Transcriptional Regulation of a Chitinase Gene by 20-Hydroxyecdysone and Starvation in the Oriental Fruit Fly, Bactrocera dorsalis
Abstract
:1. Introduction
2. Results
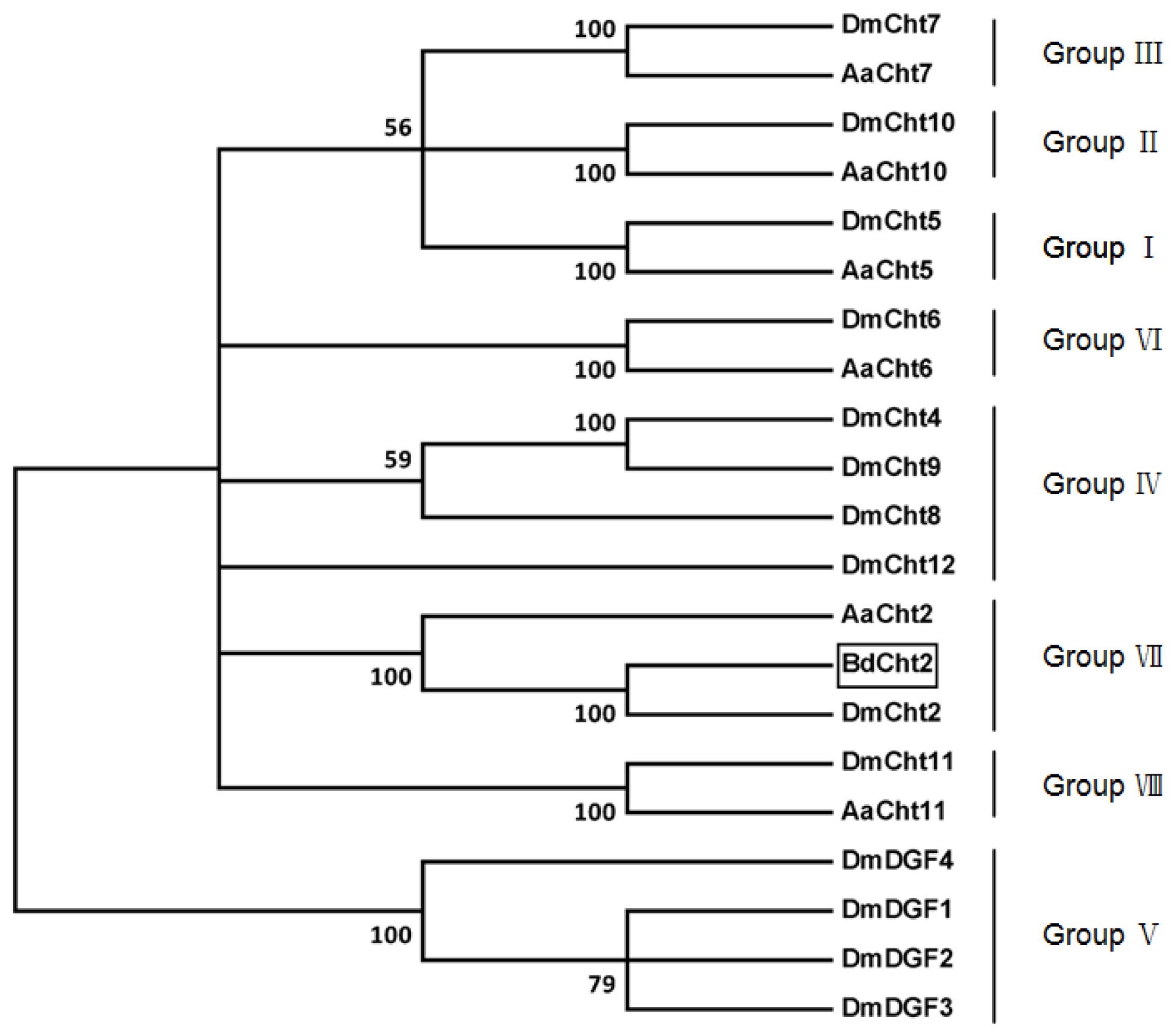
2.1. Sequence Analysis of BdCht2 cDNA
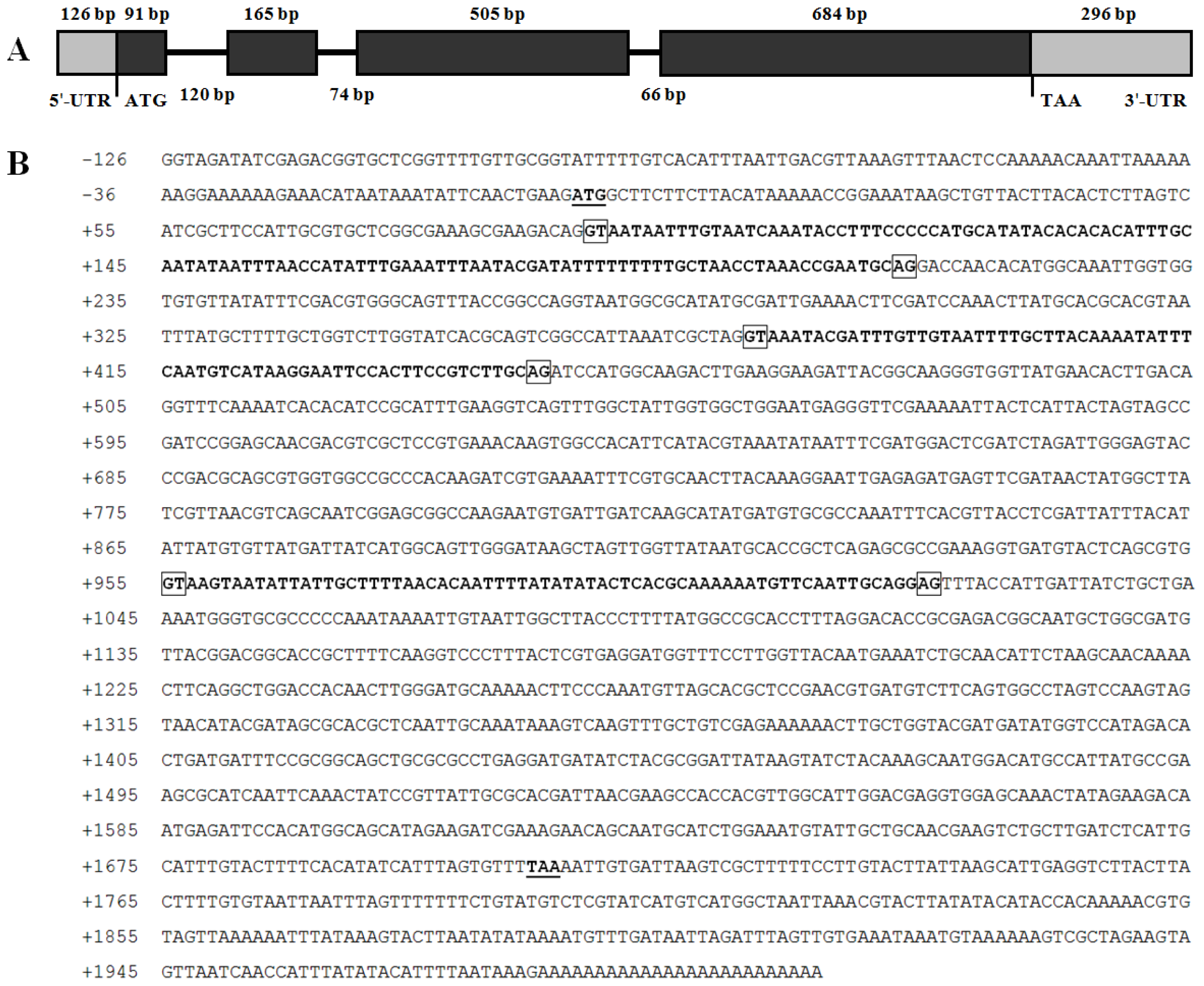
2.2. Genomic DNA Structure and 5′ Flanking Region of BdCht2
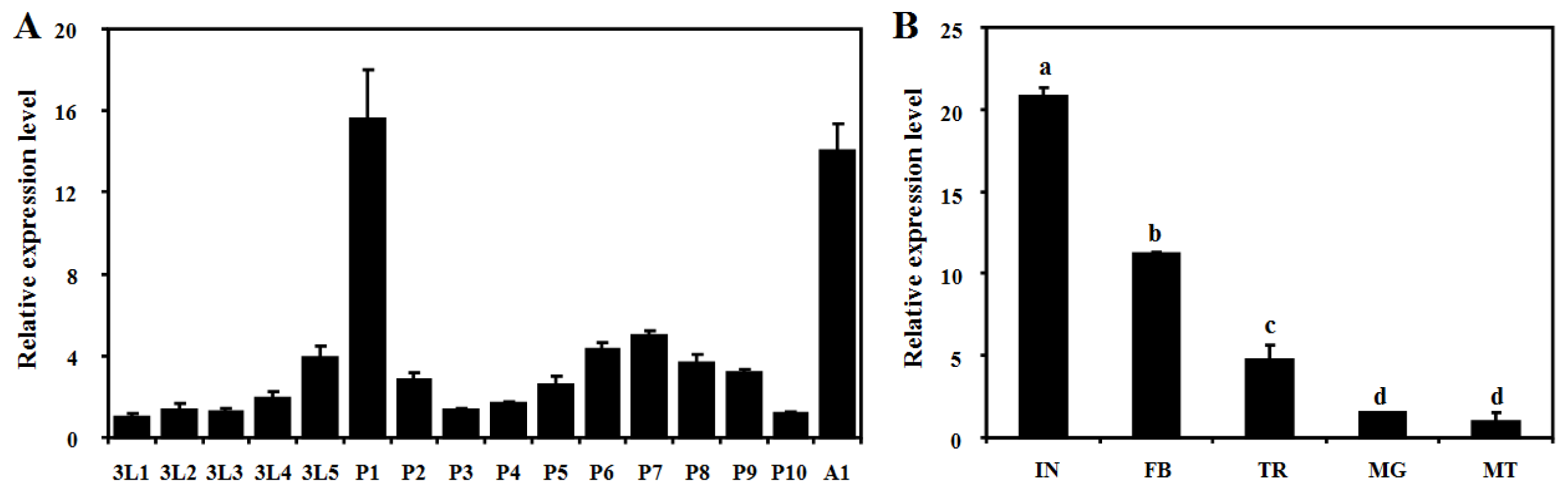
2.3. Developmental and Tissue-Specific Expression Patterns of BdCht2
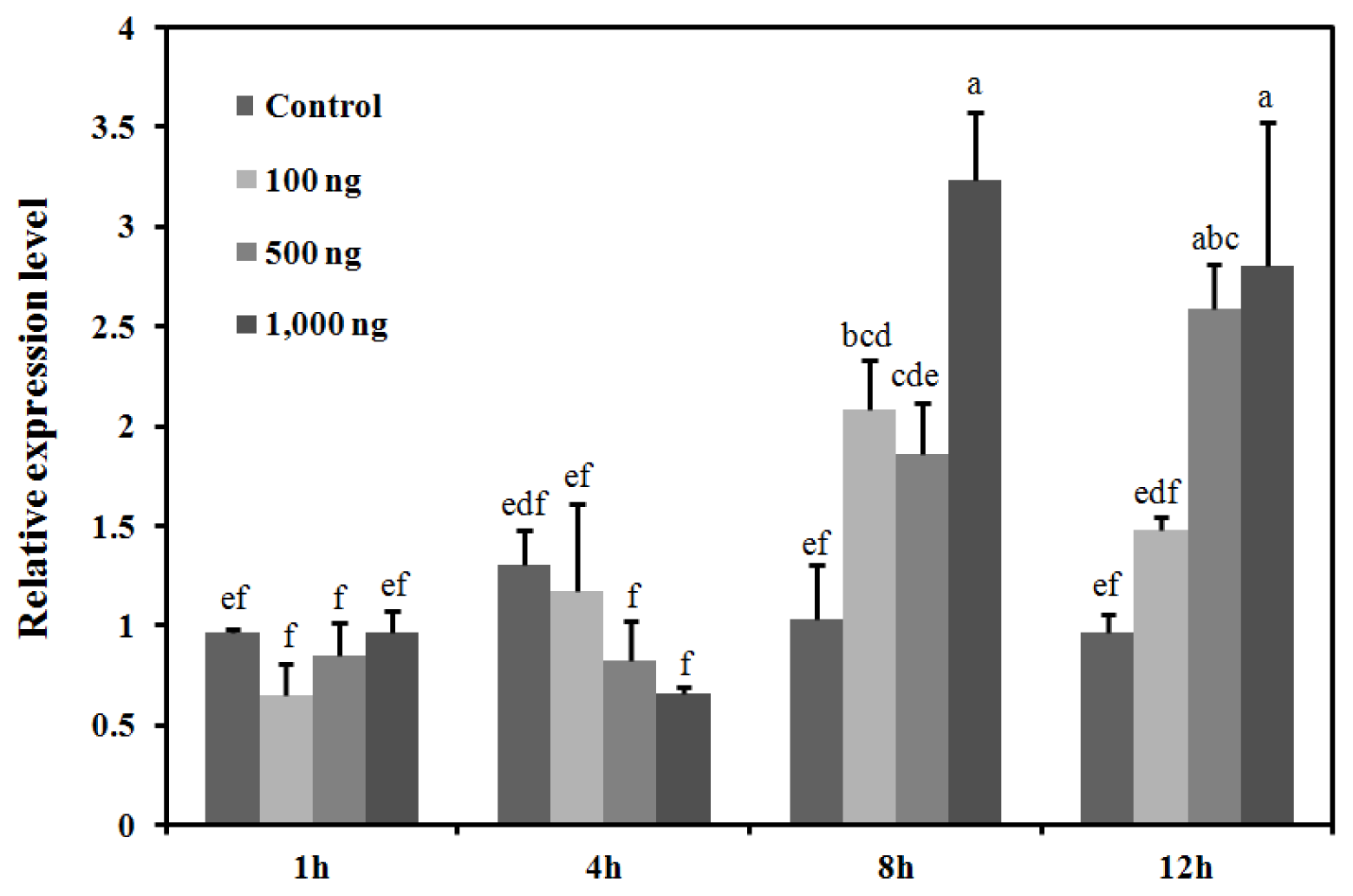
2.4. Response of the BdCht2 Expression to the Application of 20E
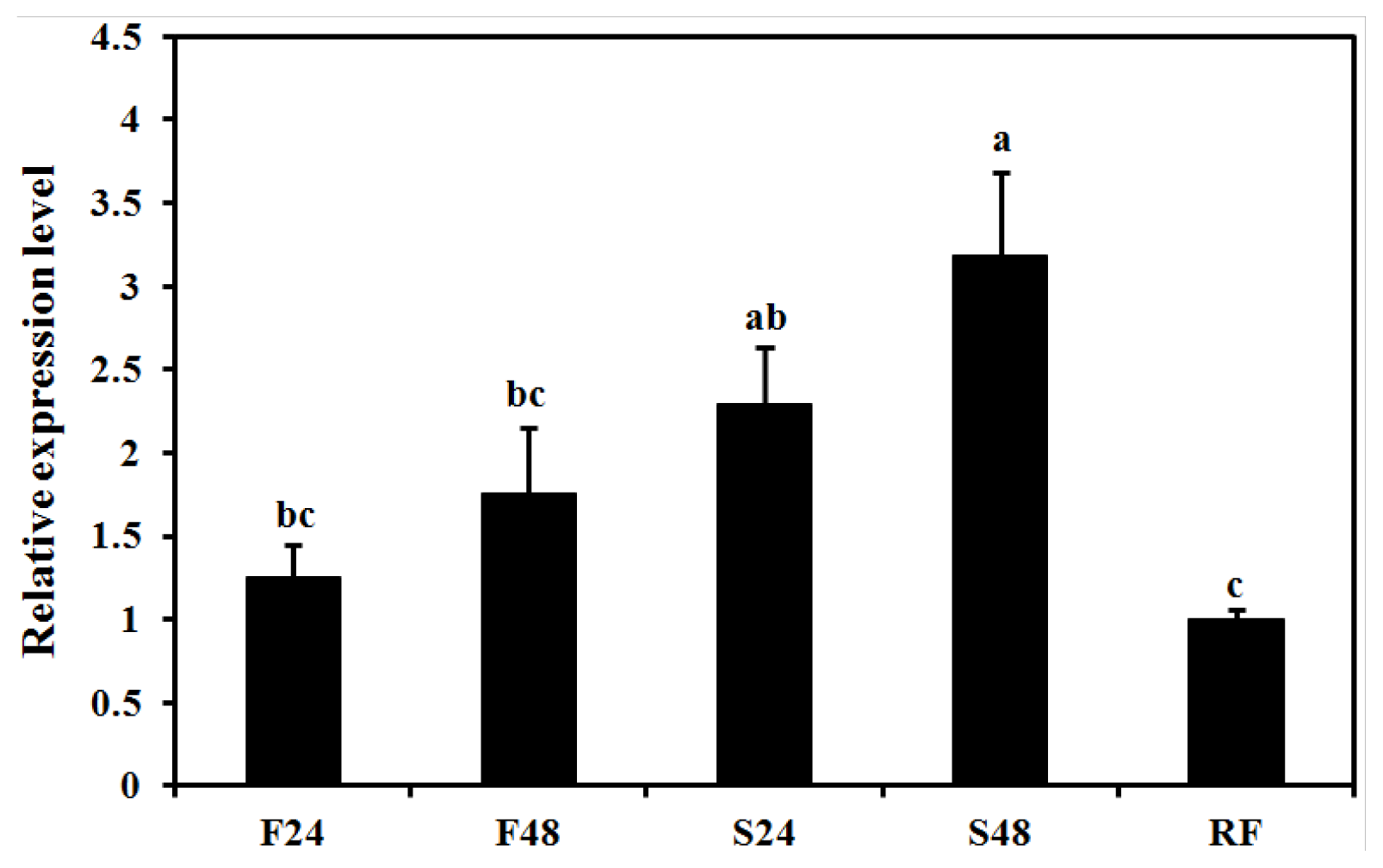
2.5. Effect of Starvation on Expression of BdCht2
3. Discussion
4. Experimental Section
4.1. Test Insect
4.2. Cloning of the Full-Length BdCht2 cDNA
4.3. Cloning of the Genomic Sequences and 5′-Flanking Region of BdCht2
4.4. Cloning and Sequencing
4.5. Bioinformatic Analysis and Phylogenetic Tree Construction
4.6. Developmental and Tissue-Specific Expression of BdCht2
4.7. 20E Treatment
4.8. Gene Expression Patterns in Feeding and Starvation Treatment
5. Conclusions


| Name sequence | Primer sequence (5′–3′) | Usage |
|---|---|---|
| BdCht2-3GSP1 | GAAGATTACGGCAAGGGTGGT | 3′-RACE |
| BdCht2-3GSP2 | CCATAGACACCGATGATTTCCG | |
| BdCht2-F | CTGAAGATGGCTTCTTCTTACATAA | cDNA Full-length confirmation |
| BdCht2-R | CAATTTTAAAACACTAAATGATATG | |
| BdCht2-SP1 | TCCGGATCGGCTACTAGTAATGAG | Cloning 5′-flanking regions |
| BdCht2-SP2 | GTCAAGTGTTCATAACCACCCTTGC | |
| Q-BdCht2-F | ATCACACATCCGCATTTGAA | Quantitative real-time PCR |
