3D-QSAR Studies on a Series of Dihydroorotate Dehydrogenase Inhibitors: Analogues of the Active Metabolite of Leflunomide
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Sets
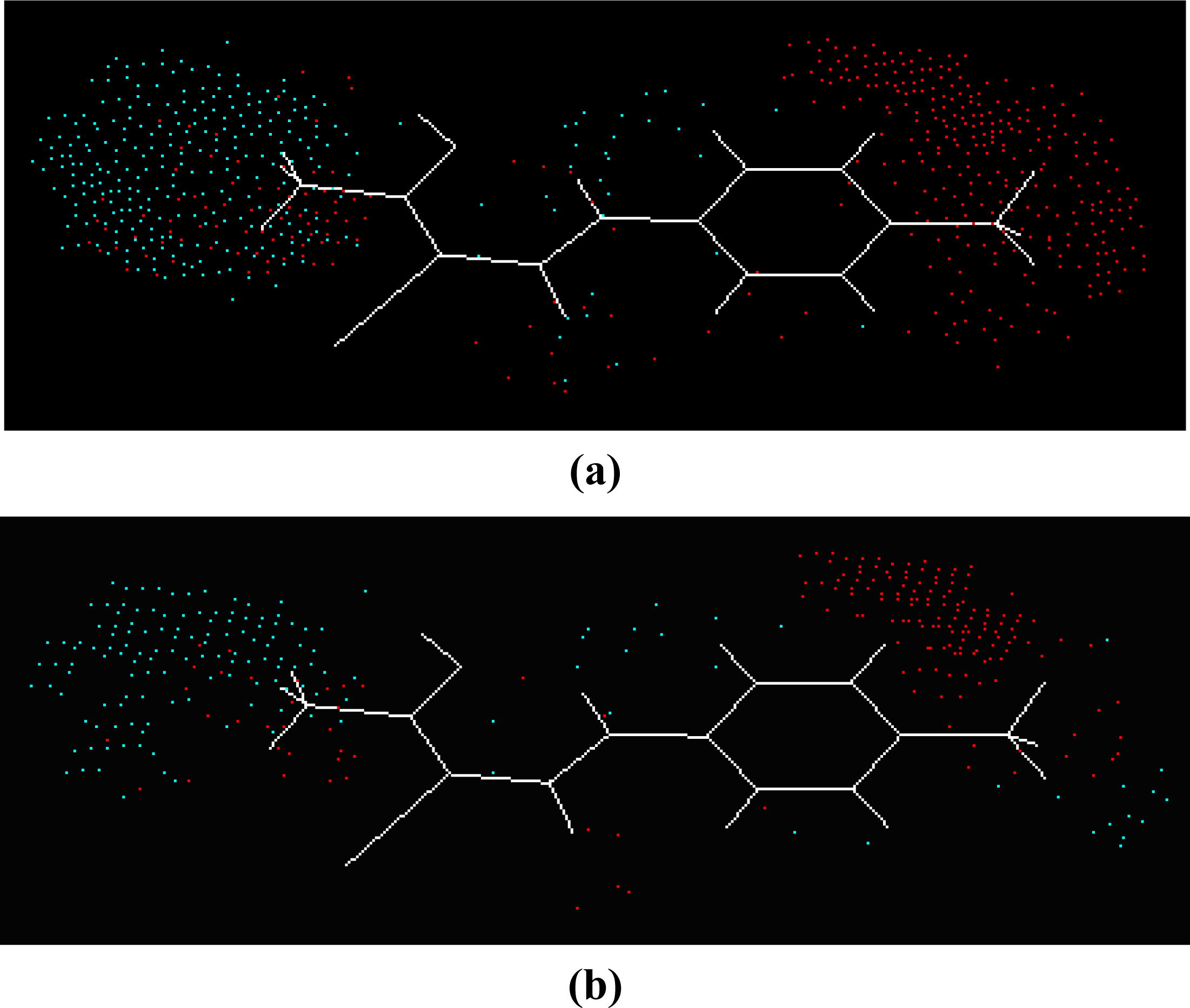
2.2. Molecular Modeling and Alignment
2.3. SOMFA 3D-QSAR Models
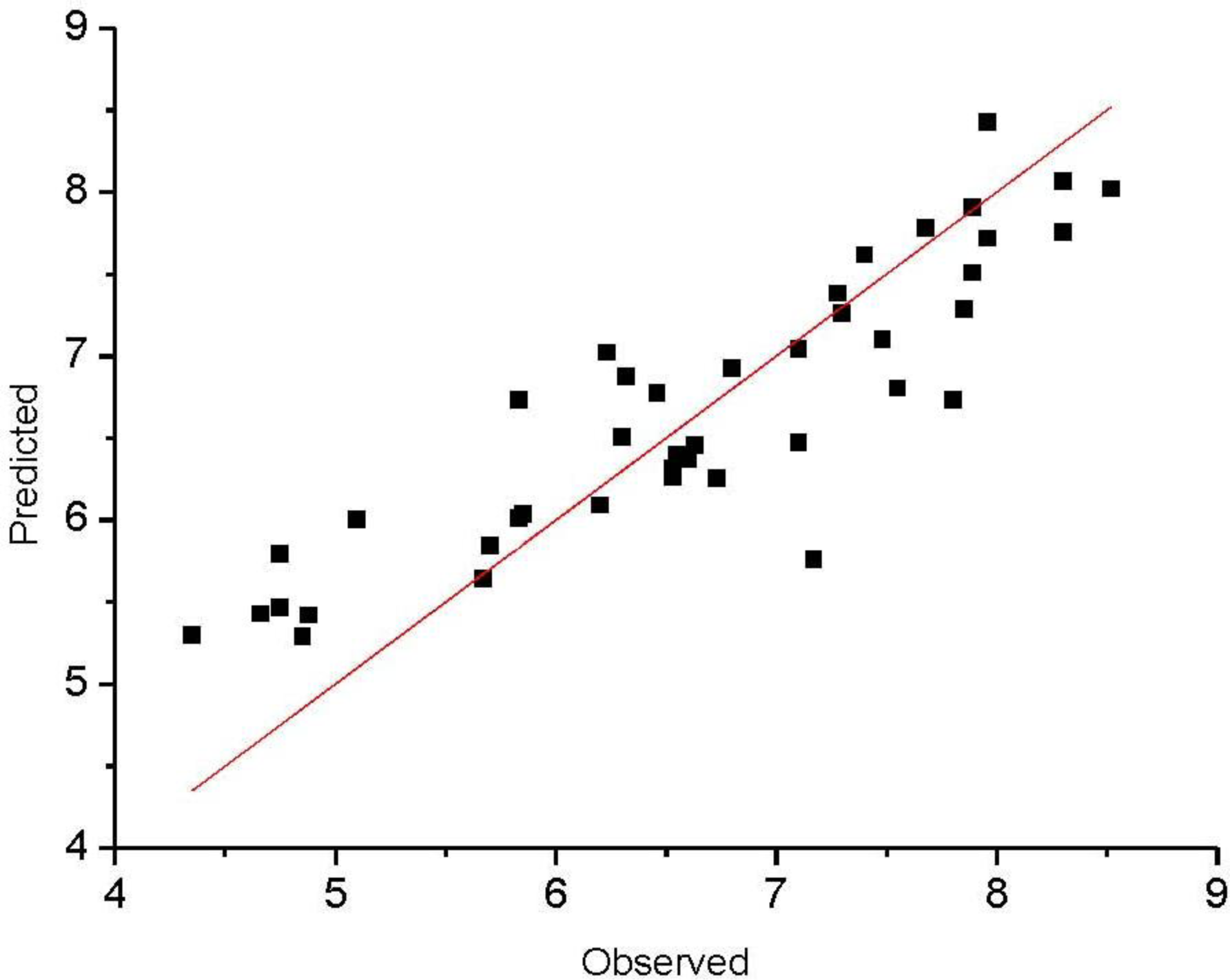
3. Results and Discussion
4. Conclusions
Acknowledgments
References
- Jones, M. Pyrimidine nucleotide biosynthesis in animals: Genes, enzymes, and regulation of UMP biosynthesis. Ann. Rev. Biochem 1980, 49, 253–279. [Google Scholar]
- Nagy, M; Lacroute, F; Thomas, D. Divergent evolution of pyrimidine biosynthesis between anaerobic and aerobic yeasts. Proc. Natl. Acad. Sci. USA 1992, 89, 8966–8970. [Google Scholar]
- Tamta, H; Mukhopadhyay, AK. Biochemical targets for malaria chemotherapy. Curr. Res. Inf. Pharm. Sci 2003, 4, 6–9. [Google Scholar]
- Copeland, RA; Marcinkeviciene, J; Haque, TS; Kopcho, LM; Jiang, W; Wang, K; Ecret, LD; Sizemore, C; Amsler, KA; Foster, L; Tadesse, S; Combs, AP; Stern, AM; Trainor, GL; Slee, A; Rogers, MJ; Hobbs, F. Helicobacter pylori-selective antibacterials based on inhibition of pyrimidine biosynthesis. J. Biol. Chem 2000, 275, 33373–33378. [Google Scholar]
