Small Regulatory RNAs in the Control of Motility and Biofilm Formation in E. coli and Salmonella
Abstract
:1. Structural Organization and Regulation of Biofilms
1.1. The Switch from Motile to Sessile Lifestyle—Inverse Regulation of Flagellar and Curli/Cellulose Control Cascades
1.2. Regulation of Other Biofilm Matrix Components: Colanic Acid and PGA
2. Regulation of Biofilm Formation by Small Noncoding RNAs
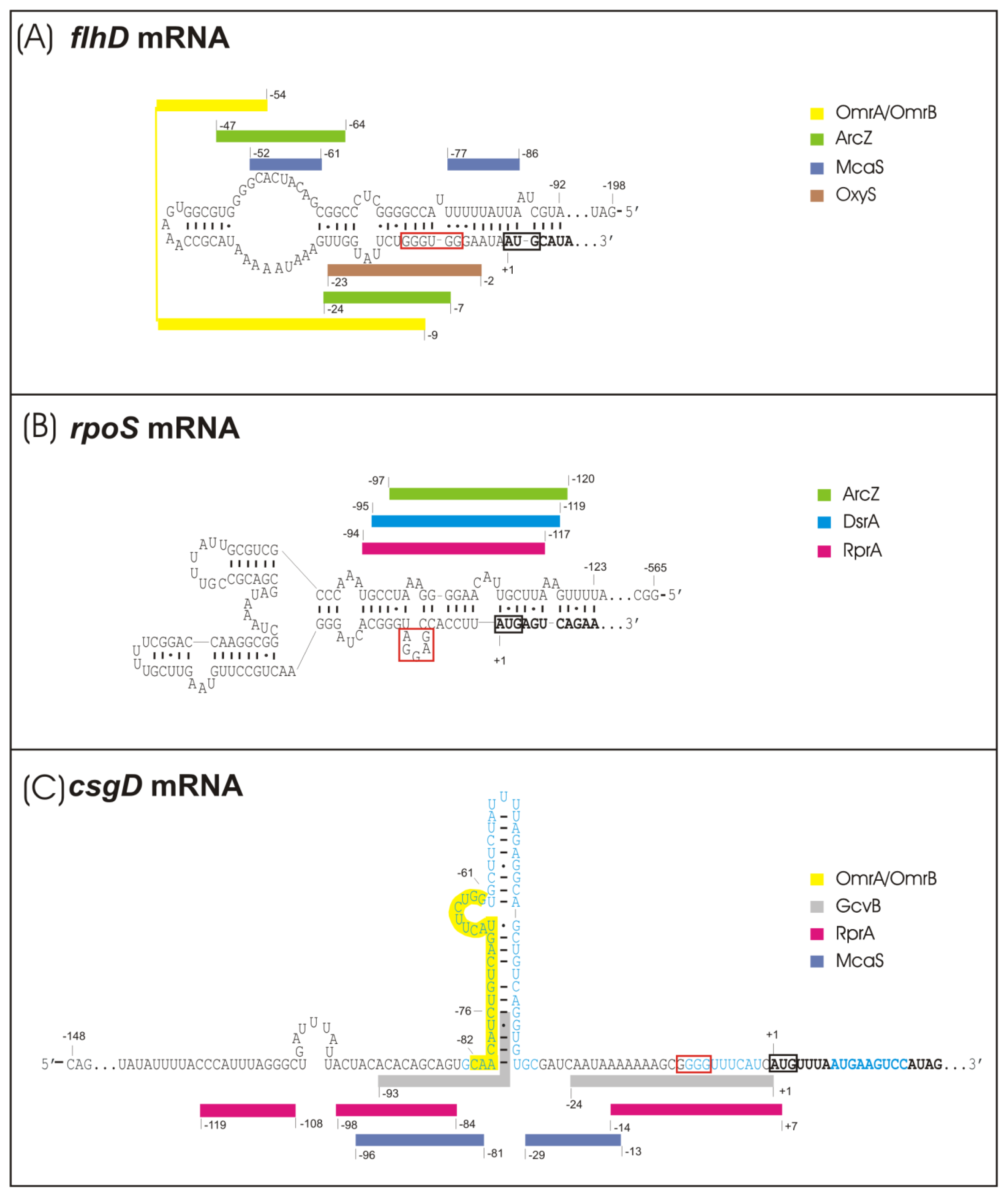
2.1. sRNA-Mediated Regulation of the Flagellar Cascade—flhDC mRNA as a Major Site for sRNA Action
2.2. sRNA-Mediated Regulation of the Curli Control Cascade—rpoS, ydaM and csgD mRNAs as sRNA Targets
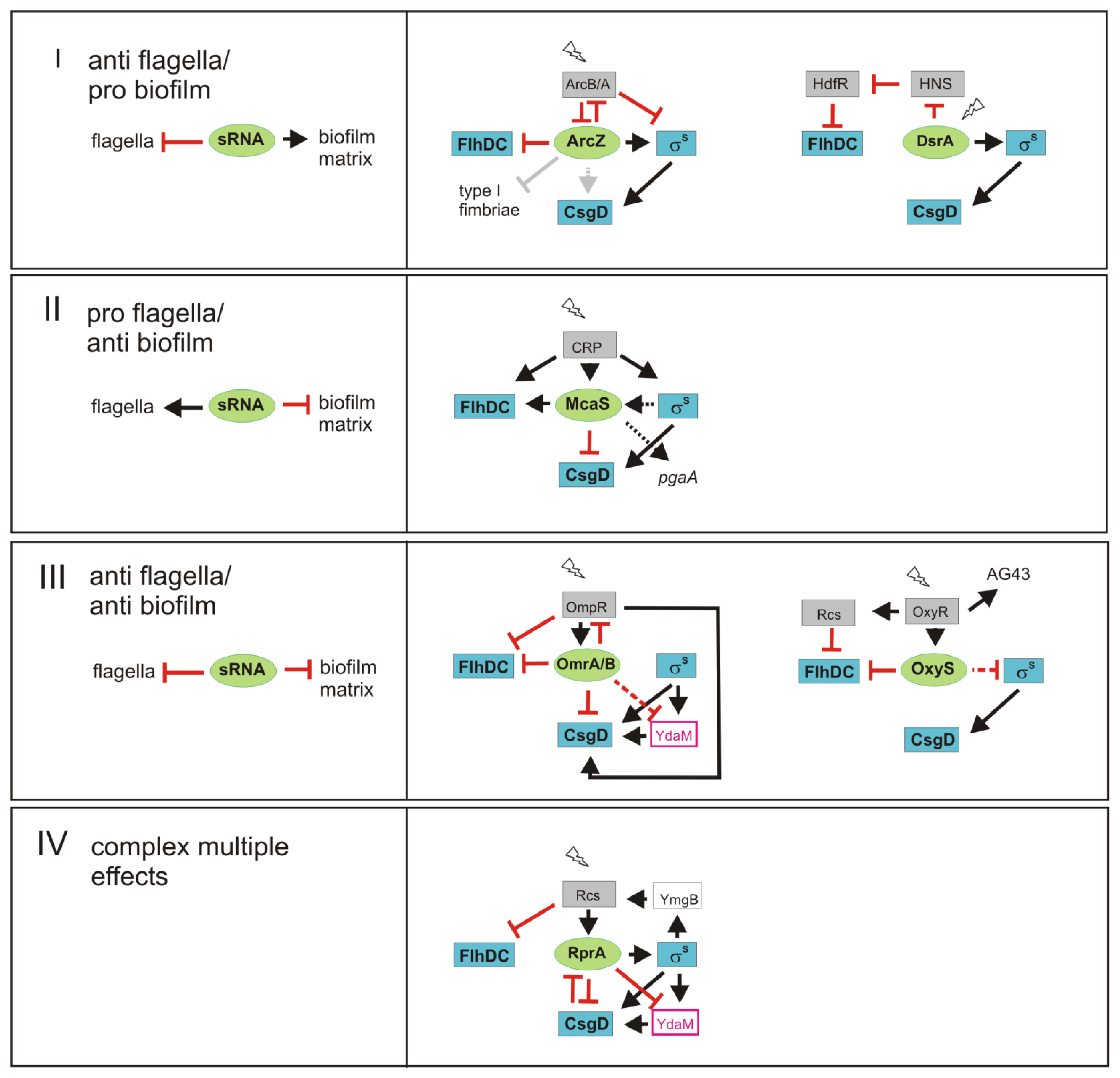
3. sRNAs Contribute to Inverse Regulation of Flagella and Biofilm Components in Different Regulatory Patterns
4. Species-Specific Variations in the Regulation by sRNAs
5. Conclusions and Perspectives
Acknowledgments
Conflict of Interest
References
- Karatan, E.; Watnick, P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev 2009, 73, 310–347. [Google Scholar]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol 2010, 8, 623–633. [Google Scholar]
- Branda, S.S.; Vik, S.; Friedman, L.; Kolter, R. Biofilms: The matrix revisited. Trends Microbiol 2005, 13, 20–26. [Google Scholar]
- Anderson, G.G.; O’Toole, G.A. Innate and induced resistance mechanisms of bacterial biofilms. Curr. Top. Microbiol. Immunol 2008, 322, 85–105. [Google Scholar]
- Danese, P.N.; Pratt, L.A.; Dove, S.L.; Kolter, R. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol 2000, 37, 424–432. [Google Scholar]
- Agladze, K.; Wang, X.; Romeo, T. Spatial periodicity of Escherichia coli K-12 biofilm microstructure initiates during a reversible, polar attachment phase of development and requires the polysaccharide adhesin PGA. J. Bacteriol 2005, 187, 8237–8246. [Google Scholar]
- Korea, C.G.; Ghigo, J.M.; Beloin, C. The sweet connection: Solving the riddle of multiple sugar-binding fimbrial adhesins in Escherichia coli: Multiple E. coli fimbriae form a versatile arsenal of sugar-binding lectins potentially involved in surface-colonisation and tissue tropism. Bioessays 2011, 33, 300–311. [Google Scholar]
- Beloin, C.; Roux, A.; Ghigo, J.M. Escherichia coli biofilms. Curr. Top. Microbiol. Immunol 2008, 322, 249–289. [Google Scholar]
- Pratt, L.A.; Kolter, R. Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol 1998, 30, 285–293. [Google Scholar]
- Wood, T.K.; Gonzalez Barrios, A.F.; Herzberg, M.; Lee, J. Motility influences biofilm architecture in Escherichia coli. Appl. Microbiol. Biotechnol 2006, 72, 361–367. [Google Scholar]