| Q-BdCht2-R | CGTCGGGTACTCCCAATCTA | |
| Q-α-Tubulin-F | CGCATTCATGGTTGATAACG | |
| Q-α-Tubulin-R | GGGCACCAAGTTAGTCTGGA | |
Acknowledgments
Conflicts of Interest
References
- Kramer, K.J.; Koga, D. Insect chitin: Physical state, synthesis, degradation and metabolic regulation. Insect Biochem 1986, 16, 851–877. [Google Scholar]
- Merzendorfer, H.; Zimoch, L. Chitin metabolism in insects: Structure, function and regulation of chitin synthases and chitinases. J. Exp. Biol 2003, 206, 4393–4412. [Google Scholar]
- Lehane, M. Peritrophic matrix structure and function. Annu. Rev. Entomol 1997, 42, 525–550. [Google Scholar]
- Kramer, K.; Muthukrishnan, S. Chitin Metabolism in Insects. In Comprehensive Molecular Insect Science; Gilbert, L.I., Iatrou, K., Gill, S., Eds.; Elsevier Press: Oxford, UK, 2005; Volume 4, pp. 111–144. [Google Scholar]
- Nation, J.L. Insect Physiology and Biochemistry, 2nd ed; CRC Press: Boca Raton, FL, USA, 2008. [Google Scholar]
- Henrissat, B.; Bairoach, A. Updating the sequence-based classification of glycosyl hydrolases. Biochem. J 1993, 316, 695–696. [Google Scholar]
- Zhu, Q.S.; Arakane, Y.; Beeman, R.W.; Kramer, K.J.; Muthukrishnan, S. Functional specialization among insect chitinase family genes revealed by RNA interference. Proc. Natl. Acad. Sci. USA 2008, 105, 6650–6655. [Google Scholar]
- Bolognesi, R.; Arakane, Y.; Muthukrishnan, S.; Kramer, K.J.; Terra, W.R.; Ferreira, C. Sequences of cDNAs and expression of genes encoding chitin synthase and chitinase in the midgut of Spodoptera frugiperda. Insect Biochem. Mol. Biol 2005, 35, 1249–1259. [Google Scholar]
