Design, Synthesis and Biological Evaluation of 3-Hydrazonoindolin-2-one Derivatives as Novel HIV-1 RNase H Inhibitors
Abstract
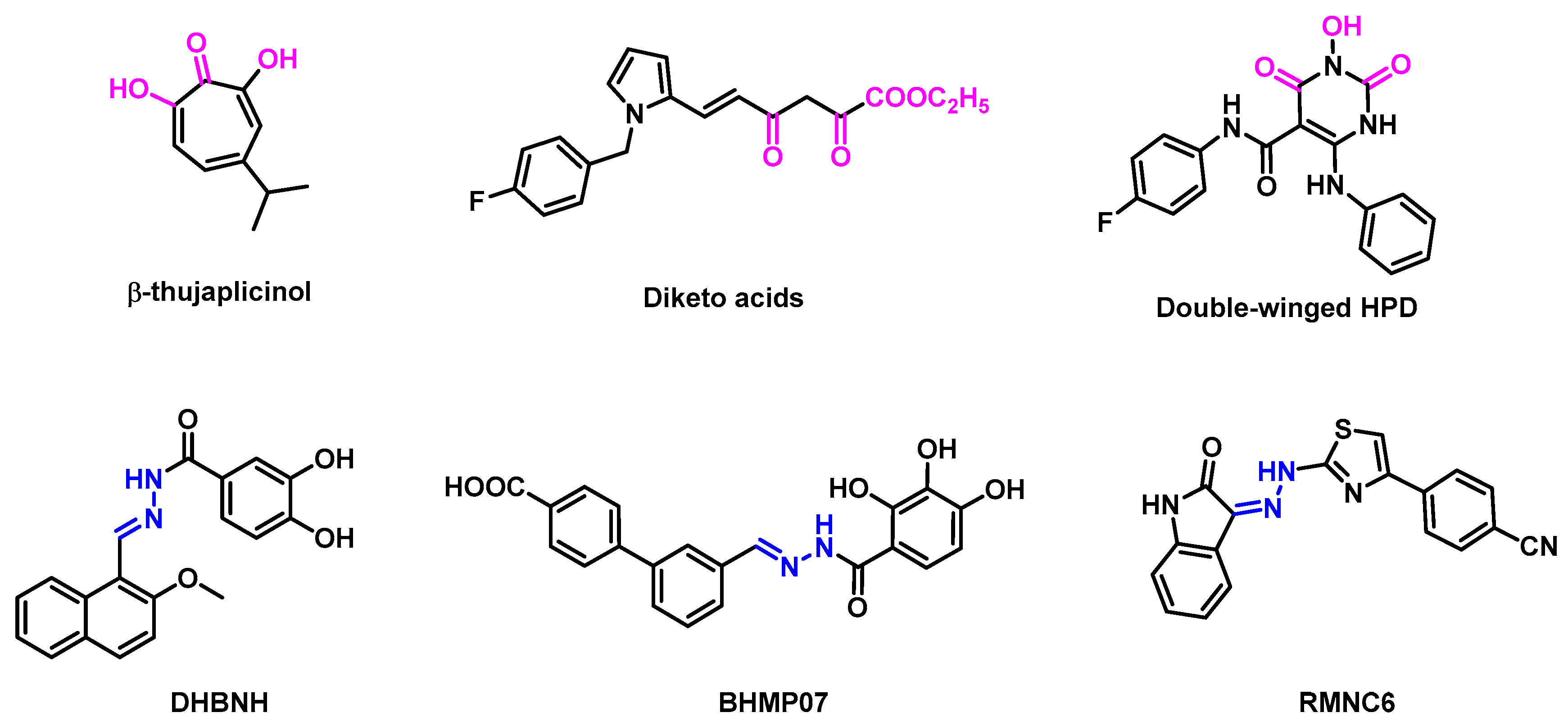
1. Introduction
2. Results and Discussion
