing to lung cancer risk directly through altered nitrosamine pharmacology—the CYP2A6 enzyme metabolically activates tobacco specific nitrosamines (TSNA) [2] and the nicotinic receptors are high-affinity binding sites for nitrosamines [3].
To explicitly explore the potential contribution of altered nitrosamine pharmacology to lung cancer risk among smokers, we have extended our investigation of gene variants to CYP2B6. CYP2B6, like CYP2A6, is expressed in the lung in addition to the liver [4,5] and metabolically activates TSNAs to DNA-reactive products [2,6]. Enzymatic activation of the potent lung procarcinogenic TSNA, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), has been linked to NNK-induced lung tumors [7], and the CYP2B6 enzyme has a high affinity for NNK [8]—roughly ten-fold higher compared to CYP2A6 [6]. CYP2A6, on the other hand, has a higher affinity for the TSNA, N′-nitrosonornicotine (NNN), than for NNK [6,9] and may preferentially metabolize NNN while CYP2B6 preferentially metabolizes NNK. In contrast to CYP2A6, CYP2B6 has a low affinity for nicotine [8] and makes a minor contribution to systemic nicotine metabolism (<20%) and characterized gene variants are not associated with differences in cigarette consumption [10,11]. Consequently, CYP2B6 gene variants are biologically positioned to influence the risk of developing lung cancer among cigarette smokers through altered metabolic activation of nitrosamines (potentially within the lung) as opposed to indirectly through cigarette consumption.
Here, we present a feasibility study of genetic variation in CYP2B6 as a potential lung cancer risk factor. To propose a role for CYP2B6 gene variants and altered nitrosamine activation, it was critical to assess whether CYP2B6 operated independently of CYP2A6 to influence lung cancer risk among smokers. The CYP2B6 and CYP2A6 genes are located in proximity on chromosome 19q23 with their 5′ regulatory regions oriented towards each other [12,13], and reports of genetic linkage, co-regulation and correlated liver expression levels [11–14] coupled with the common role of these enzymes as activators of TSNAs suggested a potential redundant role of CYP2B6 and CYP2A6 gene variants. In this exploratory analysis, our main objectives were to estimate the effect size of CYP2B6 relative to CYP2A6 and CHRNA5-A3-B4 and to establish a probable independent contribution of CYP2B6 to the risk of lung cancer among smokers and in so doing implicate a role for altered nitrosamine activation in the molecular mechanism of lung carcinogenesis.
2. Results and Discussion
CYP2B6 genotype frequencies did not deviate from the Hardy-Weinberg equilibrium overall, in controls or in cases. We found the CYP2B6*6 haplotype and the CYP2B6*4 and *9 alleles at a frequency of 24%, 2.5% and 0%, respectively, in control subjects, which is consistent with other study samples of European descent [15,16]. Less than five percent (n = 41) of the 860 participants from the original study possessed the CYP2B6*4 allele in isolation precluding an independent investigation of this allele, and thus were excluded from analyses. Table 1 summarizes the characteristics and CYP2B6*6 genotyping results of the 819 study participants included in analyses.

Table 1.
Characteristics of study subjects and genotyping results.
To assess the genetic risk for developing lung cancer, participants were separated into a high- and low-risk genotype group for each gene. We employed the same high- vs. low-risk lung cancer genotype groups for CYP2A6 and for CHRNA5-A3-B4 as before (Table 1): CYP2A6 normal vs. reduced metabolizers (those who possess at least one reduced activity allele) and CHRNA5-A3-B4 AA vs. GG/GA [1]. For CYP2B6, the CYP2B6*6 haplotype appeared to act recessively whereby only those homozygous for the CYP2B6*6 allele were afforded protection from lung cancer risk. Hence, participants with the CYP2B6*1/*1 and CYP2B6*1/*6 genotypes were pooled together into the high-risk group, while participants with the CYP2B6*6/*6 genotype comprised the low-risk group. The effect of the CYP2B6*6 variant allele relative to the CYP2B6*1 wildtype allele on the enzymatic activation of TSNAs has not yet been characterized. Our findings appear to be similar to the effects of CYP2B6*6 on the well-characterized CYP2B6 substrate efavirenz where a reduction in in vitro and in vivo metabolism and a higher incidence of side effects were only evident with the CYP2B6*6/*6 genotype [17–19]. The CYP2B6*6 allele is associated with reduced protein expression [11,20]—perhaps a substantial reduction in CYP2B6 protein (i.e., CYP2B6*6/*6) is necessary to influence lung cancer risk.