- Zhu, Q.S.; Arakane, Y.; Banerjee, D.; Beeman, R.W.; Kramer, K.J.; Muthukrishnan, S. Domain organization and phylogenetic analysis of the chitinase-like family of proteins in three species of insects. Insect Biochem. Mol. Biol 2008, 38, 452–466. [Google Scholar]
- Arakane, Y.; Muthukrishnan, S. Insect chitinase and chitinase-like proteins. Cell. Mol. Life Sci 2010, 67, 201–216. [Google Scholar]
- Zhang, D.W.; Chen, J.; Yao, Q.; Pan, Z.Q.; Chen, J.; Zhang, W.Q. Functional analysis of two chitinase genes during the pupation and eclosion stages of the beet armyworm Spodoptera exigua by RNA interference. Arch. Insect Biochem. Physiol 2012, 79, 220–234. [Google Scholar]
- Yan, J.; Cheng, Q.; Narashimhan, S.; Li, C.B.; Aksoy, S. Cloning and functional expression of a fat body-specific chitinase cDNA from the tsetse fly Glossina morsitans morsitans. Insect Biochem. Mol. Biol 2002, 32, 979–989. [Google Scholar]
- Langer, R.C.; Li, F.; Popov, V.; Kurosky, A.; Vinetz, J.M. Monoclonal antibody against the Plasmodium falciparum chitinase, PfCHT1, recognizes a malaria transmission-blocking epitope in Plasmodium gallinaceum ookinetes unrelated to the chitinase PgCHT1. Infect. Immun 2002, 70, 1581–1590. [Google Scholar]
- Kimura, S. The chitinase system in the cuticle of the silkworm Bombyx mori. Insect Biochem 1976, 6, 479–482. [Google Scholar]