- Da Re, S.; Ghigo, J.M. A CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J. Bacteriol 2006, 188, 3073–3087. [Google Scholar]
- Bokranz, W.; Wang, X.; Tschape, H.; Romling, U. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J. Med. Microbiol 2005, 54, 1171–1182. [Google Scholar]
- Zogaj, X.; Nimtz, M.; Rohde, M.; Bokranz, W.; Romling, U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol 2001, 39, 1452–1463. [Google Scholar]
- Pesavento, C.; Becker, G.; Sommerfeldt, N.; Possling, A.; Tschowri, N.; Mehlis, A.; Hengge, R. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev 2008, 22, 2434–2446. [Google Scholar]
- Pesavento, C.; Hengge, R. The global repressor FliZ antagonizes gene expression by sigmaS-containing RNA polymerase due to overlapping DNA binding specificity. Nucleic Acids Res 2012, 40, 4783–4793. [Google Scholar]
- Zhao, K.; Liu, M.; Burgess, R.R. Adaptation in bacterial flagellar and motility systems: From regulon members to “foraging”-like behavior in E. coli. Nucleic Acids Res 2007, 35, 4441–4452. [Google Scholar]
- Chevance, F.F.; Hughes, K.T. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol 2008, 6, 455–465. [Google Scholar]
- Hengge, R. The General Stress Response in Gram-Negative Bacteria. In Bacterial Stress Responses; Storz, G., Hengge, R., Eds.; ASM Press: Washington, DC, USA, 2011; pp. 251–289. [Google Scholar]
- Jenal, U.; Malone, J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet 2006, 40, 385–407. [Google Scholar]
- Hengge, R. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol 2009, 7, 263–273. [Google Scholar]
- Ryjenkov, D.A.; Simm, R.; Roemling, U.; Gomelsky, M. The PilZ domain is a receptor for the second messenger c-di-GMP: The PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem 2006, 281, 30310–30314. [Google Scholar]
- Paul, K.; Nieto, V.; Carlquist, W.C.; Blair, D.F.; Harshey, R.M. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol. Cell 2010, 38, 128–139. [Google Scholar]
- Barembruch, C.; Hengge, R. Cellular levels and activity of the flagellar sigma factor FliA of Escherichia coli are controlled by FlgM-modulated proteolysis. Mol. Microbiol 2007, 65, 76–89. [Google Scholar]
- Tomoyasu, T.; Ohkishi, T.; Ukyo, Y.; Tokumitsu, A.; Takaya, A.; Suzuki, M.; Sekiya, K.; Matsui, H.; Kutsukake, K.; Yamamoto, T. The ClpXP ATP-dependent protease regulates flagellum synthesis in Salmonella enterica serovar typhimurium. J. Bacteriol 2002, 184, 645–653. [Google Scholar]
- Boehm, A.; Kaiser, M.; Li, H.; Spangler, C.; Kasper, C.A.; Ackermann, M.; Kaever, V.; Sourjik, V.; Roth, V.; Jenal, U. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 2010, 141, 107–116. [Google Scholar]
