Cloning, Expression, and Characterization of Thermotolerant Manganese Superoxide Dismutase from Bacillus sp. MHS47
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation of the Bacillus sp. MHS47
2.2. Expression and Purification of MnSOD47
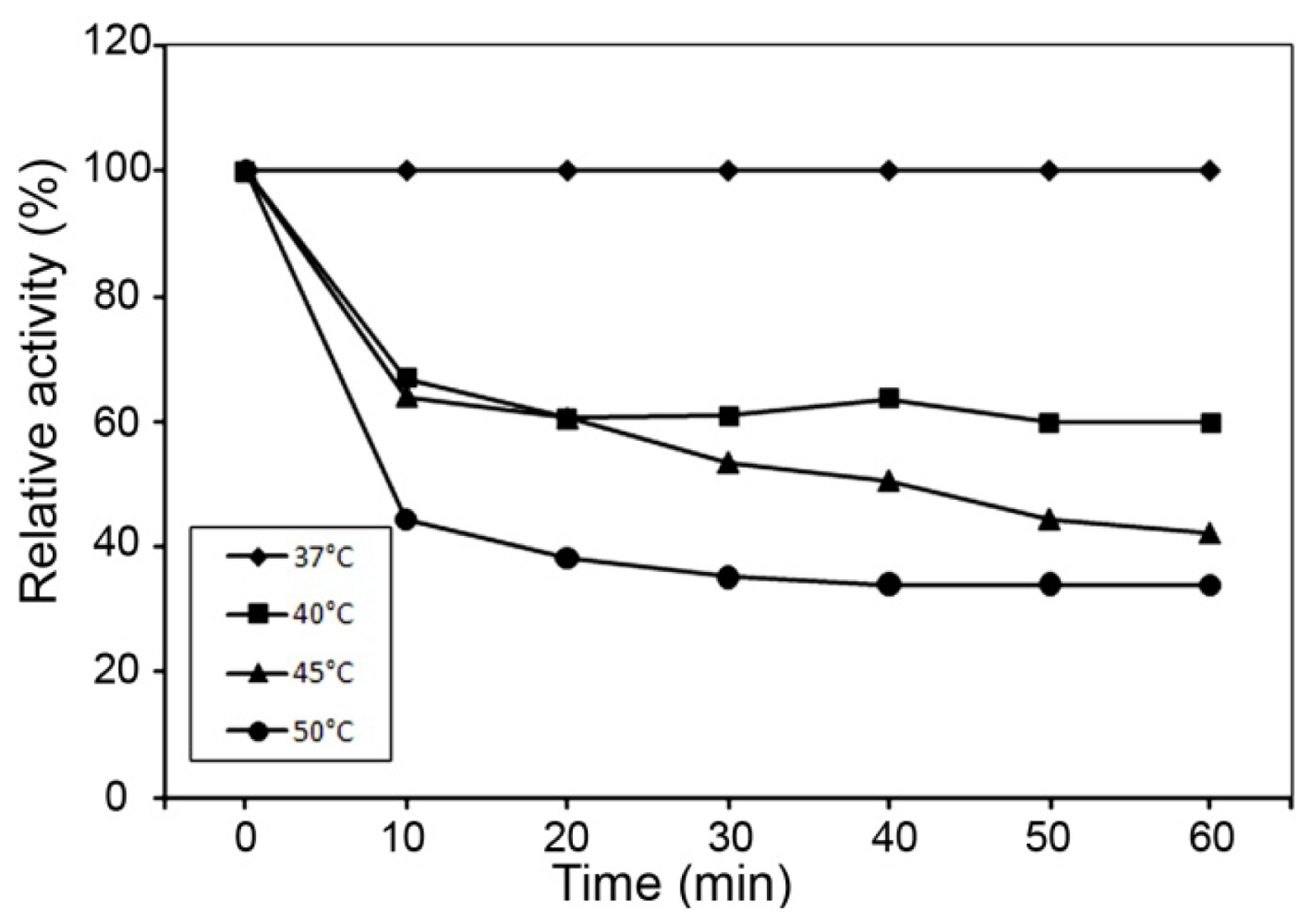
2.3. Effect of pH and Temperature on MnSOD47 Activity
2.4. Effect of Inhibitors on MnSOD47 Activity
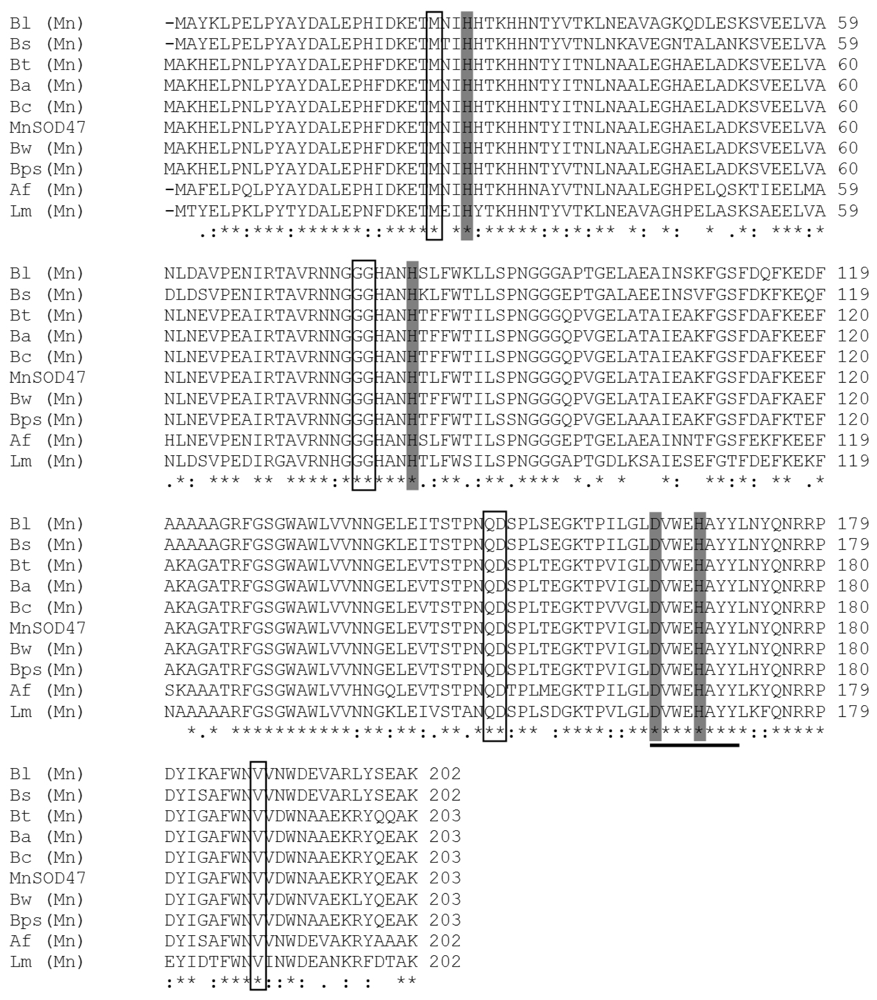
2.5. Discussion
3. Experimental Section
3.1. Bacterial Strain Collection
3.2. Genomic DNA Extraction
3.3. PCR Amplification of the 16S rDNA
3.4. PCR Amplification of the MnSOD Gene from Bacillus sp. MHS47
3.5. Cloning the PCR Fragment
3.6. DNA Sequencing of the SOD Gene
3.7. Transformation and Expression of the MnSOD47 Gene
3.8. Purification of the MnSOD47 Enzyme
3.9. SDS-PAGE
3.10. Protein Assay
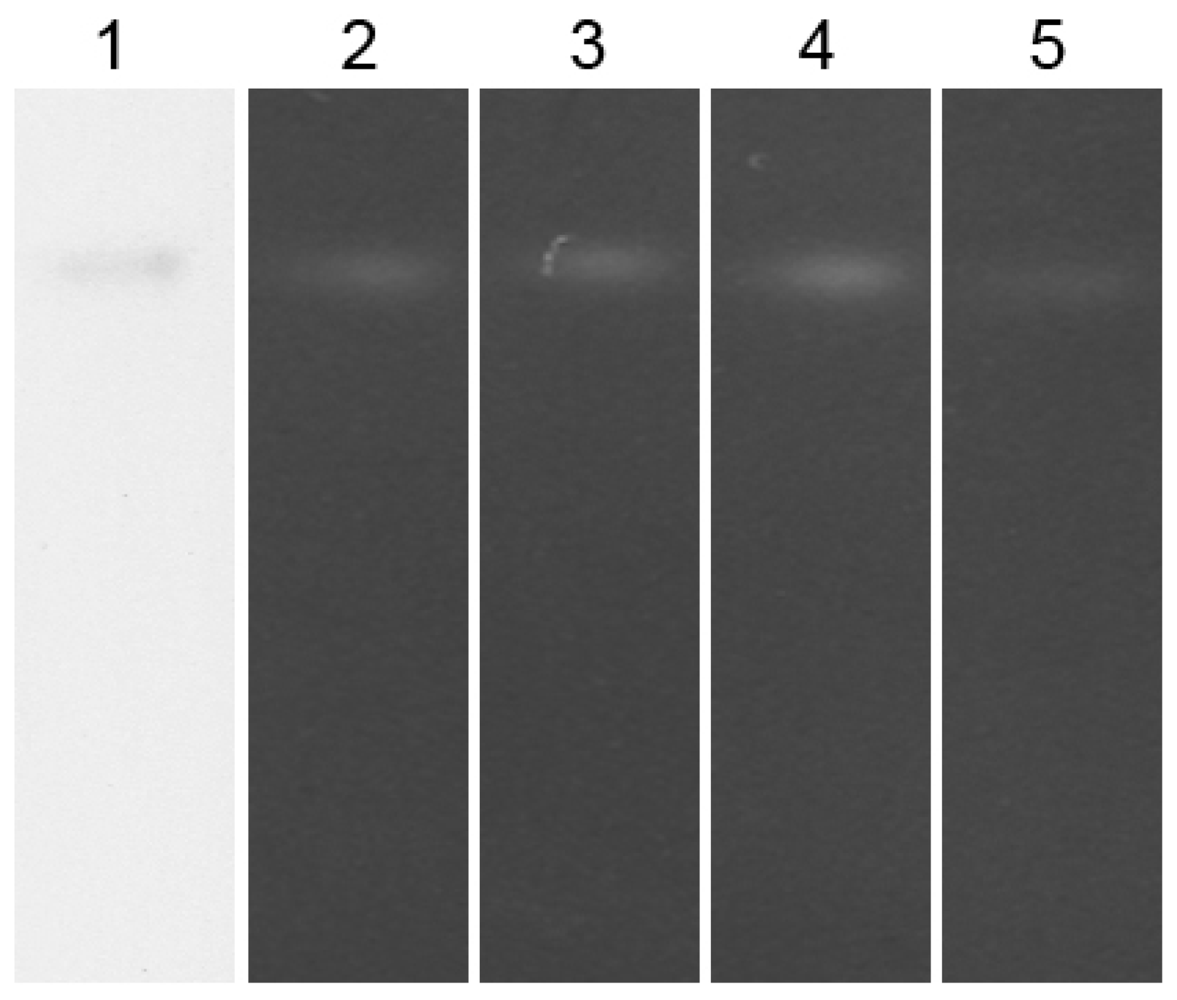
3.11. Immunoblotting Analysis
3.12. Enzyme Assay
3.13. Effect of pH and Temperature on MnSOD47 Activity
3.14. Thermostability
3.15. Enzyme Inhibitors
4. Conclusion
Acknowledgements
References
- Fridovich, I. Superoxide dismutases: Defence against endogenous superoxide radical. Ciba Found. Symp 1978, 65, 77–93. [Google Scholar]
- Lavelle, F; Michelson, AM; Dimitrijevic, L. Biological protection by superoxide dismutase. Biochem. Biophys. Res. Commun 1973, 55, 350–357. [Google Scholar]
- Paschen, W; Weser, U. Letter: Singlet oxygen decontaminating activity of erythrocuprein (superoxide dismutase). Biochim. Biophys. Acta 1973, 327, 217–222. [Google Scholar]
- Petkau, A; Chelack, WS; Pleskach, SD; Meeker, BE; Brady, CM. Radioprotection of mice by superoxide dismutase. Biochem. Biophys. Res. Commun 1975, 65, 886–893. [Google Scholar]
- Fridovich, I. Superoxide radical and superoxide dismutases. Annu. Rev. Biochem 1995, 64, 97–112. [Google Scholar]