- Morand, P; Courtin, O; Sautes, C; Westwood, R; Hercend, T; Kuo, EA; Ruuth, E. Dihydroorotate dehydrogenase is a target for the biological effects of leflunomide. Transplant. Proc 1996, 28, 3088–3091. [Google Scholar]
- McLean, JE; Neidhardt, EA; Grossman, TH; Hedstrom, L. Multiple inhibitor analysis of the Brequinar and leflunomide binding sites on human dihydroorotate dehydrogenase. Biochemistry 2001, 40, 2194–2200. [Google Scholar]
- Chen, SF; Papp, LM; Ardecky, RJ; Rao, GV; Hesson, DP; Forbes, M; Dexter, DL. Structure-activity relationship of quinoline carboxylic acids: A new class of inhibitors of dihydroorotate dehydrogenase. Biochem. Pharmaco 1990, 40, 709–714. [Google Scholar]
- Fox, RI; Herrmann, ML; Frangou, CG; Wahl, GM; Morris, RE; Strand, V; Kirschbaum, BJ. Mechanism of action for leflunomide in rheumatoid arthritis. Clin. Immunol 1999, 93, 198–208. [Google Scholar]
- Peters, GJ; Sharma, SL; Laurensse, E; Pinedo, HM. Inhibition of pyrimidine de novo synthesis by DUP-785 (NSC 368390). Investig. New Drug 1987, 5, 235–244. [Google Scholar]
- Kuo, EA; Hambleton, PT; Kay, DP; Evans, PL; Matharu, SS; Little, E; McDowall, N; Jones, CB; Hedgecock, CJR; Yea, CM; Chan, AWE; Hairsine, PW; Ager, IR; Tully, WR; Williamson, RA; Westwood, R. Synthesis, structure-activity relationships, and pharmacokinetic properties of dihydroorotate dehydrogenase inhibitiors: 2-cyano-3-cyclopropyl-3-hydroxy-N-[3′-methyl-4′-(trifluoromethyl)phenyl]propenamide and related compounds. J. Med. Chem 1996, 39, 4608–4621. [Google Scholar]
