Abstract
On the basis of the inductive QSAR descriptors we have created a neural network-based solution enabling quantification of antibacterial activity in the series of 101 synthetic cationic polypeptides (CAMEL-s). The developed QSAR model allowed 80% correct categorical classification of antibacterial potencies of the CAMEL-s both in the training and the validation sets. The accuracy of the activity predictions demonstrates that a narrow set of 3D sensitive ‘inductive’ descriptors can adequately describe the aspects of intra- and intermolecular interactions that are relevant for antibacterial activity of the cationic polypeptides. The developed approach can be further expanded for the larger sets of biologically active peptides and can serve as a useful quantitative tool for rational antibiotic design and discovery.
Introduction
QSAR models for antibiotic activity
QSAR studies of antibiotic activity represent an emerging and exceptionally important topic in the area of computed-aided drug design. Although the demand for ‘in silico’ discovery is clear in all areas of human therapeutics, the field of anti-infective drugs has a particular need for computational solutions enabling rapid identification of novel therapeutic leads. As a result, there is an urge for new antibiotics and antivirals driven by critical situations, such as the increased prevalence of multi-drug resistant bacteria and HIV/AIDS, and the emergence or re-emergence of deadly infectious diseases such as Lyme disease, West Nile virus, Hantavirus pulmonary syndrome, Norwalk-like virus, Avian influenza virus, SARS, and novel forms of Cryptococcal infection. On another hand, historically, ‘Big Pharma’ have withdrawn from the field of antimicrobial drug development in favour of more profitable areas. Consequently, very few novel antibacterial therapeutics have emerged over the last decade. At this moment, QSAR studies can help solving this problem by providing the means of rapid design and virtual screening of combinatorial anti-infective libraries, as well as for rational data mining for novel antibiotic candidates.
Few antibacterial QSAR studies have been reported up to date, which could either distinguish compounds possessing antibacterial activity from all other chemicals, or numerically reproduce antibacterial potencies in the series of closely related chemical analogues. These QSAR approaches process a variety of structure-dependent descriptors with machine learning and statistical techniques such as Artificial Neural Networks [1,2,3], Linear Discriminant Analysis, [4,5,6] Binary Logistic Regression [5], Principal Component Analysis and k-means Cluster method [7]. In some cases the results allowed the authors to introduce novel anti-infective leads, however, all of the reported QSAR solutions have been built upon already well - studied classes of traditional antibiotics. In the current work, we apply the QSAR methodology to the newest class of antibacterial therapeutics – the cationic polypeptides, which represent the latest hope in the combat against multi-drug resistant pathogens.
Cationic polypeptides as a novel class of antibacterial therapeutics
A diverse population of antimicrobial peptides (AMP-s) can be found in nature, as they are an essential component of anti-infective defence mechanisms in mammals, amphibians, insects and plants [8,9,10,11]. The majority of AMP-s share several key structural features such as short length (typically 10 to 40 amino acids), amphipathicity (i.e., a molecule has distinct cationic and hydrophobic faces), and helical or cyclic structure. In the recent years, AMP-s have drawn much attention as a potentially effective class of anti-infectious therapeutics. Considering the facts that bacterial resistance to antimicrobial peptides is infrequent [12,13,8], they are non-toxic and non-immunogenic (according to numerous reports, such as [14]), extensive research programs have been established with the aim to exploit the AMP-s as a novel stand-alone class of antibiotics.
Substantial experimental efforts have been invested into discovery and investigation of natural and synthetic cationic polypeptides possessing antibacterial, antiviral, antifungal and/or anti-tumour activities. Nevertheless, only a few very simple structure-activity studies have been reported in the literature, with the results not leading to validated QSAR models. In the current work, we have attempted to fill this gap by creating a QSAR model quantifying antibacterial activity of a broad range of rigorously investigated cationic peptides through the recently developed ‘inductive’ QSAR descriptors.
‘Inductive’ descriptors overview
The ‘inductive’ descriptors have been previously introduced, and are based on the models of inductive and steric effects, inductive electronegativity and molecular capacitance, developed in a series of papers by Cherkasov and co-authors [15,16,17,18,19]. These molecular parameters can be easily accessed from fundamental parameters of bound atoms, such as absolute electronegativities (χ), covalent radii (R) and intramolecular distances (r). The steric Rs and inductive σ* influence of n - atomic group G on a single atom j is:
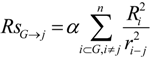
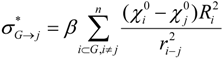
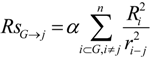
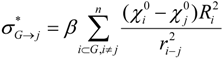
In those cases when the inductive and steric interactions occur between a given atom j and the rest of N-atomic molecule (as sub-substituent), the summation in (1) and (2) is taken over N-1 terms. Thus, the group electronegativity of (N-1)-atomic substituent around atom j is expressed as the following:
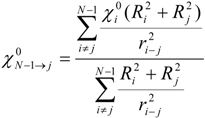
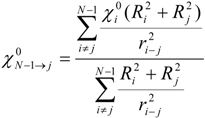
Similarly, steric and inductive effects of a singe atom onto a group of atoms (the rest of the molecule) are defined as:
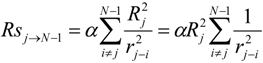
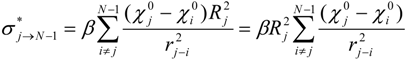
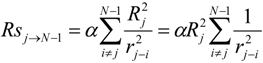
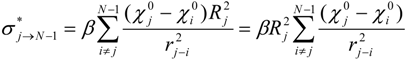
In the work [18] an iterative procedure for calculating a partial charge on j-th atom in a molecule was developed, and it is:
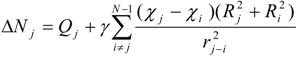 (where Qj reflects the formal charge of an toms j).
(where Qj reflects the formal charge of an toms j).
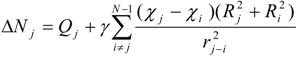
Initially, the parameter χ in (6) corresponds to χ0 - an absolute, unchanged electronegativity of an atom As the iterative calculation progresses, the equalized electronegativity χ’ gets updated according to (7).
where the local chemical hardness η0 reflects the “resistance” of electronegativity to a change of the atomic charge. The ‘inductive’ hardness ηi and softness si of a bound atom i are represented in the following manner:
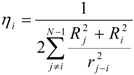
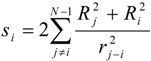
χ' ≈ χ0 + η0∆N
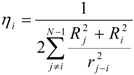
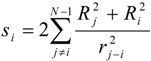
The corresponding group parameters are therefore expressed as:
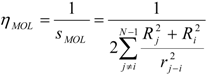
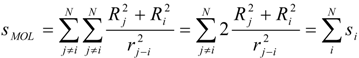
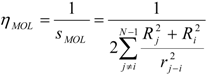
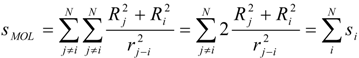
The interpretation of the physical meaning of the ‘inductive’ descriptors was developed by considering a neutral molecule as an electrical capacitor formed by charged atomic spheres [18]. This approximation relates chemical softness-hardness of bound atom(s) with the areas of the facings of an electrical capacitor radically formed by the atom(s) in a molecule (Figure 1), and correlates electronic density with capacitor-accumulated electricity.