Figure 1A presents lung cancer odds ratios for each gene alone with the low-risk genotype group serving as the reference group. Of note, the odds ratios for CYP2B6 and CYP2A6 were of similar magnitude, although the associations did not reach statistical significance. The odds ratio conferred by the CYP2B6*6 genotype is unlikely to be driven through an indirect association with CYP2A6 gene variants: we found no significant correlations between CYP2A6 and CYP2B6 gene variants (r2 = 0.00–0.01 for each pairwise comparison) when we investigated linkage disequilibrium using Haploview (v4.2) [21]. Furthermore, when we concurrently evaluated all three genes in a multivariate logistic regression model there was a negligible shift in the odds ratio for each gene (1.25 to 1.25 for CYP2B6; 1.27 to 1.26 for CYP2A6; 1.56 to 1.55 for CHRNA5-A3-B4) suggesting that variation in each gene is contributing independently to the risk of lung cancer.
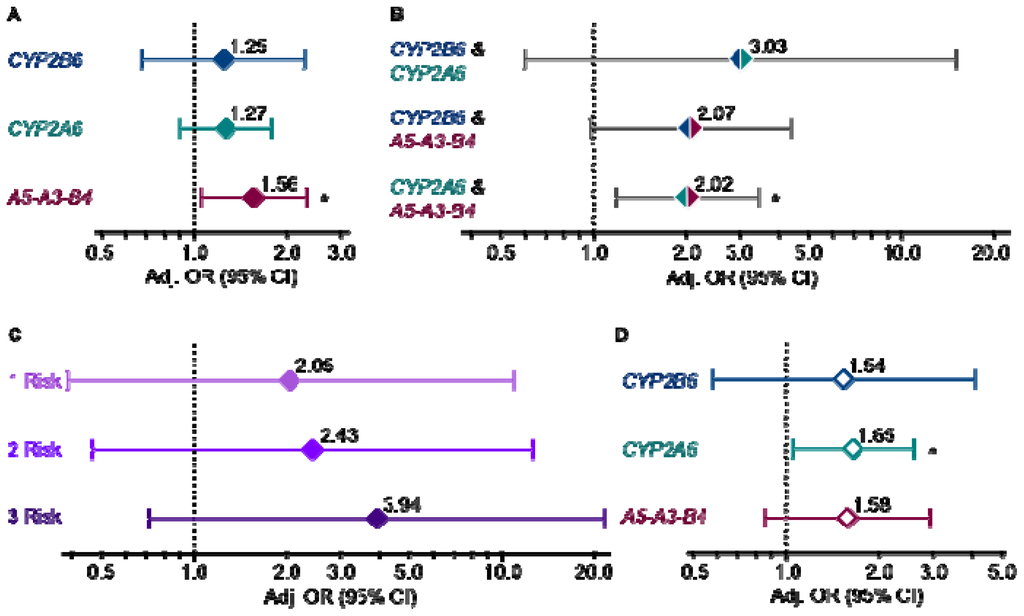
Figure 1.
(a) Lung cancer risk by CYP2B6, CYP2A6 and CHRNA5-A3-B4 genotype group for each gene alone (p = 0.47, 0.17 and 0.03); (b) each gene pair in combination (p = 0.18, 0.06 and 0.01); (c) all three genes in combination where participants were classified as having 0, 1, 2 or 3 high-risk genotypes and those with 0 high-risk genotypes served as the reference group (p = 0.40, 0.29 and 0.11); and (d) in the lighter-smoking stratum, cigarettes per day ≤ 20 (p = 0.39, 0.03 and 0.14). All odds ratios were determined by logistic regression modeling and adjusted for age, gender and log pack-years. Genotype comparisons were as follows: CYP2B6 *1/*1, *1/*6 vs. *6/*6, CYP2A6 normal vs. reduced metabolizer, CHRNA5-A3-B4 AA vs. GG/GA. *p ≤ 0.05. Refer to Supplementary Tables S1–S4 for subject numbers and unadjusted and adjusted lung cancer odds ratios for Figure 1A–D, respectively.