- Brown, P.K.; Dozois, C.M.; Nickerson, C.A.; Zuppardo, A.; Terlonge, J.; Curtiss, R., 3rd. MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2001, 41, 349–363. [Google Scholar]
- Weber, H.; Pesavento, C.; Possling, A.; Tischendorf, G.; Hengge, R. Cyclic-di-GMP-mediated signalling within the sigma network of Escherichia coli. Mol. Microbiol 2006, 62, 1014–1034. [Google Scholar]
- Ogasawara, H.; Yamamoto, K.; Ishihama, A. Regulatory role of MlrA in transcription activation of csgD, the master regulator of biofilm formation in Escherichia coli. FEMS Microbiol. Lett 2010, 312, 160–168. [Google Scholar]
- Ogasawara, H.; Yamada, K.; Kori, A.; Yamamoto, K.; Ishihama, A. Regulation of the Escherichia coli csgD promoter: Interplay between five transcription factors. Microbiology 2010, 156, 2470–2483. [Google Scholar]
- Prigent-Combaret, C.; Brombacher, E.; Vidal, O.; Ambert, A.; Lejeune, P.; Landini, P.; Dorel, C. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J. Bacteriol 2001, 183, 7213–7223. [Google Scholar]
- Gerstel, U.; Park, C.; Romling, U. Complex regulation of csgD promoter activity by global regulatory proteins. Mol. Microbiol 2003, 49, 639–654. [Google Scholar]
- Roemling, U.; Rohde, M.; Olsen, A.; Normark, S.; Reinkoster, J. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol 2000, 36, 10–23. [Google Scholar]
- Ogasawara, H.; Yamamoto, K.; Ishihama, A. Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagellar synthesis. J. Bacteriol 2011, 193, 2587–2597. [Google Scholar]
- Barnhart, M.M.; Chapman, M.R. Curli biogenesis and function. Annu. Rev. Microbiol 2006, 60, 131–147. [Google Scholar]
- Blanco, L.P.; Evans, M.L.; Smith, D.R.; Badtke, M.P.; Chapman, M.R. Diversity, biogenesis and function of microbial amyloids. Trends Microbiol 2012, 20, 66–73. [Google Scholar]
- Brombacher, E.; Dorel, C.; Zehnder, A.J.; Landini, P. The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology 2003, 149, 2847–2857. [Google Scholar]
- Roemling, U.; Sierralta, W.D.; Eriksson, K.; Normark, S. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol 1998, 28, 249–264. [Google Scholar]
- Amikam, D.; Galperin, M.Y. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 2006, 22, 3–6. [Google Scholar]
- Stout, V. Identification of the promoter region for the colanic acid polysaccharide biosynthetic genes in Escherichia coli K-12. J. Bacteriol 1996, 178, 4273–4280. [Google Scholar]
- Majdalani, N.; Gottesman, S. The Rcs phosphorelay: A complex signal transduction system. Annu. Rev. Microbiol 2005, 59, 379–405. [Google Scholar]