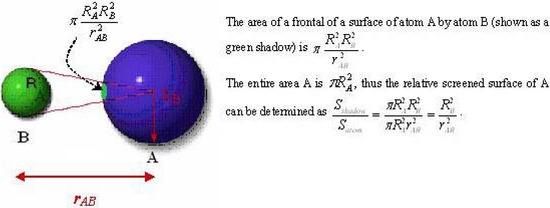
Figure 1.
Radial shielding on spherical surface of atom A by the neighbouring atom B
The validation of Cherkasov’s ‘inductive’ parameters, developed to date, has been rigorously conducted on extensive experimental datasets [15,16,17,18,19,20,21,22,23,24]. Table 1 features 50 ‘inductive’ QSAR descriptors that can be calculated in the framework of equations (1)-(11). It should be noted that in a previous study [25], these molecular parameters allowed creation of the QSAR model enabling 93% correct recognition of low-molecular weight antibacterial compounds.

Table 1.
Inductive QSAR descriptors introduced on the basis of equations (1)-(11).
| Descriptor | Characterization | Parental formula(s) |
|---|---|---|
| χ (electronegativity) – based | ||
| EO_Equalized* | Iteratively equalized electronegativity of a molecule | Calculated iteratively by (7) where charges get updated according to (6); an atomic hardness in (7) is expressed through (8) |
| Average_EO_Pos* | Arithmetic mean of electronegativities of atoms with positive partial charge | 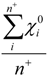 where n+ is the number of atoms i in a molecule with positive partial charge where n+ is the number of atoms i in a molecule with positive partial charge |
| Average_EO_Neg* | Arithmetic mean of electronegativities of atoms with negative partial charge | 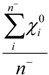 where n− is the number of atoms i in a molecule with negative partial charge where n− is the number of atoms i in a molecule with negative partial charge |
| η (hardness) – based | ||
| Global_Hardness | Molecular hardness - reversed softness of a molecule | (10) |
| Sum_Hardness* | Sum of hardnesses of atoms of a molecule | Calculated as a sum of inversed atomic softnesses in turn computed within (9) |
| Sum_Pos_Hardness | Sum of hardnesses of atoms with positive partial charge | Obtained by summing up the contributions from atoms with positive charge computed by (8) |
| Sum_Neg_Hardness | Sum of hardnesses of atoms with negative partial charge | Obtained by summing up the contributions from atoms with negative charge computed by (8) |
| Average_Hardness* | Arithmetic mean of hardnesses of all atoms of a molecule | Estimated by dividing quantity (10) by the number of atoms in a molecule |
| Average_Pos_Hardness* | Arithmetic mean of hardnesses of atoms with positive partial charge | 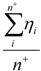 where n+ is the number of atoms i with positive partial charge. where n+ is the number of atoms i with positive partial charge. |
| Average_Neg_Hardness* | Arithmetic mean of hardnesses of atoms with negative partial charge | 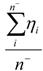 where n− is the number of atoms i with negative partial charge. where n− is the number of atoms i with negative partial charge. |
| Smallest_Pos_Hardness | Smallest atomic hardness among values for positively charged atoms | (8) |
| Smallest_Neg_Hardness | Smallest atomic hardness among values for negatively charged atoms. | (8) |
| Largest_Pos_Hardness* | Largest atomic hardness among values for positively charged atoms | (8) |
| Largest_Neg_Hardness* | Largest atomic hardness among values for negatively charged atoms | (8) |
| Hardness_of_Most_Pos* | Atomic hardness of an atom with the most positive charge | (8) |
| Hardness_of_Most_Neg | Atomic hardness of an atom with the most negative charge | (8) |
| s (softness) – based | ||
| Global_Softness | Molecular softness – sum of constituent atomic softnesses | (11) |
| Total_Pos_Softness | Sum of softnesses of atoms with positive partial charge | Obtained by summing up the contributions from atoms with positive charge computed by (9) |
| Total_Neg_Softness* | Sum of softnesses of atoms with negative partial charge | Obtained by summing up the contributions from atoms with negative charge computed by (9) |
| Average_Softness | Arithmetic mean of softnesses of all atoms of a molecule | (11) divided by the number of atoms in molecule |
| Average_Pos_Softness | Arithmetic mean of softnesses of atoms with positive partial charge | 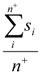 Where n+ is the number of atoms i with positive partial charge. Where n+ is the number of atoms i with positive partial charge. |
| Average_Neg_Softness | Arithmetic mean of softnesses of atoms with negative partial charge | 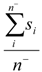 where n− is the number of atoms i with negative partial charge. where n− is the number of atoms i with negative partial charge. |
| Smallest_Pos_Softness | Smallest atomic softness among values for positively charged atoms | (9) |
| Smallest_Neg_Softness | Smallest atomic softness among values for negatively charged atoms | (9) |
| Largest_Pos_Softness | Largest atomic softness among values for positively charged atoms | (9) |
| Largest_Neg_Softness | Largest atomic softness among values for positively charged atoms | (9) |
| Softness_of_Most_Pos | Atomic softness of an atom with the most positive charge | (9) |
| Softness_of_Most_Neg | Atomic softness of an atom with the most negative charge | (9) |
| q (charge)- based | ||
| Total_Charge | Sum of absolute values of partial charges on all atoms of a molecule | 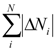 where all the contributions ∆Ni derived within (6) where all the contributions ∆Ni derived within (6) |
| Total_Charge_Formal* | Sum of charges on all atoms of a molecule (formal charge of a molecule) | Sum of all contributions (6) |
| Average_Pos_Charge* | Arithmetic mean of positive partial charges on atoms of a molecule | 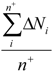 where n+ is the number of atoms i with positive partial charge where n+ is the number of atoms i with positive partial charge |
| Average_Neg_Charge* | Arithmetic mean of negative partial charges on atoms of a molecule | 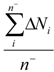 where n− is the number of atoms i with negative partial charge where n− is the number of atoms i with negative partial charge |
| Most_Pos_Charge* | Largest partial charge among values for positively charged atoms | (6) |
| Most_Neg_Charge | Largest partial charge among values for negatively charged atoms | (6) |
| σ* (inductive parameter) – based | ||