Combined gene analyses provided additional evidence that CYP2B6 was contributing independently to cancer risk. We first evaluated each gene pair by comparing participants possessing both high-risk genotypes to those with both low-risk genotypes (Figure 1B). For example, individuals in both the CYP2B6 and CYP2A6 high-risk genotype groups (CYP2B6 *1/*1, *1/*6 and CYP2A6 normal metabolizer) were compared to those in both low-risk genotype groups (CYP2B6 *6/*6 and CYP2A6 reduced metabolizer). Larger odds ratio for CYP2B6 and CYP2A6 in combination relative to either gene alone suggests that variation in these genes independently and additively contribute to lung cancer risk possibly through preferential enzymatic activation of distinct TSNAs—CYP2B6 has a high affinity for NNK, whereas CYP2A6 has a high affinity for NNN [8,9]. Comparable odds ratios for the combination of CHRNA5-A3-B4 with either CYP2B6 or CYP2A6 is also of note and may suggest that concomitant variation in a nicotine/nitrosamine pharmacokinetic and pharmacodynamic gene confers a consistent degree of lung cancer risk.
Combined variation in both CYP2B6 and CYP2A6 appeared to confer a greater increase in lung cancer risk (odds ratio of ~3.00) compared to variation in either CYP2B6 or CYP2A6 paired with variation in CHRNA5-A3-B4 (odds ratio of ~2.00). Similarly high odds ratio have been reported for the combined effect of variation in two genes encoding enzymatic activators of polycyclic aromatic hydrocarbons, MPO and CYP1A1 (odds ratio of 2.88) [22]. Genetic variation in multiple pharmacokinetic genes, particularly if activating different procarcinogens, presumably increases the load of activated procarcinogens from cigarette smoke leading to increased DNA adducts and perhaps an increased occurrence of DNA mutations that result in cellular transformation. While there have been numerous studies on the association of gene variants within cigarette smoke pharmacokinetic pathways and lung cancer, less is known regarding variation in pharmacodynamic pathways such as nicotinic receptor signaling, and it remains to be determined whether the combination of gene variants within pharmacokinetic and dynamic pathways will confer a degree of risk similar to that observed in this study.
The combined impact of variation in all three genes is presented in Figure 1C. We separated individuals into four groups based on the number of high-risk genotypes that they possessed: those with none of the high-risk genotypes (0 risk), any one (1 risk), any two (2 risk), or all three high-risk genotypes (3 risk). The 0 risk group served as the reference group. The odds ratios increased in magnitude with each increase in the number of high-risk genotypes. A post-estimation Wald test for homogeneity between the combined genotype groups (p = 0.06) also suggests that separating ever-smokers by their combined CYP2B6/CYP2A6/CHRNA5-A3-B4 genotypes stratified them by their risk for developing lung cancer.
To gain further insight into the mechanisms by which these gene variants could be contributing to lung cancer risk within the context of smoking, we performed subgroups analyses by daily cigarette use and by years of smoking. We previously noted that the impact of CYP2A6 gene variants was greater on lung cancer risk in the lighter-smoking half of this study population (cigarettes per day ≤20) [1]. We again performed a median split based on cigarette consumption (cigarettes/day ≤20 vs. >20) and findings in the lighter-smoking stratum are presented in Figure 1D. Similar to CYP2A6, the impact (odds ratio) of CYP2B6 was larger in the lighter-smoking stratum although it was not statistically significant. When we concurrently evaluated all three genes in a multivariate logistic regression model there was a negligible shift in the odds ratio for each gene (1.54 to 1.56 for CYP2B6; 1.65 to 1.64 for CYP2A6; 1.58 to 1.60 for CHRNA5-A3-B4) again suggesting that variation in each gene is contributing independently to the risk of lung cancer. Combined gene analyses also yielded larger odds ratios in the lighter-smoking stratum (data not shown). Gene effects at lower cigarette exposure have been reported for other metabolic genes such as CYP1A1, GSTM1 and MPO[23–25]. At high cigarette (carcinogen) exposure, subtle differences in nitrosamine activation based on genotype may be difficult to detect [26]. Of note, mean daily cigarette consumption within the lighter-smoking stratum was 16.6, which is similar to the reported mean of 15.1 among daily smokers in the US in 2010 [27]. Thus, risk estimates and risk factors among the lighter-smoking stratum in our study population are relevant to present day smoking habits.