- Koga, D.; Funakoshi, T.; Fujimoto, H.; Kuwano, E.E.M. Effects of 20-hydroxyecdysone and KK-42 on chitinase and beta-N-acetylglucosaminidase during the larval-pupal transformation of Bombyx mori. Insect Biochem 1991, 21, 277–284. [Google Scholar]
- Royer, V.; Fraichard, S.; Bouhin, H. A novel putative insect chitinase with multiple catalytic domains: Hormonal regulation during metamorphosis. Biochem. J 2002, 366, 921–928. [Google Scholar]
- Kramer, K.J.; Corpuz, L.; Choi, H.K.; Muthukrishnan, S. Sequence of a cDNA and expression of the gene encoding epidermal and gut chitinases of Manduca sexta. Insect Biochem. Mol. Biol 1993, 23, 691–701. [Google Scholar]
- Takahashi, M.; Kiuchi, M.; Kamimura, M. A new chitinase related gene, BmChiR1, is induced in the Bombyx mori anterior silk gland at molt and metamorphosis by ecdysteroid. Insect Biochem. Mol. Biol 2002, 32, 147–151. [Google Scholar]
- Cymborowski, B.; Bogus, M.; Beckage, N.E.; Williams, C.M.; Riddiford, L.M. Juvenile hormone titers and metabolism during starvation-induced supernumerary larval moulting of the tobacco hornworm Manduca sexta L. J. Insect Physiol 1982, 28, 129–135. [Google Scholar]
- Munyiri, F.N.; Asano, W.; Shintani, Y.; Ishikawa, Y. Threshold weight for starvation-triggered metamorphosis in the yellow-spotted longicorn beetle, Psacothea hilaris (Coleoptera: Cerambycidae). Appl. Entomol. Zool 2003, 38, 509–515. [Google Scholar]
- Khajuria, C.; Buschman, L.L.; Chen, M.S.; Muthukrishnan, S.; Zhu, K.Y. A gut-specific chitinase gene essential for regulation of chitin content of peritrophic matrix and growth of Ostrinia nubilalis larvae. Insect Biochem. Mol. Biol 2010, 40, 621–629. [Google Scholar]