| Total_Sigma_mol_i | Sum of inductive parameters σ*(molecule→atom) for all atoms within a molecule | 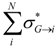 where contributions where contributions 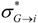 are computed by equation (2) with n=N-1 – i.e. each atom j is considered against the rest of the molecule G are computed by equation (2) with n=N-1 – i.e. each atom j is considered against the rest of the molecule G |
| Total_Abs_Sigma_mol_i | Sum of absolute values of group inductive parameters σ*(molecule→atom) for all atoms within a molecule | 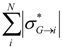 |
| Most_Pos_Sigma_mol_i* | Largest positive group inductive parameter σ*(molecule→atom) for atoms in a molecule | (2) |
| Most_Neg_Sigma_mol_i | Largest (by absolute value) negative group inductive parameter σ*(molecule→atom) for atoms in a molecule | (2) |
| Most_Pos_Sigma_i_mol | Largest positive atomic inductive parameter σ*(atom→molecule) for atoms in a molecule | (5) |
| Most_Neg_Sigma_i_mol* | Largest negative atomic inductive parameter σ*(atom→molecule) for atoms in a molecule | (5) |
| Sum_Pos_Sigma_mol_i | Sum of all positive group inductive parameters σ*( molecule →atom) within a molecule | 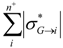 where where 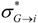 >0 and n+ is the number of N-1 atomic substituents in a molecule with positive inductive effect (electron acceptors) >0 and n+ is the number of N-1 atomic substituents in a molecule with positive inductive effect (electron acceptors) |
| Sum_Neg_Sigma_mol_i* | Sum of all negative group inductive parameters σ*( molecule →atom) within a molecule | 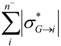 where where 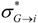 <0 and n− is the number of N-1 atomic substituents in a molecule with negative inductive effect (electron donors) <0 and n− is the number of N-1 atomic substituents in a molecule with negative inductive effect (electron donors) |
| Rs (steric parameter) – based | ||
| Largest_Rs_mol_i | Largest value of steric influence Rs(molecule→atom) in a molecule | (1) where n=N-1 - each atom j is considered against the rest of the molecule G |
| Smallest_Rs_mol_i* | Smallest value of group steric influence Rs(molecule→atom) in a molecule | (1) where n=N-1 - each atom j is considered against the rest of the molecule G |
| Largest_Rs_i_mol | Largest value of atomic steric influence Rs(atom→molecule) in a molecule | (4) |
| Smallest_Rs_i_mol | Smallest value of atomic steric influence Rs(atom→molecule) in a molecule | (4) |
| Most_Pos_Rs_mol_i | Steric influence Rs(molecule→atom) ON the most positively charged atom in a molecule | (1) |
| Most_Neg_Rs_mol_i* | Steric influence Rs(molecule→atom) ON the most negatively charged atom in a molecule | (1) |
| Most_Pos_Rs_i_mol | Steric influence Rs(atom→molecule) OF the most positively charged atom to the rest of a molecule | (4) |
| Most_Neg_Rs_i_mol | Steric influence Rs(atom→molecule) OF the most negatively charged atom to the rest of a molecule | (4) |
* – descriptors selected for building the antibiotic peptide QSAR model.
Results and Discussion
Experimental data
In the current work, we have used the ‘inductive’ descriptors to investigate structure-activity relationships in a series of antibiotic peptides called CAMEL-s. These compounds represent derivatives from the hybrid polypeptide CAMEL0 previously created by the respective fusion of the C- and N-terminus sequences of natural peptides Cecropin and Melittin. Despite the rather limited variability in amino acid sequences among these leucine-rich peptides, their antibacterial activity ranges over several orders of magnitude. It has been experimentally demonstrated that the CAMEL-s exhibit high activity against various strains (including the drug-resistant ones) of Gram-positive and Gram-negative bacteria, including Bacteroides, Bordetella, Campylobacter, Corynebacterium, Klebsiella, Listeria, Moraxella, Pastuerella, Taylorella, Yersinia, Rhodococcus, Staphylococcus and Streptococcus [26,27,28]. The minimal inhibitory concentrations for the series of 101 CAMEL-s against the listed microorganisms have been previously averaged to produce the mean antibiotic potency parameters [26,27,28]. These values extracted from the SAPD database [29] have been collected into Table 2 and subjected to QSAR analysis with the ‘inductive’ descriptors.
Factors governing bioactivity of CAMEL-s
The common mode of action of antimicrobial peptides is disruption of bacterial cell membranes via electrostatic and hydrophobic interactions [1,3,4,12,14,29,30,31,32,33,34,35,36]. It is believed that amphipatic peptides can penetrate or form pores in the cell membranes through the insertion into the lipid bilayer mediated by hydrophobic forces, while their electrostatic interaction with phospholipid headgroups leads to membrane disruption. Cationic peptides exhibit high affinity only toward negatively charged surfaces of bacterial cells while they do not tend to interact with eukaryotic cells surfaces composed mostly of zwitterionic phospholipids. It has been demonstrated that antimicrobial activity of polypeptides can be influenced by their helicity, hydrophobicity and amphipathicity [5,37,38,39,40,41,42,43,44]. Nonetheless, the exact nature of this correlation is still unclear and the understanding of the factors influencing the AMP activity is incomplete. We postulate that a set of the developed ‘inductive’ descriptors can adequately reflect those structure-dependent properties of CAMEL-s pertaining to their antibacterial activity. The reasoning for this stems from the fact that the parameters calculated within (1)–(11) cover a very broad range of proprieties of bound atoms and molecules related to their size, polarizability, electronegativity, compactness, mutual inductive and steric influence and distribution of electronic density, etc.
Descriptors calculation and selection
All 50 inductive QSAR descriptors (presented in detail in Table 1) have been calculated for all 101 CAMEL molecules under study. To compute the ‘inductive’ descriptors we have used the custom SVL-scripts implemented in the MOE package [45]. It should be noted that all of the produced parameters are 3D-sensitive and depend on the structure of polypeptides. The CAMEL-s were initially built in the alpha-helical conformations (the helicity was confirmed by a number of secondary structure predictors) which were further optimized by the MMFF9f molecular mechanic simulations.