We also performed a median split by smoking duration (years of smoking ≤38 vs. >38) to gain insight into the temporal relationship between genetic risk factors and carcinogenesis (data not shown). We observed nominally larger odds ratios for CHRNA5-A3-B4 in the shorter- vs. longer-duration stratum, 1.66 vs. 1.27, consistent with the association of variation in this chromosomal region with an earlier age of cancer onset [28]. For CYP2B6 and CYP2A6, we observed nominally larger odds ratios in the longer- vs. shorter-duration stratum, 2.05 vs. 0.75 and 1.66 vs. 1.03, respectively. Both enzymes activate nitrosamines into reactive species capable of forming DNA adducts [2,6,8] and perhaps larger odds ratios in the longer-duration stratum reflect increasing cancer risk through an accumulation of carcinogenesis-promoting genotoxic events. The influence of CYP2B6 genotype group among those smoking for a longer duration appeared to be relatively larger than for CYP2A6 genotype group. However, it bears noting that this genetic association study was not powered for multiple subgroups analyses.
Based on our results implicating CYP2B6 and CYP2A6 gene variants with the risk of developing lung cancer risk among smokers, we hypothesize that altered nitrosamine enzymatic activation contributes to the molecular mechanism of lung carcinogenesis. Evidence corroborating our interpretation include human data demonstrating that CYP2A6 inhibition in cigarette smokers was associated with reduced activation of NNK as indicated by the increased routing of NNK to the metabolite NNAL [29], and by mouse data demonstrating that inhibition of mouse CYP2A enzymes reduced the occurrence of NNK-induced adenomas [30]. Variation in CYP2A13 may also contribute to differences in nitrosamine activation and resulting cancer risk among cigarette smokers—CYP2A13 is predominantly expressed in the respiratory tract and, like CYP2B6, has a high affinity for NNK while playing a negligible role in peripheral nicotine metabolism [31,32]. A number of CYP2A13 gene variants have been discovered [33], and the CYP2A13*2 allele was associated with lung cancer risk among Chinese [34]. CYP2A13 was not genotyped within this sample due to the low prevalence of gene variants among European Americans [33].
There are a number of limitations to this study that are inherent to most genetic association studies investigating cancer risk. For example, odds ratio adjustments for cigarette exposure relied on cigarette pack-years, which was derived from self-reported average daily cigarette consumption and years of smoking and may not adequately capture inter-individual differences in carcinogen exposure over time. Statistical power was also a limitation, as anticipated based on results from our previous investigation of CYP2A6, which only reached significance in the lighter-smoking stratum [1]. Assuming a comparable effect size for CYP2B6, even analyses within the lighter-smoking stratum would be underpowered due to the smaller size of the reference CYP2B6 genotype group, CYP2B6*6/*6. Thus, reported findings were meant to be mainly investigative (hypothesis generating) in nature but strongly support further investigation of CYP2B6, particularly in smoking populations characterized by a higher prevalence of CYP2B6 gene variants and by lighter-smoking, such as African Americans and Alaska Native people [15,35]. Both African American and Alaska Native smokers experience more difficulty quitting despite lower cigarette consumption [36–39], and it was among the longer-duration and lighter-smokers where we observed larger odds ratio for CYP2B6 and CYP2A6. Among these populations, which have a disproportionately high lung cancer risk [40,41], CYP2B6 genotype may facilitate the identification of sub-groups of smokers at higher and lower cancer risk.
3. Experimental Section
We investigated the association of CYP2B6 gene variants with lung cancer risk in the case-control population in which we previously investigated CYP2A6 and CHRNA5-A3-B4[1] enabling us to account for the influence of CYP2A6 and CHRNA5-A3-B4, which we already knew to be associated with both cigarette consumption and lung cancer risk. Briefly, all study participants were current or former smokers of European descent. Healthy control subjects were frequency matched to non-small cell lung cancer case subjects by age, gender and smoking variables [1]. This study was approved by Review Boards at the M.D. Anderson Cancer Center (Houston, TX, USA) and the University of Toronto (Toronto, ON, Canada).