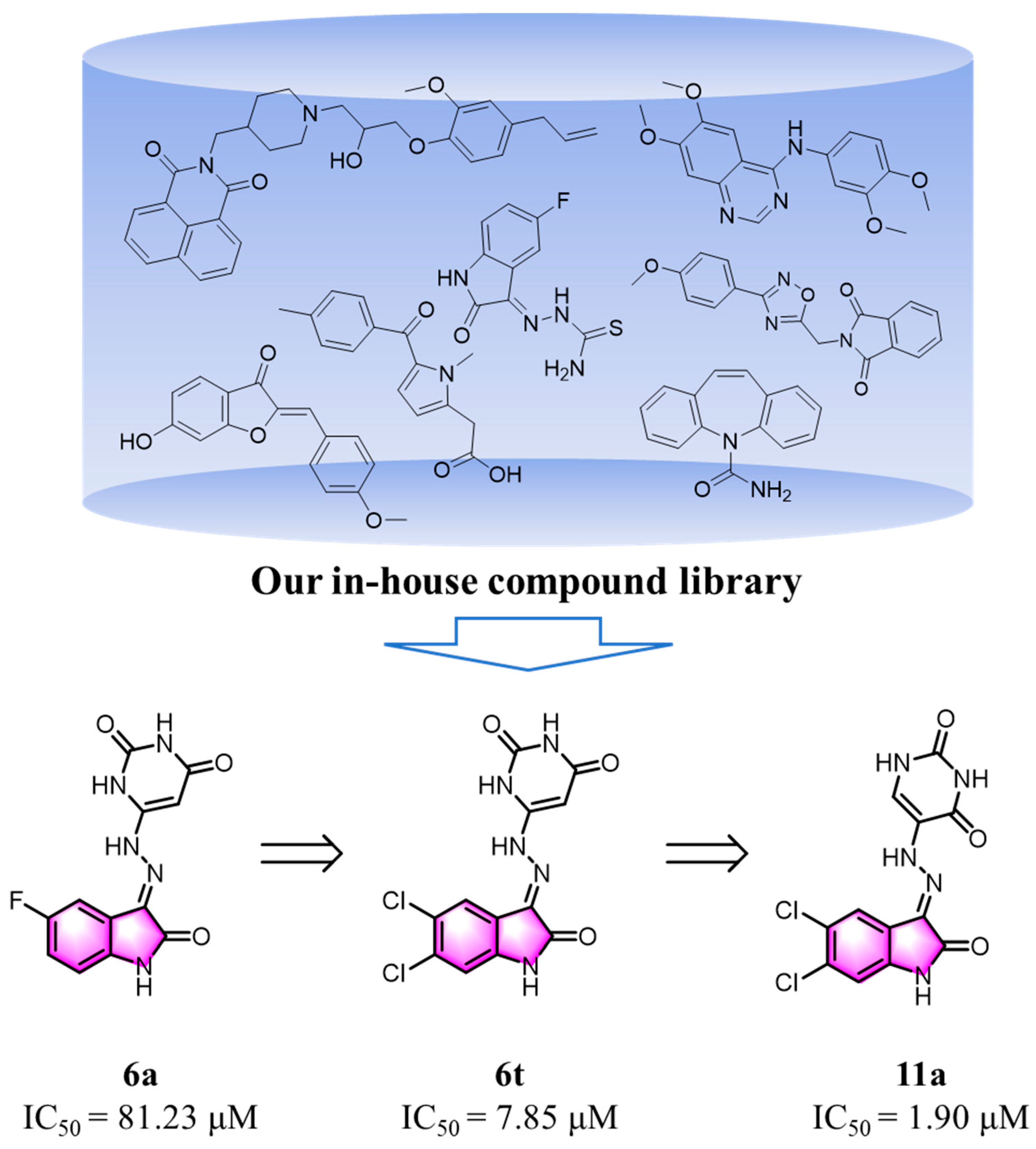
2.1. Design of New Compounds
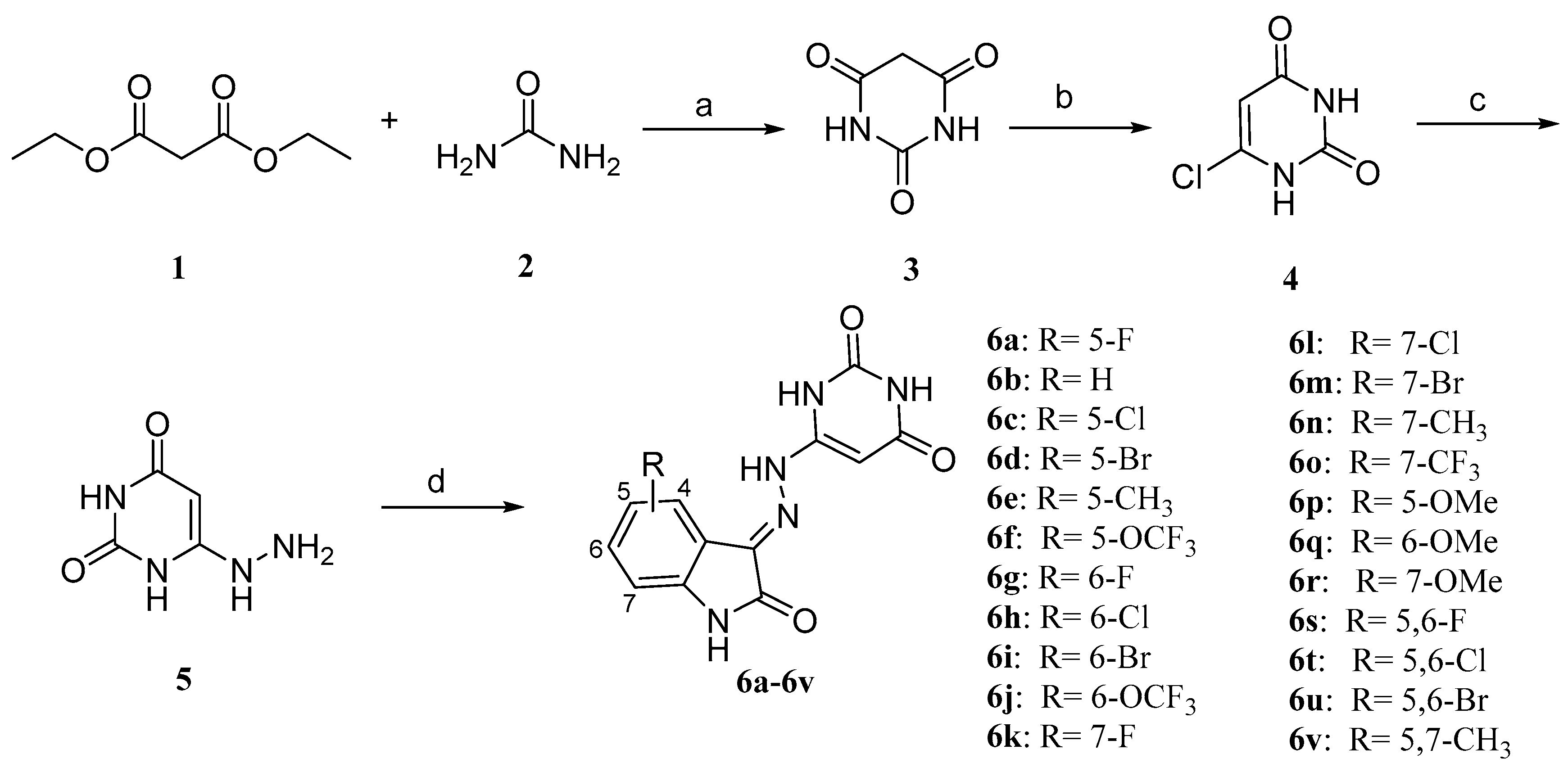
2.2. Chemistry