- Majdalani, N.; Hernandez, D.; Gottesman, S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol 2002, 46, 813–826. [Google Scholar]
- Ferrieres, L.; Clarke, D.J. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol. Microbiol 2003, 50, 1665–1682. [Google Scholar]
- Castanie-Cornet, M.P.; Cam, K.; Bastiat, B.; Cros, A.; Bordes, P.; Gutierrez, C. Acid stress response in Escherichia coli: Mechanism of regulation of gadA transcription by RcsB and GadE. Nucleic Acids Res 2010, 38, 3546–3554. [Google Scholar]
- Hagiwara, D.; Sugiura, M.; Oshima, T.; Mori, H.; Aiba, H.; Yamashino, T.; Mizuno, T. Genome-wide analyses revealing a signaling network of the RcsC-YojN-RcsB phosphorelay system in Escherichia coli. J. Bacteriol 2003, 185, 5735–5746. [Google Scholar]
- Francez-Charlot, A.; Laugel, B.; van Gemert, A.; Dubarry, N.; Wiorowski, F.; Castanie-Cornet, M.P.; Gutierrez, C.; Cam, K. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol 2003, 49, 823–832. [Google Scholar]
- Tschowri, N.; Busse, S.; Hengge, R. The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue-light response of Escherichia coli. Genes Dev 2009, 23, 522–534. [Google Scholar]
- Tschowri, N.; Lindenberg, S.; Hengge, R. Molecular function and potential evolution of the biofilm-modulating blue light-signalling pathway of Escherichia coli. Mol. Microbiol 2012, 85, 893–906. [Google Scholar]
- Jubelin, G.; Vianney, A.; Beloin, C.; Ghigo, J.M.; Lazzaroni, J.C.; Lejeune, P.; Dorel, C. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol 2005, 187, 2038–2049. [Google Scholar]
- Dorel, C.; Lejeune, P.; Rodrigue, A. The Cpx system of Escherichia coli, a strategic signaling pathway for confronting adverse conditions and for settling biofilm communities? Res. Microbiol 2006, 157, 306–314. [Google Scholar]
- Wei, B.L.; Brun-Zinkernagel, A.M.; Simecka, J.W.; Pruss, B.M.; Babitzke, P.; Romeo, T. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol 2001, 40, 245–256. [Google Scholar]
- Jonas, K.; Edwards, A.N.; Ahmad, I.; Romeo, T.; Romling, U.; Melefors, O. Complex regulatory network encompassing the Csr, c-di-GMP and motility systems of Salmonella Typhimurium. Environ. Microbiol 2010, 12, 524–540. [Google Scholar]
- Wang, X.; Dubey, A.K.; Suzuki, K.; Baker, C.S.; Babitzke, P.; Romeo, T. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol 2005, 56, 1648–1663. [Google Scholar]
- Jonas, K.; Edwards, A.N.; Simm, R.; Romeo, T.; Romling, U.; Melefors, O. The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol. Microbiol 2008, 70, 236–257. [Google Scholar]
- Boehm, A.; Steiner, S.; Zaehringer, F.; Casanova, A.; Hamburger, F.; Ritz, D.; Keck, W.; Ackermann, M.; Schirmer, T.; Jenal, U. Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress. Mol. Microbiol 2009, 72, 1500–1516. [Google Scholar]