For the current analyses, we genotyped participants for the CYP2B6*6 allele, a haplotype defined by the co-occurrence of non-synonymous single nucleotide polymorphisms in exons 4 (G516T) and 5 (A785G) [15,16]. The CYP2B6*6 PCR-based genotyping assay begins with a gene-specific amplification followed by a haplotype-specific amplification in which the co-occurrence of the exon 4 and 5 variants is detected using a forward primer targeted to the exon 4 variant and a reverse primer targeted to the exon 5 variant [16]. The CYP2B6*6 haplotype is prevalent in all world populations ranging in frequency from approximately 14% to 62% [15] and is associated with reduced protein expression and reduced in vivo activity for a number of substrates [11,15,42]. We previously genotyped participants for CYP2A6 alleles with reduced activity that are common in populations of European descent (CYP2A6*2, *4, *9, *12) and for the SNP rs1051730 G>A, which tags the loci associated with both smoking and lung cancer risk within the CHRNA5-A3-B4 cluster on chromosome 15q25.1 [1,43].
Odds ratios were estimated using logistic regression modeling, which permitted the concurrent evaluation of potential covariates such as cigarette pack-years. Reported odds ratios were adjusted for age, gender and log cigarette pack-years, a common proxy for cigarette exposure. We also adjusted odds ratios for cigarette consumption and years of smoking, separately and concurrently, but observed negligible shifts in the results. In all lung cancer risk analyses, the low-risk genotype group served as the reference group. Statistical analyses were performed with Stata Release 11 (StataCorp LP, College Station, TX, USA).
4. Conclusions
CYP2B6 gene variants are biologically positioned to influence the risk of developing lung cancer among cigarette smokers through altered enzymatic activation of nitrosamines. However, based on reports of genetic linkage and co-regulation with CYP2A6, coupled with a potential redundancy of function through substrate overlap, it was unclear whether CYP2B6 would influence lung cancer risk independently of CYP2A6. Furthermore, both enzymes are expressed within the lung and localized nitrosamine activation has been linked to nitrosamine-induced lung tumors [2,4,7].
We found evidence that variation in CYP2B6 influences lung cancer risk with a similar magnitude to CYP2A6 and that variation in these genes influences lung cancer risk independently. Preferential enzymatic activation of distinct TSNAs, NNK by CYP2B6 and NNN by CYP2A6 [8,9], may underlie the observed independent contributions of these genes to lung cancer risk. Combined analyses of concurrent variation in CYP2B6, CYP2A6 and CHRNA5-A3-B4 appeared to stratify ever-smokers by lung cancer risk supporting an important role for variation in both nicotine/nitrosamine pharmacokinetic and pharmacodynamic pathways to the risk for lung cancer among smokers.
Based on our findings, lung cancer genetic association studies among ethnic populations with a higher percentage of CYP2B6 gene variants would be beneficial to validate our observations. Furthermore, additional animal studies modulating the expression and/or activity level of nitrosamine metabolizing enzymes, such as CYP2A and CYP2B, would provide valuable insight into the biological mechanism behind our observed differences in lung cancer risk by CYP2B6 and CYP2A6 gene variants.
Supplementary Information
ijms-14-08381-s001.pdfAcknowledgments
This work was supported by the Canadian Institutes of Health Research (MOP86471 to R.F.T., Doctoral Award to C.A.W.); the Campbell Family Mental Health Research Institute, the Centre for Addiction and Mental Health and the CAMH Foundation; a University Endowed Chair in Addictions (R.F.T.); the National Institutes of Health (U01 DA020830 to R.F.T., CA55769 and CA127219 to M.R.S., U19 CA148127, CA133996 and CA121197 to C.I.A.); the Kleberg Center for Molecular Markers at M.D. Anderson Cancer Center; the Flight Attendant Medical Research Institutes; an Ashley Studentship for Research in Tobacco Control (C.A.W.); the Canada Foundation for Innovation (20289 and 16014 to R.F.T.) and the Ontario Ministry of Research and Innovation.
Conflict of Interest
R.F.T. has consulted for Novartis and McNeil. No other conflicts to disclose.