- Hassan, HM. Superoxide dismutases. Ciba Found. Symp 1980, 79, 125–142. [Google Scholar]
- Angelova, M; Dolashka-Angelova, P; Ivanova, E; Serkedjieva, J; Slokoska, L; Pashova, S; Toshkova, R; Vassilev, S; Simeonov, I; Hartmann, HJ; Stoeva, S; Weser, U; Voelter, W. A novel glycosylated Cu/Zn-containing superoxide dismutase: Production and potential therapeutic effect. Microbiology 2001, 147, 1641–1650. [Google Scholar]
- Farrow, KN; Lakshminrusimha, S; Reda, WJ; Wedgwood, S; Czech, L; Gugino, SF; Davis, JM; Russell, JA; Steinhorn, RH. Superoxide dismutase restores eNOS expression and function in resistance pulmonary arteries from neonatal lambs with persistent pulmonary hypertension. Am. J. Physiol. Lung Cell Mol. Physiol 2008, 295, L979–L987. [Google Scholar]
- Kakimoto, K; Kojima, Y; Ishii, K; Onoue, K; Maeda, H. The suppressive effect of gelatin-conjugated superoxide dismutase on disease development and severity of collagen-induced arthritis in mice. Clin. Exp. Immunol 1993, 94, 241–246. [Google Scholar]
- Laurila, JP; Castellone, MD; Curcio, A; Laatikainen, LE; Haaparanta-Solin, M; Gronroos, TJ; Marjamaki, P; Martikainen, S; Santoro, M; Laukkanen, MO. Extracellular superoxide dismutase is a growth regulatory mediator of tissue injury recovery. Mol. Ther 2009, 17, 448–454. [Google Scholar]
- Mates, JM; Sanchez-Jimenez, FM. Role of reactive oxygen species in apoptosis: Implications for cancer therapy. Int. J. Biochem. Cell Biol 2000, 32, 157–170. [Google Scholar]
- Rios, L; Cluzel, J; Vennat, JC; Menerath, JM; Doly, M. Comparison of intraocular treatment of DMTU and SOD following retinal ischemia in rats. J. Ocul. Pharmacol. Ther 1999, 15, 547–556. [Google Scholar]