- Clarke, A.R.; Armstrong, K.F.; Carmichael, A.E.; Milne, J.R.; Raghu, S.; Roderick, G.K.; Yeates, D.K. Invasive phytophagous pests arising through a recent tropical evolutionary radiation: The Bactrocera dorsalis complex of fruit flies. Annu. Rev. Entomol 2005, 50, 293–319. [Google Scholar]
- Stephens, A.E.A.; Kriticos, D.J.; Leriche, A. The current and future potential geographical distribution of the oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae). Bull. Entomol. Res 2007, 97, 369–378. [Google Scholar]
- Jin, T.; Zeng, L.; Lin, Y.; Lu, Y.; Liang, G. Insecticide resistance of the oriental fruit fly, Bactrocera dorsalis (Hendel) (Diptera: Tephritidae), in mainland China. Pest Manag. Sci 2011, 67, 370–376. [Google Scholar]
- Hsu, J.C.; Feng, H.T.; Wu, W.J.; Geib, S.; Mao, C.H.; Vontas, J. Truncated transcripts of nicotinic acetylcholine subunit gene Bdα6 are associated with spinosad resistance in Bactrocera dorsalis. Insect Biochem. Mol. Biol 2012, 42, 805–816. [Google Scholar]
- Vega, H.D.; Specht, C.A.; Liu, Y.; Robbins, P.W. Chitinases are a multi-gene family in Aedes, Anopheles and Drosophila. Insect Mol. Biol 1998, 7, 233–239. [Google Scholar]
- Feix, M.; Gloggler, S.; Londershausen, M.; Weidemann, W.; Spindler, K.D.; Spindler-Barth, M. A cDNA encoding a chitinase from the epithelial cell line of Chironomus tentans (Insecta, Diptera) and its functional expression. Arch. Insect Biochem. Physiol 2000, 45, 24–36. [Google Scholar]