- Jonas, K.; Melefors, O. The Escherichia coli CsrB and CsrC small RNAs are strongly induced during growth in nutrient-poor medium. FEMS Microbiol. Lett 2009, 297, 80–86. [Google Scholar]
- Froehlich, K.S.; Papenfort, K.; Berger, A.A.; Vogel, J. A conserved RpoS-dependent small RNA controls the synthesis of major porin OmpD. Nucleic Acids Res 2012, 40, 3623–3640. [Google Scholar]
- Benjamin, J.A.; Desnoyers, G.; Morissette, A.; Salvail, H.; Masse, E. Dealing with oxidative stress and iron starvation in microorganisms: An overview. Can. J. Physiol. Pharmacol 2010, 88, 264–272. [Google Scholar]
- Vogel, J.; Papenfort, K. Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol 2006, 9, 605–611. [Google Scholar]
- Gogol, E.B.; Rhodius, V.A.; Papenfort, K.; Vogel, J.; Gross, C.A. Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc. Natl. Acad. Sci. USA 2011, 108, 12875–12880. [Google Scholar]
- Mika, F.; Busse, S.; Possling, A.; Berkholz, J.; Tschowri, N.; Sommerfeldt, N.; Pruteanu, M.; Hengge, R. Targeting of csgD by the small regulatory RNA RprA links stationary phase, biofilm formation and cell envelope stress in Escherichia coli. Mol. Microbiol 2012, 84, 51–65. [Google Scholar]
- Thomason, M.K.; Fontaine, F.; de Lay, N.; Storz, G. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol. Microbiol 2012, 84, 17–35. [Google Scholar]
- De Lay, N.; Gottesman, S. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol. Microbiol 2012, 86, 524–538. [Google Scholar]
- Monteiro, C.; Papenfort, K.; Hentrich, K.; Ahmad, I.; le Guyon, S.; Reimann, R.; Grantcharova, N.; Romling, U. Hfq and Hfq-dependent small RNAs are major contributors to multicellular development in Salmonella enterica serovar Typhimurium. RNA Biol 2012, 9, 489–502. [Google Scholar]
- Jorgensen, M.G.; Nielsen, J.S.; Boysen, A.; Franch, T.; Moller-Jensen, J.; Valentin-Hansen, P. Small regulatory RNAs control the multi-cellular adhesive lifestyle of Escherichia coli. Mol. Microbiol 2012, 84, 36–50. [Google Scholar]
- Gottesman, S.; Storz, G. Bacterial small RNA regulators: Versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef]
- Storz, G.; Vogel, J.; Wassarman, K.M. Regulation by small RNAs in bacteria: Expanding frontiers. Mol. Cell 2011, 43, 880–891. [Google Scholar]
- Beisel, C.L.; Storz, G. Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol. Rev 2010, 34, 866–882. [Google Scholar]
- Romeo, T.; Vakulskas, C.A.; Babitzke, P. Post-transcriptional regulation on a global scale: Form and function of Csr/Rsm systems. Environ. Microbiol 2012, 15, 313–324. [Google Scholar]