- Teoh, ML; Fitzgerald, MP; Oberley, LW; Domann, FE. Overexpression of extracellular superoxide dismutase attenuates heparanase expression and inhibits breast carcinoma cell growth and invasion. Cancer Res 2009, 69, 6355–6363. [Google Scholar]
- Trotti, A. Toxicity antagonists in cancer therapy. Curr. Opin. Oncol 1997, 9, 569–578. [Google Scholar]
- Yunoki, M; Kawauchi, M; Ukita, N; Noguchi, Y; Nishio, S; Ono, Y; Asari, S; Ohmoto, T; Asanuma, M; Ogawa, N. Effects of lecithinized superoxide dismutase on traumatic brain injury in rats. J. Neurotrauma 1997, 14, 739–746. [Google Scholar]
- Zemlyak, I; Nimon, V; Brooke, S; Moore, T; McLaughlin, J; Sapolsky, R. Gene therapy in the nervous system with superoxide dismutase. Brain Res 2006, 1088, 12–18. [Google Scholar]
- Zhong, W; Oberley, LW; Oberley, TD; St Clair, DK. Suppression of the malignant phenotype of human glioma cells by overexpression of manganese superoxide dismutase. Oncogene 1997, 14, 481–490. [Google Scholar]
- Emregul, E. Development of a new biosensor for superoxide radicals. Anal. Bioanal. Chem 2005, 383, 947–954. [Google Scholar]
- Li, DC; Gao, J; Li, YL; Lu, J. A thermostable manganese-containing superoxide dismutase from the thermophilic fungus Thermomyces lanuginosus. Extremophiles 2005, 9, 1–6. [Google Scholar]
- Amo, T; Atomi, H; Imanaka, T. Biochemical properties and regulated gene expression of the superoxide dismutase from the facultatively aerobic hyperthermophile Pyrobaculum calidifontis. J. Bacteriol 2003, 185, 6340–6347. [Google Scholar]
- Lim, JH; Yu, YG; Han, YS; Cho, S; Ahn, BY; Kim, SH; Cho, Y. The crystal structure of an Fe-superoxide dismutase from the hyperthermophile Aquifex pyrophilus at 1.9 A resolution: structural basis for thermostability. J. Mol. Biol 1997, 270, 259–274. [Google Scholar]