- Ren, SJ; Wu, SK; Lien, EJ. Dihydroorotate dehydrogenase inhibitors: Quantitative structure-activity relationship analysis. Pharm. Res 1998, 15, 286–295. [Google Scholar]
- Leban, J; Saeb, W; Garcia, G; Baumgartner, R; Kramer, B. Discovery of a novel series of DHODH inhibitors by a docking procedure and QSAR refinement. Bioorg. Med. Chem. Lett 2004, 14, 55–58. [Google Scholar]
- Ojha, PK; Roy, K. Chemometric modeling, docking and in silico design of triazolopyrimidine-based dihydroorotate dehydrogenase inhibitors as antimalarials. Eur. J. Med. Chem 2010, 45, 4645–4656. [Google Scholar]
- Robinson, DD; Winn, PJ; Lyne, PD; Richards, WG. Self-organizing molecular field analysis: A tool for structure-activity studies. J. Med. Chem 1999, 42, 573–583. [Google Scholar]
- Cramer, RD, III; Patteerson, DE; Bunce, JD. Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J. Am. Chem. Soc 1988, 110, 5959–5967. [Google Scholar]
- Kremmer, JM. What I would like to know about leflunomide. J. Rheumatol 2004, 31, 1029–1031. [Google Scholar]
- Thompson, MA. ArgusLab 4.0.1. Planaria Software LLC: Seattle, WA, USA, 2001. Available online: http://www.arguslab.com (accessed on 3 May 2011).