- Bossi, L.; Schwartz, A.; Guillemardet, B.; Boudvillain, M.; Figueroa-Bossi, N. A role for Rho-dependent polarity in gene regulation by a noncoding small RNA. Genes Dev 2012, 26, 1864–1873. [Google Scholar]
- Vogel, J.; Luisi, B.F. Hfq and its constellation of RNA. Nat. Rev. Microbiol 2011, 9, 578–589. [Google Scholar]
- Udekwu, K.I.; Darfeuille, F.; Vogel, J.; Reimegard, J.; Holmqvist, E.; Wagner, E.G. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev 2005, 19, 2355–2366. [Google Scholar]
- Papenfort, K.; Pfeiffer, V.; Mika, F.; Lucchini, S.; Hinton, J.C.; Vogel, J. SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol 2006, 62, 1674–1688. [Google Scholar]
- Johansen, J.; Rasmussen, A.A.; Overgaard, M.; Valentin-Hansen, P. Conserved small non-coding RNAs that belong to the sigmaE regulon: Role in down-regulation of outer membrane proteins. J. Mol. Biol 2006, 364, 1–8. [Google Scholar]
- Soper, T.J.; Woodson, S.A. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA 2008, 14, 1907–1917. [Google Scholar]
- Holmqvist, E.; Reimegard, J.; Sterk, M.; Grantcharova, N.; Romling, U.; Wagner, E.G. Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis. EMBO J 2010, 29, 1840–1850. [Google Scholar]
- Mandin, P.; Gottesman, S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J 2010, 29, 3094–3107. [Google Scholar]
- Majdalani, N.; Cunning, C.; Sledjeski, D.; Elliott, T.; Gottesman, S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. USA 1998, 95, 12462–12467. [Google Scholar]
- Papenfort, K.; Said, N.; Welsink, T.; Lucchini, S.; Hinton, J.C.; Vogel, J. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol. Microbiol 2009, 74, 139–158. [Google Scholar]
- Guillier, M.; Gottesman, S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol 2006, 59, 231–247. [Google Scholar]
- Altuvia, S.; Weinstein-Fischer, D.; Zhang, A.; Postow, L.; Storz, G. A small, stable RNA induced by oxidative stress: Role as a pleiotropic regulator and antimutator. Cell 1997, 90, 43–53. [Google Scholar]
- Zhang, A.; Altuvia, S.; Tiwari, A.; Argaman, L.; Hengge-Aronis, R.; Storz, G. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J 1998, 17, 6061–6068. [Google Scholar]
- Liu, H.; Wang, X.; Wang, H.D.; Wu, J.; Ren, J.; Meng, L.; Wu, Q.; Dong, H.; Wu, J.; Kao, T.Y.; et al. Escherichia coli noncoding RNAs can affect gene expression and physiology of Caenorhabditis elegans. Nat. Commun. 2012, 3, 1073. [Google Scholar]
- Opdyke, J.A.; Kang, J.G.; Storz, G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol 2004, 186, 6698–6705. [Google Scholar]