- Shinoda, T.; Kobayashi, J.; Matsui, M.; Chinzei, Y. Cloning and functional expression of a chitinase cDNA from the common cutworm, Spodoptera litura, using a recombinant baculovirus lacking the virus-encoded chitinase gene. Insect Biochem. Mol. Biol 2001, 31, 521–532. [Google Scholar]
- Zheng, Y.; Zheng, S.; Cheng, X.; Ladd, T.; Lingohr, E.J.; Krell, P.J.; Arif, B.M.; Retnakaran, A.; Feng, Q.L. A molt-associated chitinase cDNA from the spruce budworm Choristoneura fumiferana. Insect Biochem. Mol. Biol 2002, 32, 1813–1823. [Google Scholar]
- Ahmad, T.; Rajagopal, R.; Bhatnagar, R.K. Molecular characterization of chitinase from polyphagous pest Helicoverpa armigera. Biochem. Biophys. Res. Comm 2003, 310, 188–195. [Google Scholar]
- Genta, F.A.; Blanes, L.; Cristofoletti, P.T.; do Lago, C.L.; Terra, W.R.; Ferreira, C. Purification, characterization and molecular cloning of the major chitinase from Tenebrio molitor larval midgut. Insect Biochem. Mol. Biol 2006, 36, 789–800. [Google Scholar]
- Choo, Y.M.; Lee, K.S.; Kim, B.Y.; Kim, D.H.; Yoon, H.J.; Sohn, H.D.; Jin, B.R. A gut-specific chitinase from the mulberry longicorn beetle, Apriona germari (Coleoptera: Cerambycidae): cDNA cloning, gene structure, expression and enzymatic activity. Eur. J. Entomol 2007, 104, 173–180. [Google Scholar]
- Yang, W.J.; Xu, K.K.; Cong, L.; Wang, J.J. Identification, mRNA expression, and functional analysis of chitin synthase 1 gene and its two alternative splicing variants in oriental fruit fly Bactrocera dorsalis. Int. J. Biol. Sci 2013, 9, 331–342. [Google Scholar]