- Liu, S; Neidhardt, EA; Grossman, TH; Ocain, T; Clardy, J. Structures of human dihydroorotate dehydrogenase in complex with antiproliferative agents. Struct. Fold. Des 2000, 8, 25–33. [Google Scholar]
- Pedretti, A; Villa, L; Vistoli, G. VEGA: A Versatile program to convert, handle and visualize molecular structures on windows-based PCs. J. Mol. Graph. Model 2002, 21, 47–49. [Google Scholar]
- Li, SL; Zheng, Y; Yin, WG. Three dimensional quantitative structure-activity relationship studies on a series of typical tricycle compounds. Beijing Huagong Daxue Xuebao 2007, 34, 249–253. (in Chinese). [Google Scholar]
- Li, SL; Zheng, Y. Self-organizing molecular field analysis on a new series of COX-2 selective inhibitors: 1,5-diarylimidazoles. Int. J. Mol. Sci 2006, 7, 220–229. [Google Scholar]
- SPSS software version 16. IBM Corporation: Somers, NY, USA, 2007. Available online: http://www.spss.com/ (accessed on 3 May 2011).





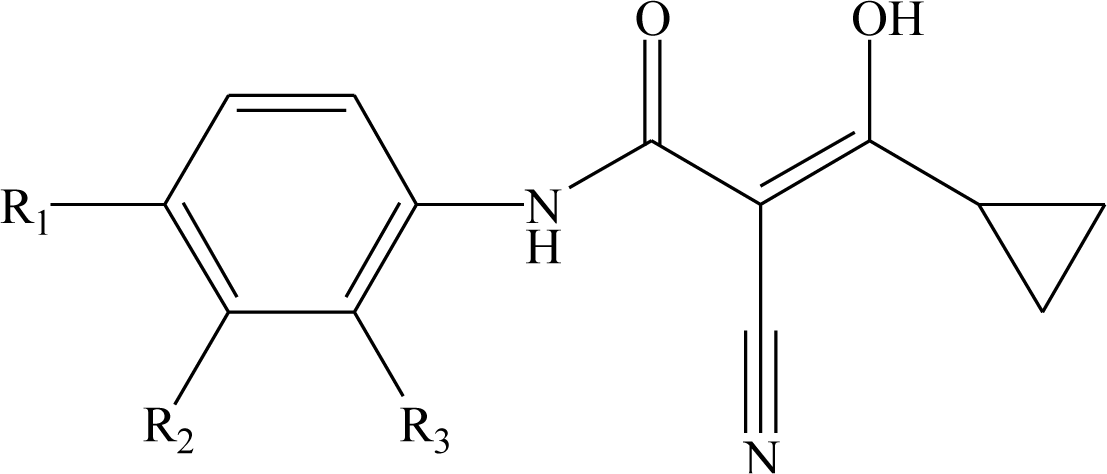 | |||||||
|---|---|---|---|---|---|---|---|
| Compd No. | R1 | R2 | R3 | Compd No. | R1 | R2 | R3 |
| 1 | H | H | H | 29 | Cl | CH3 | H |
| 2 | CH3 | H | H | 30* | Cl | H | CH3 |
| 3 | CF3 | H | H | 31 | CH3 | Cl | H |
| 4 | H | CF3 | H | 32 | Br | CH3 | H |
| 5* | Cl | H | H | 33 | CN | CH3 | H |
| 6 | H | Cl | H | 34 | CF3S | CH3 | H |
| 7 | H | H | Cl | 35* | CF3O | CH3 | H |
| 8 | Br | H | H | ||||
| 9 | CN | H | H | 36 | 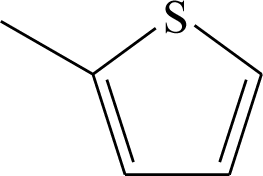 | H | H |
| 10* | -CH2CN | H | H | ||||
| 11 | CF3S | H | H | ||||
| 12 | CF3SO | H | H | 37 | 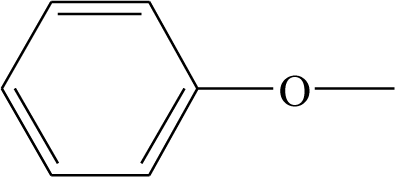 | H | H |
| 13 | CF3SO2 | H | H | ||||
| 14 | CH3S | H | H | ||||