- Tramonti, A.; de Canio, M.; de Biase, D. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: Transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol. Microbiol 2008, 70, 965–982. [Google Scholar]
- Opdyke, J.A.; Fozo, E.M.; Hemm, M.R.; Storz, G. RNase III participates in GadY-dependent cleavage of the gadX-gadW mRNA. J. Mol. Biol 2011, 406, 29–43. [Google Scholar]
- Liu, M.Y.; Gui, G.; Wei, B.; Preston, J.F., 3rd; Oakford, L.; Yuksel, U.; Giedroc, D.P.; Romeo, T. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem 1997, 272, 17502–17510. [Google Scholar]
- Suzuki, K.; Wang, X.; Weilbacher, T.; Pernestig, A.K.; Melefors, O.; Georgellis, D.; Babitzke, P.; Romeo, T. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J. Bacteriol 2002, 184, 5130–5140. [Google Scholar]
- Boehm, A.; Vogel, J. The csgD mRNA as a hub for signal integration via multiple small RNAs. Mol. Microbiol 2012, 84, 1–5. [Google Scholar]
- Weber, H.; Polen, T.; Heuveling, J.; Wendisch, V.F.; Hengge, R. Genome-wide analysis of the general stress response network in Escherichia coli: SigmaS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol 2005, 187, 1591–1603. [Google Scholar]
- Soper, T.; Mandin, P.; Majdalani, N.; Gottesman, S.; Woodson, S.A. Positive regulation by small RNAs and the role of Hfq. Proc. Natl. Acad. Sci. USA 2010, 107, 9602–9607. [Google Scholar]
- Sledjeski, D.D.; Gupta, A.; Gottesman, S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J 1996, 15, 3993–4000. [Google Scholar]
- Sledjeski, D.; Gottesman, S. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 1995, 92, 2003–2007. [Google Scholar]
- Lease, R.A.; Cusick, M.E.; Belfort, M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl. Acad. Sci. USA 1998, 95, 12456–12461. [Google Scholar]
- Barth, M.; Marschall, C.; Muffler, A.; Fischer, D.; Hengge-Aronis, R. Role for the histone-like protein H-NS in growth phase-dependent and osmotic regulation of sigmaS and many sigmaS-dependent genes in Escherichia coli. J. Bacteriol 1995, 177, 3455–3464. [Google Scholar]
- Ko, M.; Park, C. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J. Mol. Biol 2000, 303, 371–382. [Google Scholar]
- Majdalani, N.; Chen, S.; Murrow, J.; St John, K.; Gottesman, S. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol 2001, 39, 1382–1394. [Google Scholar]
- Vianney, A.; Jubelin, G.; Renault, S.; Dorel, C.; Lejeune, P.; Lazzaroni, J.C. Escherichia coli tol and rcs genes participate in the complex network affecting curli synthesis. Microbiology 2005, 151, 2487–2497. [Google Scholar]
- Mika, F.; Hengge, R. A two-component phosphotransfer network involving ArcB, ArcA, and RssB coordinates synthesis and proteolysis of sigmaS (RpoS) in E. coli. Genes Dev 2005, 19, 2770–2781. [Google Scholar]