- Seatovic, S; Gligic, L; Radulovic, Z; Jankov, RM. Purification and partial characterization of superoxide dismutase from the thermophilic bacteria Thermothrix sp. J. Serb. Chem. Soc 2004, 69, 9–16. [Google Scholar]
- Ursby, T; Adinolfi, BS; Al-Karadaghi, S; de Vendittis, E; Bocchini, V. Iron superoxide dismutase from the archaeon Sulfolobus solfataricus: Analysis of structure and thermostability. J. Mol. Biol 1999, 286, 189–205. [Google Scholar]
- Wang, X; Yang, H; Ruan, L; Liu, X; Li, F; Xu, X. Cloning and characterization of a thermostable superoxide dismutase from the thermophilic bacterium Rhodothermus sp. XMH10. J. Ind. Microbiol. Biotechnol 2008, 35, 133–139. [Google Scholar]
- Whittaker, MM; Whittaker, JW. Recombinant superoxide dismutase from a hyperthermophilic archaeon, Pyrobaculum aerophilium. J. Biol. Inorg. Chem 2000, 5, 402–408. [Google Scholar]
- Whittaker, MM; Whittaker, JW. A glutamate bridge is essential for dimer stability and metal selectivity in manganese superoxide dismutase. J. Biol. Chem 1998, 273, 22188–22193. [Google Scholar]
- Cheng, W; Tung, YH; Chiou, TT; Chen, JC. Cloning and characterisation of mitochondrial manganese superoxide dismutase (mtMnSOD) from the giant freshwater prawn Macrobrachium rosenbergii. Fish Shellfish Immunol 2006, 21, 453–466. [Google Scholar]
- Graeff-Wohlleben, H; Killat, S; Banemann, A; Guiso, N; Gross, R. Cloning and characterization of an Mn-containing superoxide dismutase (SodA) of Bordetella pertussis. J. Bacteriol 1997, 179, 2194–2201. [Google Scholar]
- Parker, MW; Blake, CC. Iron- and manganese-containing superoxide dismutases can be distinguished by analysis of their primary structures. FEBS Lett 1988, 229, 377–382. [Google Scholar]
- Babitha, MP; Prakash, HS; Shekar, SH. Purification and partial characterization of manganese superoxide dismutase from downy mildew resistant pearl millet seedlings inoculated with Sclerospora graminicola. Plant Sci 2002, 163, 917–924. [Google Scholar]