| 15* | CH3SO | H | H | 38 |  | H | H |
| 16 | CH3SO2 | H | H | ||||
| 17 | CF3O | H | H | ||||
| 18 | CH3O | H | H | 39 | 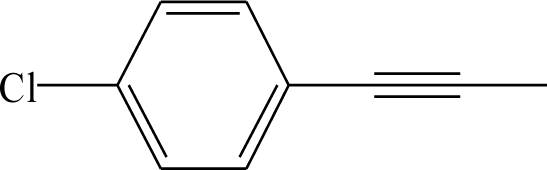 | H | H |
| 19 | OH | H | H | ||||
| 20* | NO2 | H | H | ||||
| 21 | H2N | H | H | 40* |  | H | H |
| 22 | CH3CO | H | H | ||||
| 23 | H2NCO | H | H | ||||
| 24 | HOOC- | H | H | 41 | 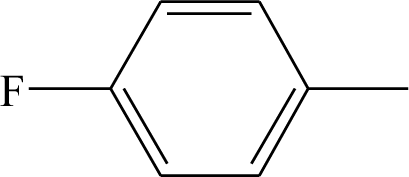 | H | H |
| 25* | CH3O2C- | H | H | ||||
| 26 | CF3 | CH3 | H | ||||
| 27 | CF3 | C2H5 | H | 42 | 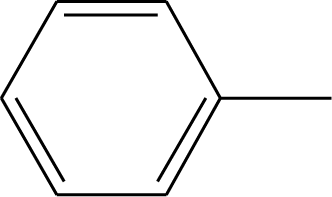 | H | H |
| 28 | C2F5 | CH3 | H | ||||
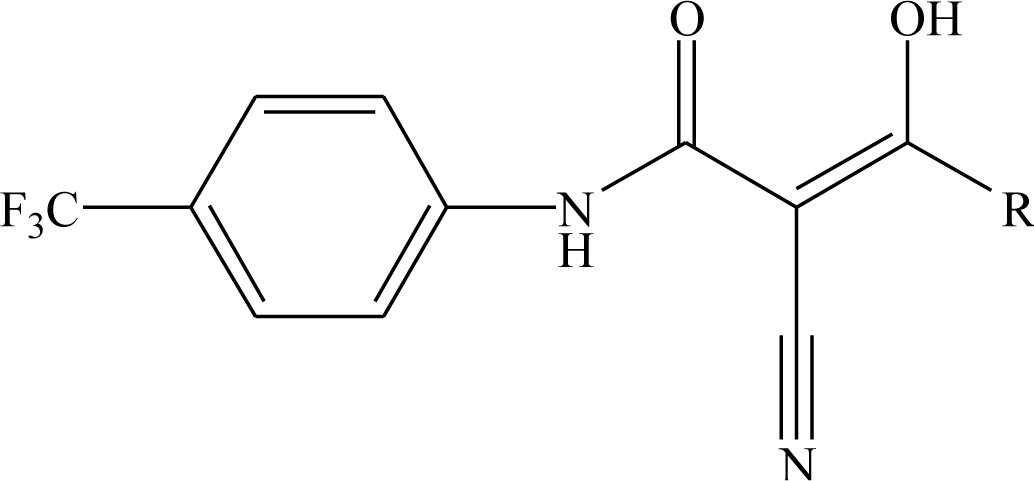 | |||
|---|---|---|---|
| Compd No. | R | Compd No. | R |
| 43 | -CH3 | 48 | 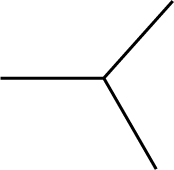 |
| 3 | 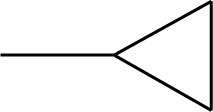 | 49 | 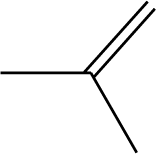 |
| 44 | 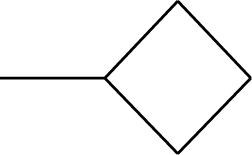 | 50* |  |
| 45* | 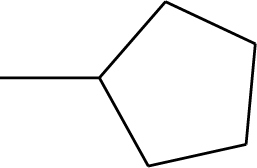 | 51 | 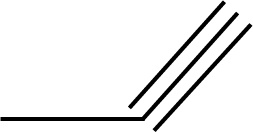 |
| 46 |  | 52 | 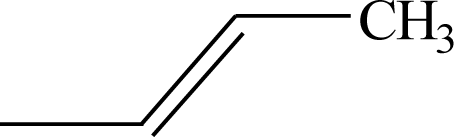 |
| 47 | 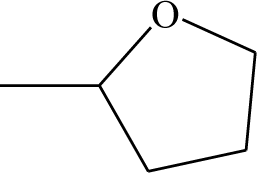 | 53* | 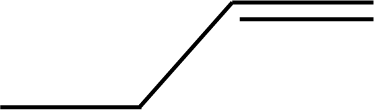 |
 | |||
|---|---|---|---|
| Alignment No. | 1st atom | 2nd atom | 3rd atom |
| 1 | 1 | 2 | 3 |
| 2 | 2 | 4 | 5 |
| Rat DHODH | Mouse DHODH | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All analogues | Aromatic substituted analogues | Side chain 3-substituted analogues | All analogues | Aromatic substituted analogues | Side chain 3-substituted analogues | |||||||
| Align. | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 2 |
| r2 | 0.658 | 0.687 | 0.758 | 0.665 | 0.778 | 0.897 | 0.517 | 0.572 | 0.554 | 0.657 | 0.697 | 0.859 |