It should be mentioned here, that some inductive descriptors may reflect related or similar molecular/atomic properties and can be correlated in certain cases (even though the analytical representation of those descriptors does not directly imply their co-linearity). Moreover, most of the CAMEL-s have very similar three-dimensional structures and, therefore, special precautions were taken in selecting the appropriate ‘inductive’ descriptors for the QSAR model. Hence, to eliminate the cross-correlation among the independent variables, we pre-computed pairwise regressions between all pairs of the 50 QSAR parameters for CAMEL-s. We subsequently removed those descriptors that linearly correlated with R≥0.9. As a result of this procedure, only 20 parameters were selected for further simulation (for more information refer to the Legend of Table 1). The descriptors are: Average Electronegativities of the Negatively/Positively Charged Atoms, Molecular (equalized) Electronegativity, Total Formal Charge, Average Atomic Hardness, Sum of Atomic Hardnesses, Average Negative/Positive Charges, Largest Positive Charge, Average Atomic Hardnesses of Negatively/Positively Charged Atoms, Hardness of the Most Positively Charged Atom, Largest Hardness among the Negatively/Positively Charged Atoms, Sum of Softnesses of Negatively Charged Atoms, Steric Effect on the Most Negatively Charged Atom, Most Negative Inductive Constant of an Atom in Molecule, Largest Positive Inductive Effect on an Atom in Molecule, The Smallest Steric Effect on a Atom in Molecule, Sum of all Negative Inductive Effects on Atoms in Molecule.
The averaged values of these 20 indices were also separately calculated for antibacterial CAMEL-s conventionally sub-divided into three activity groups: very active (with the mean potency above 4), mild (potency between 2 and 4) and moderate antibacterials (potency < 2). The ‘inductive’ descriptors averaged within these three groups are plotted in Figure 2. Although the curves for moderate and very active peptides are very close for the most part, all three categories of CAMEL-s can be clearly distinguished by the selected QSAR parameters.
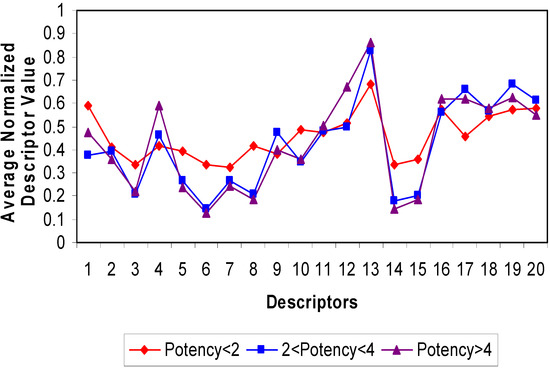
Figure 2.
Distribution of the averaged values of the ‘inductive’ indices among ‘Very active’, ‘Moderately active’ and ‘Mild’ CAMEL polypeptides under investigation.
Therefore, it is reasonable to assume that the ‘inductive’ QSAR descriptors can effectively be used for the numerical quantification of the average potency of the CAMEL-s.
Composition of the training and the testing (validation) sets
In order to relate the ‘inductive’ descriptors to the experimental mean potencies of the peptides, we have employed the method of Artificial Neural Networks (ANN). Machine-learning approaches, particularly ANN, represent one of the essential parts of the modern QSAR, and the detailed description of the corresponding methodologies can be found elsewhere [e.g., in 46].
For this study, we chose the standard back-propagation configuration for the ANN and we used the Stuttgart Neural Network Simulator package [47] to implement the model. For effective training of the network (primarily to avoid overfitting), we used the training sets of 91 compounds randomly selected as 90 percent of the available CAMEL-s. Such random sampling has been performed 20 times and, thus, 20 independent QSAR models have been created in order to evaluate the average predictive ability of the method. One of the training sets with 91 CAMEL peptides is presented in Table 2.

Table 2.
Training Set of Camels in the 90/10 Split: the Mean Experimental Potencies vs. Predicted Potencies Using a Neural Network with Eight Hidden Nodes. The experimental potencies are average potencies against 24 Gram-positive and Gram-negative bacterial strains. The Camels are sorted according to ascending experimental potencies.
| Identity | Peptide sequence (NH2 corresponds to the amidated C-terminus group) | Experimental Potency | Predicted Potency |
| CAMEL118 | KWKLFlgIlAVLKVL-NH2 | 0.159 | 0.335 |
| CAMEL17 | KWnLngnInAVLKVL-NH2 | 0.209 | 0.176 |
| CAMEL38 | KWKgeleIeAeLKVL-NH2 | 0.376 | 0.172 |
| CAMEL107 | gWKLglKIlnVLKVL-NH2 | 0.496 | 1.216 |
| CAMEL20 | KWKLFKKnnnnnKhn-NH2 | 0.498 | 1.49 |
| CAMEL116 | KWhLFllIlAVLKVL-NH2 | 0.514 | 0.405 |
| CAMEL34 | KrgLFKKgGAVLKgL-NH2 | 0.528 | 1.227 |
| CAMEL18 | KWhLrnKIGAVrnnL-NH2 | 0.537 | 1.183 |
| CAMEL16 | KhKLFKKIGAhrKrn-NH2 | 0.553 | 1.445 |