- Ekanayake, PM; Kang, HS; de Zyosa, M; Jee, Y; Lee, YH; Lee, J. Molecular cloning and characterization of Mn-superoxide dismutase from disk abalone (Haliotis discus discus). Comp. Biochem. Physiol. B Biochem. Mol. Biol 2006, 145, 318–324. [Google Scholar]
- Lan, X; Zhang, X; Hu, J; Shimosaka, M. Cloning, expression, and characterization of a chitinase from the chitinolytic bacterium Aeromonas hydrophila strain SUWA-9. Biosci. Biotechnol. Biochem 2006, 70, 2437–2442. [Google Scholar]
- Motoshima, H; Minagawa, E; Tsukasaki, F; Kaminogawa, S. Cloning and nucleotide sequencing of genes encoding Mn-superoxide dismutase and class II fumarase from Thermus aquaticus YT-1. J Ferment Bioeng 1998, 86, 21–27. [Google Scholar]
- Ken, CF; Lee, CC; Duan, KJ; Lin, CT. Unusual stability of manganese superoxide dismutase from a new species, Tatumella ptyseos ct: Its gene structure, expression, and enzyme properties. Protein Expr. Purif 2005, 40, 42–50. [Google Scholar]
- Misra, HP; Fridovich, I. Inhibition of superoxide dismutases by azide. Arch. Biochem. Biophys 1978, 189, 317–322. [Google Scholar]
- Song, NN; Zheng, Y; E, SJ; Li, DC. Cloning, expression, and characterization of thermostable manganese superoxide dismutase from Thermoascus aurantiacus var. levisporus. J. Microbiol 2009, 47, 123–130. [Google Scholar]
- Ritalahti, KM; Loffler, FE. Populations implicated in anaerobic reductive dechlorination of 1,2-dichloropropane in highly enriched bacterial communities. Appl. Environ. Microbiol 2004, 70, 4088–4095. [Google Scholar]
- Laemmli, UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar]
- Beauchamp, C; Fridovich, I. Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal. Biochem 1971, 44, 276–287. [Google Scholar]