- Zhang, J.Z.; Zhang, X.; Arakane, Y.; Muthukrishnan, S.; Kramer, K.J.; Ma, E.B.; Zhu, K.Y. Comparative genomic analysis of chitinase and chitinase-like genes in the African malaria mosquito (Anopheles gambiae). PLoS One 2011, 6, e19899. [Google Scholar]
- Shen, Z.; Jacobs-Lorena, M. Characterization of a novel guts pecific chitinase gene from the human malaria vector Anopheles gambiae. J. Biol. Chem 1997, 272, 28895–28900. [Google Scholar]
- Ramalho-Ortigao, J.M.; Traub-Cseko, Y.M. Molecular characterization of Llchit1, a midgut chitinase cDNA from the eishmaniasis vector Lutzomyia longipalpis. Insect Biochem. Mol. Biol 2003, 33, 279–287. [Google Scholar]
- Riddiford, L.M.; Hiruma, K.; Zhou, X.; Nelson, C.A. Insights into the molecular basis of the hormonal control of molting and metamorphosis from Manduca sexta and Drosophila melanogaster. Insect Biochem. Mol. Biol 2003, 33, 1327–1338. [Google Scholar]
- Cong, L.; Yang, W.J.; Shen, G.M.; Dou, W.; Wang, J.J. Molecular characterization of the cDNA encoding ecdysone receptor isoform B1 and its expression in the oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae). Fla. Entomol 2012, 95, 650–658. [Google Scholar]
- Shen, G.M.; Dou, W.; Niu, J.Z.; Jiang, H.B.; Yang, W.J.; Jia, F.X.; Hu, F.; Cong, L.; Wang, J.J. Transcriptome analysis of the oriental fruit fly (Bactrocera dorsalis). PLoS One 2011, 6, e29127. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol 2011, 28, 2731–2739. [Google Scholar]