| CAMEL39 | hWhLhKhrGArhKVL-NH2 | 0.677 | 1.184 |
| CAMEL134 | gWeLgeeIlnVLKVL-NH2 | 0.708 | 0.16 |
| CAMEL115 | KWhLFlKIlAVLKVL-NH2 | 0.741 | 1.646 |
| CAMEL50 | KWKLFKKhGnVrKVL-NH2 | 0.771 | 1.747 |
| CAMEL10 | KnKrnKKIGAVLKVL-NH2 | 0.848 | 1.216 |
| CAMEL14 | KhnLFKgIGAVLlVL-NH2 | 0.922 | 1.121 |
| CAMEL51 | KWKLFKKIGnrnKVL-NH2 | 0.947 | 1.631 |
| CAMEL113 | lWKLFlhIlAVLKVL-NH2 | 0.962 | 0.162 |
| CAMEL26 | KnKLeKKIGAVLKVL-NH2 | 1.027 | 1.339 |
| CAMEL52 | KWKLgKgIGAVgKVL-NH2 | 1.033 | 1.08 |
| CAMEL58 | KWKLFnrIGhnrKVn-NH2 | 1.049 | 1.739 |
| CAMEL137 | gWrLFrgIrAVLnVL-NH2 | 1.074 | 0.296 |
| CAMEL54 | KWgLFKnIGAVLhVn-NH2 | 1.156 | 0.223 |
| CAMEL57 | rWKLnnnIGArLKVL-NH2 | 1.206 | 0.977 |
| CAMEL33 | hWKLFKKIGhVnKrL-NH2 | 1.34 | 1.937 |
| CAMEL60 | hWKrFlrIGhnLnVn-NH2 | 1.495 | 1.385 |
| CAMEL11 | KWKLFKKIGgVggVL-NH2 | 1.593 | 2.133 |
| CAMEL120 | gWKLFlKIlAVLKVL-NH2 | 1.598 | 0.785 |
| CAMEL13 | gWKLFKnrGAVLKhL-NH2 | 1.605 | 2.444 |
| CAMEL8 | KWKLFnKrGAVLKVL-NH2 | 1.605 | 2.404 |
| CAMEL19 | rWKnFKnIrAnLrVL-NH2 | 1.742 | 2.017 |
| CAMEL56 | KWKLFgKnGrnLlVL-NH2 | 1.814 | 0.826 |
| CAMEL138 | gWrLFKgIrAVLnVL-NH2 | 1.826 | 1.157 |
| CAMEL41 | KWKLFKKgavlkvlt-NH2 | 1.891 | 3.518 |
| CAMEL47 | KWKLFKKrnAVLKVL-NH2 | 1.964 | 2.497 |
| CAMEL44 | KWKLFKKIGAnLKVL-NH2 | 2.07 | 3.032 |
| CAMEL12 | KWKLFKrIGAVhKrL-NH2 | 2.242 | 1.675 |
| CAMEL53 | hWKLFKKIhAVrKhL-NH2 | 2.244 | 1.495 |
| CAMEL112 | KWKLFlhIlAVLKVL-NH2 | 2.245 | 1.178 |
| CAMEL32 | KWKLFrKIGAVhrVL-NH2 | 2.306 | 2.906 |
| CAMEL29 | lWKLFKKhGAVLKVL-NH2 | 2.422 | 4.74 |
| CAMEL36 | KWhLnKrIhAVLKrL-NH2 | 2.587 | 2.129 |
| CAMEL1 | KWKLFKKgigavlkv-NH2 | 2.649 | 1.83 |
| CAMEL35 | KWKLFrrIGAVLKhr-NH2 | 2.706 | 1.816 |
| CAMEL30 | KrKrFrKIGAVLKVL-NH2 | 2.747 | 1.849 |
| CAMEL15 | KWKLFKlrGrVrKVL-NH2 | 2.879 | 3.728 |
| CAMEL31 | KWKLFKKIGlgLgVL-NH2 | 3.148 | 2.325 |
| CAMEL2 | KWKLFKKlkvlttgl-NH2 | 3.246 | 3.974 |
| CAMEL126 | lWrLlKhIlrVLKVL-NH2 | 3.259 | 3.974 |
| CAMEL55 | KWlLFKKIGAVLlnh-NH2 | 3.425 | 3.229 |
| CAMEL37 | lrKLFKKIrAVLlVr-NH2 | 3.617 | 3.407 |
| CAMEL125 | lWrLlKKIlrVLKVL-NH2 | 3.822 | 4.82 |
| CAMEL119 | gWKLFKlIGAVLKVL-NH2 | 3.86 | 3.429 |
| CAMEL28 | KWKLgKKIGAVLgVL-NH2 | 3.946 | 3.055 |
| CAMEL114 | KWKLFhKIlAVLKVL-NH2 | 4.105 | 4.843 |
| CAMEL61 | KWKLFKKavlkvltt-NH2 | 4.105 | 3.995 |
| CAMEL111 | KWKLFhlIGAVLKVL-NH2 | 4.165 | 3.484 |
| CAMEL49 | nWKLFhKIGAVLKVL-NH2 | 4.187 | 4.819 |
| CAMEL25 | KWKLrKKIGAVLKVL-NH2 | 4.262 | 3.762 |
| CAMEL43 | KWKgFKKIGAVLKVL-NH2 | 4.319 | 4.359 |
| CAMEL105 | gWKLgKKIGrVLKVL-NH2 | 4.336 | 5.555 |
| CAMEL124 | KWKLFKlIrAVLKVL-NH2 | 4.533 | 5.078 |
| CAMEL7 | KWKLFKKIGAVLhnL-NH2 | 4.534 | 3.614 |
| CAMEL121 | gWKgFKKIGrVLKVL-NH2 | 4.759 | 5.465 |
| CAMEL27 | KWKLFKKIGAVLnrL-NH2 | 4.869 | 4.987 |
| CAMEL3 | KWgLFKKIGAVLKVL-NH2 | 4.989 | 3.87 |
| CAMEL81 | KWKLFKKvlkVLttg-NH2 | 5.297 | 4.455 |
| CAMEL106 | gWKLFKKIGrVLKVL-NH2 | 5.318 | 5.577 |
| CAMEL127 | gWKLFKKIGrVLrVL-NH2 | 5.47 | 5.13 |
| CAMEL130 | lWKLFKKIrrlLKVL-NH2 | 5.49 | 4.547 |
| CAMEL128 | lWKLFKKIGrVLKVL-NH2 | 5.493 | 5.375 |
| CAMEL131 | lWKLFrKIrrlLrVL-NH2 | 5.504 | 5.682 |
| CAMEL104 | gWKLgKKIlrVLrVL-NH2 | 5.562 | 5.771 |
| CAMEL108 | KWKLgKKIlnVLKVL-NH2 | 5.566 | 4.905 |
| CAMEL109 | gWrLgKKIlrVLKVL-NH2 | 5.572 | 5.7 |
| CAMEL123 | lWKLFKKIrrVLrVL-NH2 | 5.614 | 5.656 |
| CAMEL0 | KWKLFKKIGAVLKVL-NH2 | 5.712 | 5.14 |
| CAMEL42 | hWKLFKKIGAVLKVL-NH2 | 5.712 | 4.706 |
| CAMEL102 | gWKLgKKIlrVLKVL-NH2 | 5.725 | 5.674 |
| CAMEL6 | nWKLFKKIGAVLKVL-NH2 | 5.803 | 5.205 |
| CAMEL23 | KWhLFKKIGAVLKVL-NH2 | 5.81 | 4.7 |
| CAMEL101 | KWKLgKKIlrVLKVL-NH2 | 5.845 | 5.465 |
| CAMEL103 | gWKLglKIlrVLKVL-NH2 | 5.879 | 5.238 |
| CAMEL22 | gWKLFKKIGAVLKVL-NH2 | 5.946 | 5.363 |
| CAMEL110 | gWKLgKKIlnVLKVL-NH2 | 6.043 | 5.637 |
| CAMEL129 | lWKLFKKInrVLKVL-NH2 | 6.045 | 5.212 |
| CAMEL4 | KWKLFhKIGAVLKVL-NH2 | 6.072 | 4.877 |
| CAMEL24 | KWKLFKhIGAVLKVL-NH2 | 6.167 | 4.88 |
| CAMEL132 | gWKLgKhIlnVLKVL-NH2 | 6.182 | 5.035 |
| CAMEL48 | KWKLgKKIGAVLKVL-NH2 | 6.323 | 5.026 |
| CAMEL136 | vWrLiKKIlrVfKgL-NH2 | 6.613 | 5.374 |
| CAMEL135 | gWrLiKKIlrVfKgL-NH2 | 6.665 | 5.559 |
The remaining 10 polypeptides, featured in Table 3, were used as the corresponding testing group to access the method’s predictive ability. For each polypeptide in the training and testing sets, we have transformed 20 network input descriptors into the normalized values varying from 0 to 1. Similarly, the output parameters from the ANN (mean antibacterial potencies) were normalized to [0:1] range.