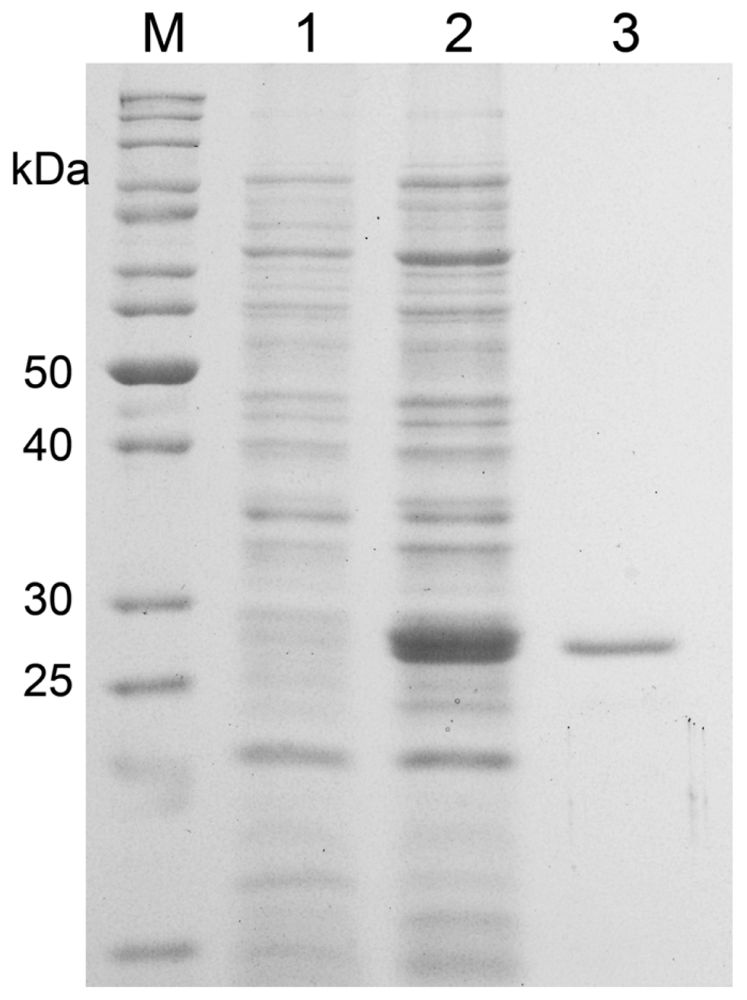


| Purification | Total Volume (mL) | Total Protein (mg) | Total Activity (U) | Specific Activity (U/mg) | Protein Recovery (%) | Purification (fold) |
|---|---|---|---|---|---|---|
| Crude Extract Non-Induce | 25 | 15.31 | 1701 | 111.10 | - | - |
| Crude Extract Induce | 25 | 14.77 | 3648 | 252.13 | 100 | 1 |
| Ni-NTA | 4.5 | 0.56 | 1981 | 3537.75 | 54.3 | 14 |
© 2011 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Areekit, S.; Kanjanavas, P.; Khawsak, P.; Pakpitchareon, A.; Potivejkul, K.; Chansiri, G.; Chansiri, K. Cloning, Expression, and Characterization of Thermotolerant Manganese Superoxide Dismutase from Bacillus sp. MHS47. Int. J. Mol. Sci. 2011, 12, 844-856. https://doi.org/10.3390/ijms12010844
Areekit S, Kanjanavas P, Khawsak P, Pakpitchareon A, Potivejkul K, Chansiri G, Chansiri K. Cloning, Expression, and Characterization of Thermotolerant Manganese Superoxide Dismutase from Bacillus sp. MHS47. International Journal of Molecular Sciences. 2011; 12(1):844-856. https://doi.org/10.3390/ijms12010844
Chicago/Turabian StyleAreekit, Supatra, Pornpimon Kanjanavas, Paisarn Khawsak, Arda Pakpitchareon, Kajeenart Potivejkul, Gaysorn Chansiri, and Kosum Chansiri. 2011. "Cloning, Expression, and Characterization of Thermotolerant Manganese Superoxide Dismutase from Bacillus sp. MHS47" International Journal of Molecular Sciences 12, no. 1: 844-856. https://doi.org/10.3390/ijms12010844
APA StyleAreekit, S., Kanjanavas, P., Khawsak, P., Pakpitchareon, A., Potivejkul, K., Chansiri, G., & Chansiri, K. (2011). Cloning, Expression, and Characterization of Thermotolerant Manganese Superoxide Dismutase from Bacillus sp. MHS47. International Journal of Molecular Sciences, 12(1), 844-856. https://doi.org/10.3390/ijms12010844