| rCV2 | 0.636 | 0.664 | 0.739 | 0.641 | 0.715 | 0.865 | 0.485 | 0.545 | 0.516 | 0.620 | 0.610 | 0.822 |
| F | 98.428 | 112.251 | 125.687 | 79.723 | 35.180 | 87.269 | 52.608 | 65.608 | 47.310 | 72.892 | 23.106 | 61.314 |
| s | 0.651 | 0.623 | 0.528 | 0.621 | 0.604 | 0.412 | 0.657 | 0.619 | 0.628 | 0.551 | 0.590 | 0.402 |
| c1 | 0.695 | 0.766 | 0.650 | 0.769 | 0.800 | 0.987 | 0.531 | 0.625 | 0.429 | 0.660 | 0.934 | 0.918 |
| rpred2 | 0.818 | 0.717 | 0.571 | 0.549 | 0.972 | 0.981 | 0.657 | 0.679 | 0.512 | 0.693 | 0.993 | 0.991 |
| Compd | Rat DHODH | Mouse DHODH | ||||
|---|---|---|---|---|---|---|
| log(1/IC50) | log(1/IC50) | |||||
| Observed | Predicted | Residuala | Observed | Predicted | Residuala | |
| 1 | 5.699 | 5.846 | −0.146 | 5.801 | 5.888 | −0.088 |
| 2 | 6.631 | 6.454 | 0.175 | 6.541 | 6.182 | 0.358 |
| 3 | 7.678 | 7.783 | −0.103 | 7.328 | 7.385 | −0.055 |
| 4 | 6.320 | 6.873 | −0.553 | 6.280 | 6.545 | −0.265 |
| 6 | 6.465 | 6.772 | −0.312 | 5.523 | 6.447 | −0.927 |
| 7 | 4.876 | 5.421 | −0.541 | 4.780 | 5.501 | −0.721 |
| 8 | 7.102 | 6.474 | 0.625 | 7.444 | 6.336 | 1.103 |
| 9 | 7.276 | 7.383 | −0.103 | 7.377 | 7.114 | 0.265 |
| 11 | 8.301 | 8.067 | 0.232 | 7.000 | 6.723 | 0.277 |
| 12 | 7.796 | 6.736 | 1.063 | 6.380 | 5.957 | 0.422 |
| 13 | 8.523 | 8.018 | 0.501 | 7.051 | 6.885 | 0.164 |
| 14 | 7.886 | 7.509 | 0.380 | 6.352 | 6.350 | −0.002 |
| 16 | 6.801 | 6.923 | −0.124 | 4.821 | 6.026 | −1.206 |
| 17 | 8.301 | 7.754 | 0.545 | 6.762 | 6.681 | 0.079 |
| 18 | 6.730 | 6.254 | 0.475 | 5.429 | 5.684 | −0.254 |
| 19 | 5.100 | 6.006 | −0.906 | − | − | − |
| 21 | 4.660 | 5.428 | −0.768 | 4.500 | 5.275 | −0.776 |