Table 3.
Validation (testing) Set of Camels in the 90/10 Split: Experimental Mean Potencies vs. Predicted Potencies by a Neural Network with Eight Hidden Nodes.
| Identity | A.A. Sequence | Experimental Potency | Predicted Potency |
|---|---|---|---|
| CAMEL59 | KgKggKKgGrggKVL-NH2 | 1.077 | 1.030 |
| CAMEL40 | gWlLhrnIGnVLhrL-NH2 | 1.387 | 1.167 |
| CAMEL5 | KWKLFKKnGAVLKVL-NH2 | 1.408 | 2.076 |
| CAMEL139 | gWKLFKgIrAVLnVL-NH2 | 1.497 | 1.524 |
| CAMEL117 | lWhLFlKIlAVLKVL-NH2 | 1.515 | 0.194 |
| CAMEL140 | gWrLlKKIleVLKVL-NH2 | 4.136 | 1.203 |
| CAMEL45 | KWKnFKKIGAVLKVL-NH2 | 4.249 | 4.149 |
| CAMEL122 | lWKLFKKIrrVLKVL-NH2 | 6.142 | 5.593 |
| CAMEL9 | KWrLFKnIGAVLKVL-NH2 | 6.292 | 2.917 |
| CAMEL46 | KWKLFKgIrAVLKVL-NH2 | 6.45 | 4.825 |
QSAR model for CAMEL-s antibiotic potency
To quantify the antibacterial potencies of the CAMEL-s from the training sets, for each of them we built ANN consisting of 20 input, 8 hidden and 1 output nodes (as indicated on Figure 3).
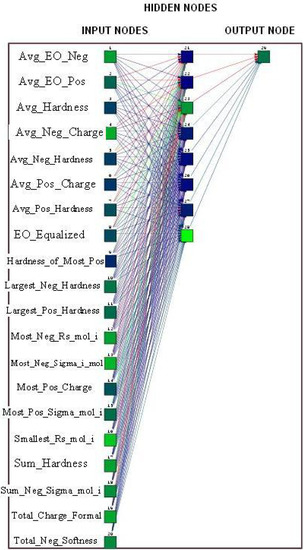
Figure 3.
Configuration of the Artificial Neural Network with 20 input, 8 hidden and 1 output nodes used in the study.
During the learning phase, the normalized values of the selected 20 ‘inductive’ descriptors for each training peptide are fed to the ANN along with the experimental potencies. The relationship between the 20 input parameters and potency is established by recursively correcting the weights attributed to each parameter in such a way that the training potencies converge to the experimental potencies. The training was conducted with the learning rate of 0.8 and the learning update threshold of 0.2, while the input patterns were shuffled, and the initial weights were randomly assigned between 0 and 1. Random noise ranging from -0.002 to 0.002 was added to the ANN inputs to avoid the entrapment of the learning function in a local minimum. The theoretical antibacterial activities of 91 CAMEL-s from the representative training sample estimated during the learning phase are shown in Table 2 and plotted against the experimental numbers in Figure 4.
Figure 4.
Predicted Potencies vs. Experimental Mean Potencies in the Training Set.
As it can be seen from the data, the ‘inductive’ descriptors allowed reproducing the average antibacterial activity of 91 CAMEL-s in the presented set of the training compounds with rather good accuracy. Considering that the examined potencies represent some averaged, not well standardized properties, the resulting QSAR predictions can be viewed as very adequate. To investigate the predictive power of the developed ANN-based solution, and to ensure that no overtraining occurred, we examined the network’s performance on the testing compounds from Table 3. The normalized patterns of the independent variables for 10 CAMEL-s not used for the learning phase were passed through the trained ANN. The pre-estimated node–associated weights of the networks were used to compute the theoretical potencies of the validation compounds. The resulting output parameters collected in Table 3 demonstrate that with the exception of two peptides (CAMEL-s 9 and 140), the predicted CAMEL potencies accurately reproduce the experimental data, thereby validating the QSAR model generated here. It should be noted that similar ANN performance has been observed on all 20 training/testing random pairs of peptide datasets (the results for compounds from Table 2 and Table 3 are actually one of the least accurate among the studied).
To assess the predictive ability of the developed approach in the categorical context (which is more appropriate for such non-standardized data with considerable uncertainty), we have also transformed the continuous outputs from the training and testing procedures into the discrete categorical format. Using the previously outlined conditional classification of the studied polypeptides, the performance of the neural network was assessed by comparing the categorical classification of the outputs from the neural network with the categories corresponding to the experimental potencies. For the cases in which the experimental potency and the predicted potency correspond to the same activity category the prediction of activity was considered to be correct.
Based on this simple assessment, the correct predictions by the developed QSAR model can be considered as 79% accurate for the presented training set (72 out of 91 CAMEL-s were correctly assigned) and 80% accurate for the testing set in Table 3 (8 correct predictions out of 10). The significant deviations of the predicted potencies from the experimental potencies for two validation peptides – CAMEL 9 and CAMEL 140, indicate that the ANN-based approach underestimated the potency of these compounds.
Thus, the computed mean potency of CAMEL9 (2.917) is much lower than the corresponding experimental activity (6.292). This can, perhaps, be attributed to the non-exact character of the potency parameters and the model’s discrepancies. Similarly, for CAMEL140, we predicted its potency to be 1.203 rather than 4.136. In this case, the error could be attributed to the very uncharacteristic composition of this peptide. It contains the negatively charged glutamic acid (E) residue which could not be adequately captured by the neural network which is trained on the set of peptides not containing this structural feature (only one peptide from the training set has E in the sequence). Despite the occasional misclassification, the developed ANN predictor has not missed by more than one class of antibacterial activity or, in other words, it did not place any peptides with high activity to the class of mild therapeutics and vice versa.