References
- Wassenaar, C.A.; Dong, Q.; Wei, Q.; Amos, C.I.; Spitz, M.R.; Tyndale, R.F. Relationship between CYP2A6 and CHRNA5-CHRNA3-CHRNB4 variation and smoking behaviors and lung cancer risk. J. Natl. Cancer Inst 2011, 103, 1342–1346. [Google Scholar]
- Rossini, A.; de Almeida Simao, T.; Albano, R.M.; Pinto, L.F. CYP2A6 polymorphisms and risk for tobacco-related cancers. Pharmacogenomics 2008, 9, 1737–1752. [Google Scholar]
- Schuller, H.M. Is cancer triggered by altered signalling of nicotinic acetylcholine receptors? Nat. Rev. Cancer 2009, 9, 195–205. [Google Scholar]
- Gervot, L.; Rochat, B.; Gautier, J.C.; Bohnenstengel, F.; Kroemer, H.; de Berardinis, V.; Martin, H.; Beaune, P.; de Waziers, I. Human CYP2B6: Expression, inducibility and catalytic activities. Pharmacogenetics 1999, 9, 295–306. [Google Scholar]
- Anttila, S.; Raunio, H.; Hakkola, J. Cytochrome P450-mediated pulmonary metabolism of carcinogens: Regulation and cross-talk in lung carcinogenesis. Am. J. Respir. Cell Mol. Biol 2011, 44, 583–590. [Google Scholar]
- Jalas, J.R.; Hecht, S.S.; Murphy, S.E. Cytochrome P450 enzymes as catalysts of metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, a tobacco specific carcinogen. Chem. Res. Toxicol 2005, 18, 95–110. [Google Scholar]
- Weng, Y.; Fang, C.; Turesky, R.J.; Behr, M.; Kaminsky, L.S.; Ding, X. Determination of the role of target tissue metabolism in lung carcinogenesis using conditional cytochrome P450 reductase-null mice. Cancer Res 2007, 67, 7825–7832. [Google Scholar]
- Dicke, K.E.; Skrlin, S.M.; Murphy, S.E. Nicotine and 4-(methylnitrosamino)-1-(3-pyridyl)-butanone metabolism by cytochrome P450 2B6. Drug. Metab. Dispos 2005, 33, 1760–1764. [Google Scholar]
- Wong, H.L.; Murphy, S.E.; Hecht, S.S. Cytochrome P450 2A-catalyzed metabolic activation of structurally similar carcinogenic nitrosamines: N′-nitrosonornicotine enantiomers, N-nitrosopiperidine, and N-nitrosopyrrolidine. Chem. Res. Toxicol 2005, 18, 61–69. [Google Scholar]
- Lee, A.M.; Jepson, C.; Shields, P.G.; Benowitz, N.; Lerman, C.; Tyndale, R.F. CYP2B6 genotype does not alter nicotine metabolism, plasma levels, or abstinence with nicotine replacement therapy. Cancer Epidemiol. Biomark. Prev 2007, 16, 1312–1314. [Google Scholar]
- Al Koudsi, N.; Tyndale, R.F. Hepatic CYP2B6 is altered by genetic, physiologic, and environmental factors but plays little role in nicotine metabolism. Xenobiotica 2010, 40, 381–392. [Google Scholar]
- Haberl, M.; Anwald, B.; Klein, K.; Weil, R.; Fuss, C.; Gepdiremen, A.; Zanger, U.M.; Meyer, U.A.; Wojnowski, L. Three haplotypes associated with CYP2A6 phenotypes in Caucasians. Pharmacogenet. Genomics 2005, 15, 609–624. [Google Scholar]
- Hoffman, S.M.; Nelson, D.R.; Keeney, D.S. Organization, structure and evolution of the CYP2 gene cluster on human chromosome 19. Pharmacogenetics 2001, 11, 687–698. [Google Scholar]
- Johnstone, E.; Benowitz, N.; Cargill, A.; Jacob, R.; Hinks, L.; Day, I.; Murphy, M.; Walton, R. Determinants of the rate of nicotine metabolism and effects on smoking behavior. Clin. Pharmacol. Ther 2006, 80, 319–330. [Google Scholar]
- Zanger, U.M.; Klein, K.; Saussele, T.; Blievernicht, J.; Hofmann, M.H.; Schwab, M. Polymorphic CYP2B6: Molecular mechanisms and emerging clinical significance. Pharmacogenomics 2007, 8, 743–759. [Google Scholar]
- Lee, A.M.; Jepson, C.; Hoffmann, E.; Epstein, L.; Hawk, L.W.; Lerman, C.; Tyndale, R.F. CYP2B6 genotype alters abstinence rates in a bupropion smoking cessation trial. Biol. Psychiatr 2007, 62, 635–641. [Google Scholar]