| 22 | 7.167 | 5.759 | 1.410 | 5.420 | 5.450 | −0.030 |
| 23 | 4.851 | 5.292 | −0.442 | − | − | − |
| 24 | 5.830 | 6.012 | −0.182 | 5.429 | 5.703 | −0.273 |
| 26 | 7.854 | 7.290 | 0.559 | 7.260 | 6.698 | 0.561 |
| 27 | 7.398 | 7.617 | −0.217 | 7.149 | 6.984 | 0.165 |
| 28 | 7.959 | 8.423 | −0.463 | 6.550 | 7.119 | −0.567 |
| 29 | 7.4819 | 7.104 | 0.375 | 7.400 | 6.697 | 0.703 |
| 31 | 7.1029 | 7.043 | 0.056 | 6.550 | 6.543 | 0.007 |
| 32 | 7.3019 | 7.260 | 0.039 | 7.201 | 6.813 | 0.387 |
| 33 | 7.553 | 6.803 | 0.746 | 7.444 | 6.875 | 0.564 |
| 34 | 7.959 | 7.718 | 0.241 | 6.750 | 6.729 | 0.021 |
| 36 | 6.200 | 6.092 | 0.107 | 5.599 | 5.299 | 0.300 |
| 37 | 6.530 | 6.312 | 0.217 | 6.201 | 5.678 | 0.521 |
| 38 | 6.229 | 7.021 | −0.791 | 6.250 | 6.438 | −0.188 |
| 39 | 4.750 | 5.469 | −0.720 | 5.301 | 5.380 | −0.081 |
| 41 | 5.670 | 5.640 | 0.030 | 5.070 | 5.090 | −0.020 |
| 42 | 5.830 | 6.735 | −0.905 | 5.801 | 5.856 | −0.055 |
| 43 | 7.886 | 7.906 | −0.016 | 7.161 | 7.431 | −0.270 |
| 44 | 6.550 | 6.402 | 0.147 | 6.680 | 6.274 | 0.406 |
| 46 | 4.750 | 5.791 | −1.041 | 4.932 | 5.664 | −0.732 |
| 47 | 4.350 | 5.299 | −0.950 | 5.100 | 5.488 | −0.388 |
| 48 | 6.530 | 6.261 | 0.268 | 7.036 | 6.210 | 0.826 |
| 49 | 6.600 | 6.371 | 0.229 | 6.710 | 6.276 | 0.434 |
| 51 | 6.301 | 6.505 | −0.205 | 5.680 | 6.153 | −0.473 |
| 52 | 5.851 | 6.039 | −0.189 | 5.370 | 5.743 | −0.373 |
| Compd | Rat DHODH | Mouse DHODH | ||||
|---|---|---|---|---|---|---|
| log(1/IC50) | log(1/IC50) | |||||
| Observed | Predicted | Residuala | Observed | Predicted | Residuala | |
| 5 | 7.201 | 6.882 | 0.318 | 7.444 | 6.569 | 0.871 |