The accuracy of the created QSAR model can, possibly, be further improved by pre-processing the data for most adequate training and testing sets selection [48] or by using more powerful machine learning techniques. To summarize the section, it is possible to conclude, that the developed QSAR model operating by the ‘inductive’ descriptors and utilizing the ANN algorithm can accurately quantify the antibacterial potency of the studies synthetic cationic polypeptides and can effectively place them into groups of active, moderate and mild anti-infective compounds.
Conclusions and Further Directions
The evolution of bacterial strains into multi-drug resistant organisms progresses at an alarming rate. For this reason, it is crucial to discover novel non-specific antibiotics (such as cationic polypeptides) that are active against a number of different strains of microbes, including the resistant ones. The role of QSAR models for the antimicrobial polypeptides cannot be overestimated as such predictive solutions can significantly rationalize the selection, design and refinement efforts for these drugs. The developed QSAR approach utilizing the ‘inductive’ descriptors and based on the Artificial Neural Network algorithm can be used for these purposes and can be further expanded to cover a wider range of cationic peptides active against pathogens.
The approach can also be enhanced by utilizing purely statistical techniques in conjunction with the inductive QSAR descriptors which allows interpreting contributions from individual structural factors to the potency of the AMP-s. Despite the fact that the developed ANN-based method does not currently allow us to exactly evaluate the contributions of the individual QSAR descriptors, it is clear that the employed ‘inductive’ parameters adequately reflect those aspects of intra- and intermolecular interactions which govern antibacterial activity of the cationic polypeptides.
Hence, the developed methodology can further be applied to other important classes of cationic peptides, such as those active against viruses, fungi or tumours, and can provide excellent computational guidance for discovery of novel and potent therapeutic leads.
Acknowledgements
The authors thank Dr. David Wade for the CAMEL-s activity data and for useful suggestion in preparation of the manuscript.
References
- Jaen-Oltra, J.; Salabert-Salvador, M.T.; Garcia-March, F.J.; Perez-Giminez, F.; Tomas-Vert, F. Artificial Neural Network Applied to Prediction of Fluorquinolone Antibacterial Activity by Topological Methods. J. Med. Chem. 2000, 43, 1143–1148. [Google Scholar]
- Garcia-Domenech, R.; de Julian-Ortiz, J.V. Antimicrobial Activity Characterization in a Heterogeneous Group of Compounds. J. Chem. Inf. Comp. Sci. 1998, 38, 445–449. [Google Scholar]
- Tomas-Vert, F.; Perez-Gimenez, F.; Salabert-Salvador, M.T.; Garcia-March, F.J.; Jaen-Oltra, J. Atrificial Neural Network Applied to the Discrimination of Antibacterial Activity by Topological Methods. J. Molec. Struct. (Theochem). 2000, 504, 249–259. [Google Scholar]
- Mishra, R.K.; Garcia-Domenech, R.; Galvez, J. Getting Discriminant Function of Antibacterial Activity from Physicochemical and Topological Parameters. J. Chem. Inf. Comp. Sci. 2001, 41, 387–393. [Google Scholar]
- Cronin, M.T.D.; Aptula, A.O.; Dearden, J.C.; Duffy, J.C.; Netzeva, T.I.; Patel, H.; Rowe, P.H.; Schultz, T.W.; Worth, A.P.; Voutzoulidis, K.; Schuurmann, G. Structure-Based Classification of Antibacterial Activity. J. Chem. Inf. Comp. Sci. 2002, 42, 869–878. [Google Scholar]
- Gozalbez, R.; Galvez, J.; Moreno, A.; Garcia-Domenech, R. Discovery of New Antimalarial Compounds by use of Molecular Connectivity Techniques. J. Pharm. Pharmacol. 1999, 51, 111–117. [Google Scholar]
- Molina, E.; Diaz, H.G.; Gonzalez, M.P.; Rodriguez, E.; Uriarte, E. Designing Antibacterial Compounds through a Topological Substructural Approach. J. Chem. Inf. Comp. Sci. 2004, 44, 515–521. [Google Scholar]
- Sima, P.; Trebichavsky, I.; Sigler, K. Mammalian antibiotic peptides. Folia Microbiol. 2003, 48, 123–137. [Google Scholar]
- Miele, R.; Bjorklund, G.; Barra, D.; Simmaco, M.; Engstrom, Y. Involvement of Rel factors in the expression of antimicrobial peptide genes in amphibian. Eur. J. Biochem. 2001, 268, 443–449. [Google Scholar]
- Simmaco, M.; Mignogna, G.; Barra, D. Antimicrobial peptides from amphibian skin: what do they tell us? Biopolymers 1998, 47, 435–450. [Google Scholar]
- Khush, R.S.; Leulier, F.; Lemaitre, B. Drosophila immunity: two paths to NF-κB. Trends Immunol. 2001, 22, 260–264. [Google Scholar]
- Hancock, R.E. Concerns regarding resistance to self-proteins. Microbiology. 2003, 149, 3343–3344. [Google Scholar]
- Marshall, S.H.; Arenas, G. Antimicrobial peptides: A natural alternative to chemical antibiotics and a potential for applied biotechnology. Electron. J. Biotechno. 2003, 6, 271–284. [Google Scholar]
- Hancock, R.E.; Chapple, D.S. Peptide Antibiotics. Antimicrob. Agents Chemother. 1999, 43, 1317–1323. [Google Scholar]
- Cherkasov, A.R.; Galkin, V.I.; Cherkasov, R.A. A New Approach to the Theoretical Estimation of Inductive Constants. J. Phys. Org. Chem. 1998, 11, 437–447. [Google Scholar]
- Cherkasov, A.R.; Galkin, V.I.; Cherkasov, R.A. "Inductive" Electronegativity Scale. J. Molec. Struct. (Theochem) 1999, 489, 43–46. [Google Scholar]
- Cherkasov, A.R.; Galkin, V.I.; Cherkasov, R.A. "Inductive" Electronegativity Scale. 2. "Inductive" Analog of Chemical Hardness. J. Molec. Struct. (Theochem) 2000, 497, 115–123. [Google Scholar]
- Cherkasov, A. Inductive Electronegativity Scale. Iterative Calculation of Inductive Partial Charges. J. Chem. Inf. Comp. Sci. 2003, 43, 2039–2047. [Google Scholar]