- Malpica, R.; Franco, B.; Rodriguez, C.; Kwon, O.; Georgellis, D. Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc. Natl. Acad. Sci. USA 2004, 101, 13318–13323. [Google Scholar]
- Wassarman, K.M.; Repoila, F.; Rosenow, C.; Storz, G.; Gottesman, S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev 2001, 15, 1637–1651. [Google Scholar]
- Mika, F.; Hengge, R. Freie Universität Berlin: Germany, unpublished work.
- Roemling, U.; Bian, Z.; Hammar, M.; Sierralta, W.D.; Normark, S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol 1998, 180, 722–731. [Google Scholar]
- Muffler, A.; Traulsen, D.D.; Lange, R.; Hengge-Aronis, R. Posttranscriptional osmotic regulation of the sigmaS subunit of RNA polymerase in Escherichia coli. J. Bacteriol 1996, 178, 1607–1613. [Google Scholar]
- Altuvia, S.; Zhang, A.; Argaman, L.; Tiwari, A.; Storz, G. The Escherichia coli OxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J 1998, 17, 6069–6075. [Google Scholar]
- Latasa, C.; Garcia, B.; Echeverz, M.; Toledo-Arana, A.; Valle, J.; Campoy, S.; Garcia-del Portillo, F.; Solano, C.; Lasa, I. Salmonella biofilm development depends on the phosphorylation status of RcsB. J. Bacteriol 2012, 194, 3708–3722. [Google Scholar]
- Overgaard, M.; Johansen, J.; Moller-Jensen, J.; Valentin-Hansen, P. Switching off small RNA regulation with trap-mRNA. Mol. Microbiol 2009, 73, 790–800. [Google Scholar]
- Figueroa-Bossi, N.; Valentini, M.; Malleret, L.; Fiorini, F.; Bossi, L. Caught at its own game: Regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev 2009, 23, 2004–2015. [Google Scholar]

© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Mika, F.; Hengge, R. Small Regulatory RNAs in the Control of Motility and Biofilm Formation in E. coli and Salmonella. Int. J. Mol. Sci. 2013, 14, 4560-4579. https://doi.org/10.3390/ijms14034560
Mika F, Hengge R. Small Regulatory RNAs in the Control of Motility and Biofilm Formation in E. coli and Salmonella. International Journal of Molecular Sciences. 2013; 14(3):4560-4579. https://doi.org/10.3390/ijms14034560
Chicago/Turabian StyleMika, Franziska, and Regine Hengge. 2013. "Small Regulatory RNAs in the Control of Motility and Biofilm Formation in E. coli and Salmonella" International Journal of Molecular Sciences 14, no. 3: 4560-4579. https://doi.org/10.3390/ijms14034560
APA StyleMika, F., & Hengge, R. (2013). Small Regulatory RNAs in the Control of Motility and Biofilm Formation in E. coli and Salmonella. International Journal of Molecular Sciences, 14(3), 4560-4579. https://doi.org/10.3390/ijms14034560