- Xu, C.; Ogburn, E.T.; Guo, Y.; Desta, Z. Effects of the CYP2B6*6 allele on catalytic properties and inhibition of CYP2B6 in vitro: Implication for the mechanism of reduced efavirenz metabolism and other CYP2B6 substrates in vivo. Drug Metab. Dispos 2012, 40, 717–725. [Google Scholar]
- Yimer, G.; Amogne, W.; Habtewold, A.; Makonnen, E.; Ueda, N.; Suda, A.; Worku, A.; Haefeli, W.E.; Burhenne, J.; Aderaye, G.; et al. High plasma efavirenz level and CYP2B6*6 are associated with efavirenz-based HAART-induced liver injury in the treatment of naive HIV patients from Ethiopia: A prospective cohort study. Pharmacogenomics J 2011, 12, 499–506. [Google Scholar]
- Rotger, M.; Tegude, H.; Colombo, S.; Cavassini, M.; Furrer, H.; Decosterd, L.; Blievernicht, J.; Saussele, T.; Gunthard, H.F.; Schwab, M.; et al. Predictive value of known and novel alleles of CYP2B6 for efavirenz plasma concentrations in HIV-infected individuals. Clin. Pharmacol. Ther 2007, 81, 557–566. [Google Scholar]
- Desta, Z.; Saussele, T.; Ward, B.; Blievernicht, J.; Li, L.; Klein, K.; Flockhart, D.A.; Zanger, U.M. Impact of CYP2B6 polymorphism on hepatic efavirenz metabolism in vitro. Pharmacogenomics 2007, 8, 547–558. [Google Scholar]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar]
- Larsen, J.E.; Colosimo, M.L.; Yang, I.A.; Bowman, R.; Zimmerman, P.V.; Fong, K.M. CYP1A1 Ile462Val and MPO G-463A interact to increase risk of adenocarcinoma but not squamous cell carcinoma of the lung. Carcinogenesis 2006, 27, 525–532. [Google Scholar]
- Alexandrie, A.K.; Nyberg, F.; Warholm, M.; Rannug, A. Influence of CYP1A1, GSTM1, GSTT1, and NQO1 genotypes and cumulative smoking dose on lung cancer risk in a Swedish population. Cancer Epidemiol. Biomark. Prev 2004, 13, 908–914. [Google Scholar]
- Ishibe, N.; Wiencke, J.K.; Zuo, Z.F.; McMillan, A.; Spitz, M.; Kelsey, K.T. Susceptibility to lung cancer in light smokers associated with CYP1A1 polymorphisms in Mexican- and African-Americans. Cancer Epidemiol. Biomark. Prev 1997, 6, 1075–1080. [Google Scholar]
- Schabath, M.B.; Spitz, M.R.; Hong, W.K.; Delclos, G.L.; Reynolds, W.F.; Gunn, G.B.; Whitehead, L.W.; Wu, X. A myeloperoxidase polymorphism associated with reduced risk of lung cancer. Lung Cancer 2002, 37, 35–40. [Google Scholar]
- Schwartz, A.G.; Prysak, G.M.; Bock, C.H.; Cote, M.L. The molecular epidemiology of lung cancer. Carcinogenesis 2007, 28, 507–518. [Google Scholar]
- King, B.; Dube, S.; Kaufmann, R.; Shaw, L.; Pechacek, T. Vital signs: Current cigarette smoking among adults aged ≥18 years—United States, 2005–2010. Morb. Mortal. Wkly. Rep 2011, 60, 1207–1212. [Google Scholar]
- Lips, E.H.; Gaborieau, V.; McKay, J.D.; Chabrier, A.; Hung, R.J.; Boffetta, P.; Hashibe, M.; Zaridze, D.; Szeszenia-Dabrowska, N.; Lissowska, J.; et al. Association between a 15q25 gene variant, smoking quantity and tobacco-related cancers among 17,000 individuals. Int. J. Epidemiol 2010, 39, 563–577. [Google Scholar]
- Sellers, E.M.; Ramamoorthy, Y.; Zeman, M.V.; Djordjevic, M.V.; Tyndale, R.F. The effect of methoxsalen on nicotine and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) metabolism in vivo. Nicotine Tob. Res 2003, 5, 891–899. [Google Scholar]
- Takeuchi, H.; Saoo, K.; Yokohira, M.; Ikeda, M.; Maeta, H.; Miyazaki, M.; Yamazaki, H.; Kamataki, T.; Imaida, K. Pretreatment with 8-methoxypsoralen, a potent human CYP2A6 inhibitor, strongly inhibits lung tumorigenesis induced by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in female A/J mice. Cancer Res 2003, 63, 7581–7583. [Google Scholar]