- Cherkasov, A.; Sprous, D.; Chen, R. 3D Correlation Analysis - New Method of Quantification of Substituent Effect. J. Phys. Chem. A. 2003, 107, 9695–9704. [Google Scholar]
- Cherkasov, A.; Jonsson, M. Substituent Effect on Thermo-chemical Properties of Free Radicals. New Substituent Scales for C-Centred Radicals. J. Chem. Inf. Comp. Sci. 1998, 38, 1151–1156. [Google Scholar]
- Cherkasov, A.; Jonsson, M. Substituent Effect on Thermochemical Properties of Free Radicals. Physical Interpretation of the Substituent Effect. J. Chem. Inf. Comp. Sci. 1999, 39, 1057–1063. [Google Scholar]
- Cherkasov, A.R.; Jonsson, M.; Galkin, V.I. A Novel Approach to the Analysis of Substituent Effects. Quantitative Interpretation of Ionization Potentials and Gas Basicity of Amines. J. Mol. Graph. Model. 1999, 17, 28–43. [Google Scholar]
- Cherkasov, A.; Jonsson, M. Estimation of Homolytic CH Bond Dissociation Enthalpies. J. Chem. Inf. Comp. Sci. 2000, 40, 1222–1226. [Google Scholar]
- Galkin, V.I.; Cherkasov, A.R.; Cherkasov, R.A. Modelling of Substituents Electronic and Steric Effects. Effective Analysis of Organoelement and Organophosphorus Reactivity. Phosphorus, Silicon, Sulphur. 1999, 146, 329–332. [Google Scholar]
- Cherkasov, A. Inductive QSAR Descriptors. 1. Distinguishing Compounds with Antibacterial Activity by Artificial Neural Networks. Submitted. J. Chem. Inf. Comp. Sci. 2004, 44. [Google Scholar]
- Mee, R.P.; Auton, T.R.; Morgan, P.J. Design of active analogs of a 15-residue peptide using D-optimal design, QSAR and a combinatorial search algorithm. J. Pept. Res. 1997, 49, 89–102. [Google Scholar]
- Edlund, C.; Hedberg, M.; Engström, Å.; Flock, J.-I.; Wade, D. Anti-anaerobic activity of a cecropin-melittin peptide. Clin. Microbiol. Infection 1998, 4, 181–185. [Google Scholar]
- Oh, H.; Hedberg, M.; Wade, D.; Edlund, C. Activities of synthetic hybrid peptides against bacteria: aspects of methodology and stability. Antimicrob. Agents Chemother. 2000, 44, 68–72. [Google Scholar]
- Oren, Z.; Shai, Y. Mode of action of linear amphipathic α-helical antimicrobial peptides. Biopolymers 1999, 47, 451–463. [Google Scholar]
- Bechinger, B. The structure, dynamics and orientation of antimicrobial peptides in membranes by multidimensional solid-state NMR spectroscopy. Biochim. Biophys. Acta 1999, 1462, 157–183. [Google Scholar]
- Blondelle, S.E.; Lohner, K.; Aguilar, M.I. Lipid-induced conformation and lipid-binding properties of cytolytic and antimicrobial peptides: determination and biological specificity. Biochim. Biophys. Acta 1999, 1462, 89–108. [Google Scholar]
- Epand, R.F.; Umezawa, N.; Porter, E.A.; Gellman, S.H.; Epand, R.M. Interactions of the antimicrobial β-peptide β-17 with phospholipids vesicles differ from membrane interactions of magainins. Eur. J. Biochem. 2003, 270, 1240–1248. [Google Scholar]
- Hancock, R.E.; Lehrer, R. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 1998, 16, 82–88. [Google Scholar]
- Shai, Y. Mechanism of the binding, insertion and destabilization of phospholipids bilayer membranes by α-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta 1999, 1462, 55–70. [Google Scholar]
- Takeshima, K.; Chikushi, A.; Lee, K.K.; Yonehara, S.; Matsuzaki, K. Translocation of analogues of the antimicrobial peptides magainin and buforin across human cell membranes. J. Biol. Chem. 2003, 278, 1310–1315. [Google Scholar]
- Vogt, T.C.B.; Bechinger, B. The interactions of histidine-containing amphipathic helical peptide antibiotics with lipid bilayers. J Biol. Chem. 1999, 274, 29115–29121. [Google Scholar]
- Baker, M.A.; Maloy, W.L.; Zasloff, M.; Jacob, L.S. Anticancer efficacy of Magainin2 and analogue peptides. Cancer Res. 1993, 53, 3052–3057. [Google Scholar]
- Dathe, M.; Schumann, M.; Wieprecht, T. Peptide helicity and membrane surface charge modulate the balance of electrostatic and hydrophobic interactions with lipid bilayers and biological membranes. Biochemistry 1996, 35, 12612–12622. [Google Scholar]
- Dathe, M.; Wieprecht, T. Structural features of helical antimicrobial peptides: their potential to modulate activity on model membranes and biological cells. Biochim. Biophys. Acta 1999, 1462, 71–87. [Google Scholar]
- Epand, R.M.; Vogel, H.J. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta 1999, 1462, 11–28. [Google Scholar]
- Hwang, P.M.; Vogel, H.J. Structure-function relationships of antimicrobial peptides. Biochem. Cell Biol. 1998, 76, 235–246. [Google Scholar]
- Peck-Miller, K.A.; Blake, J.; Cosand, W.L.; Darveau, R.P.; Fell, H.P. Structure-activity analysis of the antitumor and hemolytic properties of the amphiphilic alpha-helical peptide, C18G. Int. J. Pept. Prot. Res. 1994, 44, 143–151. [Google Scholar]
- Prenner, E.J.; Lewis, R.N.A.H.; McElhaney, R.N. The interaction of the antimicrobial peptide gramicidin S with lipid bilayer model and biological membranes. Biochim. Biophys. Acta 1999, 1462, 201–221. [Google Scholar]
- Shin, S.Y.; Lee, S.H.; Yang, S.T.; Park, E.J.; Lee, D.G.; Lee, M.K.; Eom, S.H.; Song, W.K.; Kim, Y.; Hahm, K.S.; Kim, J.I. Antibacterial, antitumor and hemolytic activities of alpha-helical antibiotic peptide, P18 and its analogs. J. Pept. Res. 2001, 58, 504–514. [Google Scholar]
- Chemical Computation Group Inc. Montreal (Canada). Molecular Operational Environment 2004.
- Zupan, J.; Gasteiger, J. Neural Networks in Chemistry and Drug Design; 2nd Ed.; Wiley: New York, 1999. [Google Scholar]
- SNNS: Stuttgart Neural Network Simulator; Version 4.0; University of Stuttgart: Stuttgart (Germany), 1995.
- Hawkins, D.; Basak, S.; Mills, D. Assessing Model Fit by Cross Validation. J. Chem. Inf. Comp. Sci. 2003, 43, 579–586. [Google Scholar]
© 2004 by MDPI (http://www.mdpi.org). Reproduction is permitted for noncommercial purposes.