| 10 | 5.343 | 6.928 | −1.588 | 4.429 | 6.185 | −1.755 |
| 15 | 6.080 | 7.117 | −1.037 | 4.650 | 6.340 | −1.690 |
| 20 | 7.678 | 7.376 | 0.304 | 7.301 | 6.998 | 0.302 |
| 25 | 6.801 | 6.593 | 0.207 | 5.951 | 6.084 | −0.134 |
| 30 | 5.903 | 5.710 | 0.190 | 5.429 | 5.794 | −0.364 |
| 35 | 7.745 | 8.034 | −0.294 | 6.750 | 6.847 | −0.097 |
| 40 | 6.750 | 6.632 | 0.118 | 6.201 | 5.895 | 0.305 |
| 45 | 4.500 | 5.313 | −0.813 | 4.550 | 5.392 | −0.842 |
| 50 | 7.638 | 6.461 | 1.179 | 6.750 | 6.086 | 0.664 |
| 53 | 6.971 | 6.298 | 0.672 | 7.201 | 6.174 | 1.026 |
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Li, S.-L.; He, M.-Y.; Du, H.-G. 3D-QSAR Studies on a Series of Dihydroorotate Dehydrogenase Inhibitors: Analogues of the Active Metabolite of Leflunomide. Int. J. Mol. Sci. 2011, 12, 2982-2993. https://doi.org/10.3390/ijms12052982
Li S-L, He M-Y, Du H-G. 3D-QSAR Studies on a Series of Dihydroorotate Dehydrogenase Inhibitors: Analogues of the Active Metabolite of Leflunomide. International Journal of Molecular Sciences. 2011; 12(5):2982-2993. https://doi.org/10.3390/ijms12052982
Chicago/Turabian StyleLi, Shun-Lai, Mao-Yu He, and Hong-Guang Du. 2011. "3D-QSAR Studies on a Series of Dihydroorotate Dehydrogenase Inhibitors: Analogues of the Active Metabolite of Leflunomide" International Journal of Molecular Sciences 12, no. 5: 2982-2993. https://doi.org/10.3390/ijms12052982
APA StyleLi, S.-L., He, M.-Y., & Du, H.-G. (2011). 3D-QSAR Studies on a Series of Dihydroorotate Dehydrogenase Inhibitors: Analogues of the Active Metabolite of Leflunomide. International Journal of Molecular Sciences, 12(5), 2982-2993. https://doi.org/10.3390/ijms12052982