- Shen, G.M.; Jiang, H.B.; Wang, X.N.; Wang, J.J. Evaluation of endogenous references for gene expression profiling in different tissues of the oriental fruit fly Bactrocera dorsalis (Dipter: Tephritidae). BMC Mol. Biol 2010, 11, 76. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yang, W.-J.; Xu, K.-K.; Zhang, R.-Y.; Dou, W.; Wang, J.-J. Transcriptional Regulation of a Chitinase Gene by 20-Hydroxyecdysone and Starvation in the Oriental Fruit Fly, Bactrocera dorsalis. Int. J. Mol. Sci. 2013, 14, 20048-20063. https://doi.org/10.3390/ijms141020048
Yang W-J, Xu K-K, Zhang R-Y, Dou W, Wang J-J. Transcriptional Regulation of a Chitinase Gene by 20-Hydroxyecdysone and Starvation in the Oriental Fruit Fly, Bactrocera dorsalis. International Journal of Molecular Sciences. 2013; 14(10):20048-20063. https://doi.org/10.3390/ijms141020048
Chicago/Turabian StyleYang, Wen-Jia, Kang-Kang Xu, Rui-Ying Zhang, Wei Dou, and Jin-Jun Wang. 2013. "Transcriptional Regulation of a Chitinase Gene by 20-Hydroxyecdysone and Starvation in the Oriental Fruit Fly, Bactrocera dorsalis" International Journal of Molecular Sciences 14, no. 10: 20048-20063. https://doi.org/10.3390/ijms141020048
APA StyleYang, W.-J., Xu, K.-K., Zhang, R.-Y., Dou, W., & Wang, J.-J. (2013). Transcriptional Regulation of a Chitinase Gene by 20-Hydroxyecdysone and Starvation in the Oriental Fruit Fly, Bactrocera dorsalis. International Journal of Molecular Sciences, 14(10), 20048-20063. https://doi.org/10.3390/ijms141020048