- Bao, Z.; He, X.Y.; Ding, X.; Prabhu, S.; Hong, J.Y. Metabolism of nicotine and cotinine by human cytochrome P450 2A13. Drug Metab. Dispos 2005, 33, 258–261. [Google Scholar]
- Ding, X.; Kaminsky, L.S. Human extrahepatic cytochromes P450: Function in xenobiotic metabolism and tissue-selective chemical toxicity in the respiratory and gastrointestinal tracts. Annu. Rev. Pharmacol. Toxicol 2003, 43, 149–173. [Google Scholar]
- Cauffiez, C.; Lo-Guidice, J.M.; Quaranta, S.; Allorge, D.; Chevalier, D.; Cenee, S.; Hamdan, R.; Lhermitte, M.; Lafitte, J.J.; Libersa, C.; et al. Genetic polymorphism of the human cytochrome CYP2A13 in a French population: Implication in lung cancer susceptibility. Biochem. Biophys. Res. Commun 2004, 317, 662–669. [Google Scholar]
- Wang, H.; Tan, W.; Hao, B.; Miao, X.; Zhou, G.; He, F.; Lin, D. Substantial reduction in risk of lung adenocarcinoma associated with genetic polymorphism in CYP2A13, the most active cytochrome P450 for the metabolic activation of tobacco-specific carcinogen NNK. Cancer Res 2003, 63, 8057–8061. [Google Scholar]
- Binnington, M.J.; Zhu, A.Z.; Renner, C.C.; Lanier, A.P.; Hatsukami, D.K.; Benowitz, N.L.; Tyndale, R.F. CYP2A6 and CYP2B6 genetic variation and its association with nicotine metabolism in South Western Alaska Native people. Pharmacogenet Genomics 2012, 22, 429–440. [Google Scholar]
- Boles, M.; Rohde, K.; He, H.; Maher, J.E.; Stark, M.J.; Fenaughty, A.; O’Connor, T. Effectiveness of a tobacco quitline in an indigenous population: A comparison between Alaska Native people and other first-time quitline callers who set a quit date. Int. J. Circumpolar Health 2009, 68, 170–181. [Google Scholar]
- Fu, S.S.; Kodl, M.M.; Joseph, A.M.; Hatsukami, D.K.; Johnson, E.O.; Breslau, N.; Wu, B.; Bierut, L. Racial/Ethnic disparities in the use of nicotine replacement therapy and quit ratios in lifetime smokers ages 25 to 44 years. Cancer Epidemiol. Biomark. Prev 2008, 17, 1640–1647. [Google Scholar]
- Murray, R.P.; Connett, J.E.; Buist, A.S.; Gerald, L.B.; Eichenhorn, M.S. Experience of Black participants in the Lung Health Study smoking cessation intervention program. Nicotine Tob. Res 2001, 3, 375–382. [Google Scholar]
- Smith, J.J.; Ferucci, E.D.; Dillard, D.A.; Lanier, A.P. Tobacco use among Alaska Native people in the EARTH study. Nicotine Tob Res 2010, 12, 839–844. [Google Scholar]
- Lanier, A.P.; Kelly, J.J.; Maxwell, J.; McEvoy, T.; Homan, C. Cancer in Alaska Native people, 1969–2003. Alaska Med 2006, 48, 30–59. [Google Scholar]
- Haiman, C.A.; Stram, D.O.; Wilkens, L.R.; Pike, M.C.; Kolonel, L.N.; Henderson, B.E.; Le Marchand, L. Ethnic and racial differences in the smoking-related risk of lung cancer. N. Engl. J. Med 2006, 354, 333–342. [Google Scholar]
- Hofmann, M.H.; Blievernicht, J.K.; Klein, K.; Saussele, T.; Schaeffeler, E.; Schwab, M.; Zanger, U.M. Aberrant splicing caused by single nucleotide polymorphism c.516 G>T [Q172H], a marker of CYP2B6*6, is responsible for decreased expression and activity of CYP2B6 in liver. J. Pharmacol. Exp. Ther 2008, 325, 284–292. [Google Scholar]
- Saccone, N.L.; Culverhouse, R.C.; Schwantes-An, T.H.; Cannon, D.S.; Chen, X.; Cichon, S.; Giegling, I.; Han, S.; Han, Y.; Keskitalo-Vuokko, K.; et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: A meta-analysis and comparison with lung cancer and COPD. PLoS Genet 2010, 6, e1001053. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).