2.3. Evaluation of Biological Activities
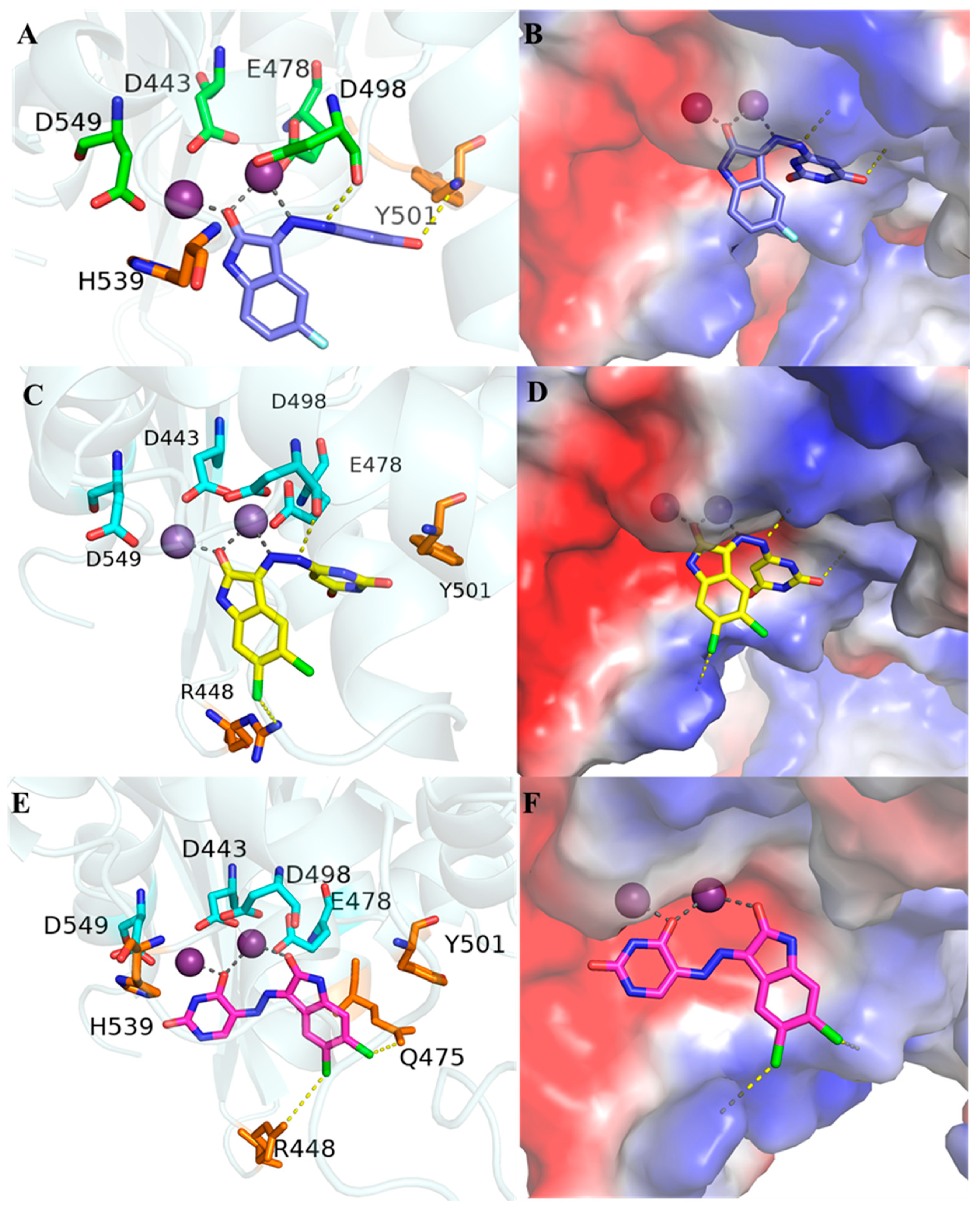
2.4. Molecular Modeling
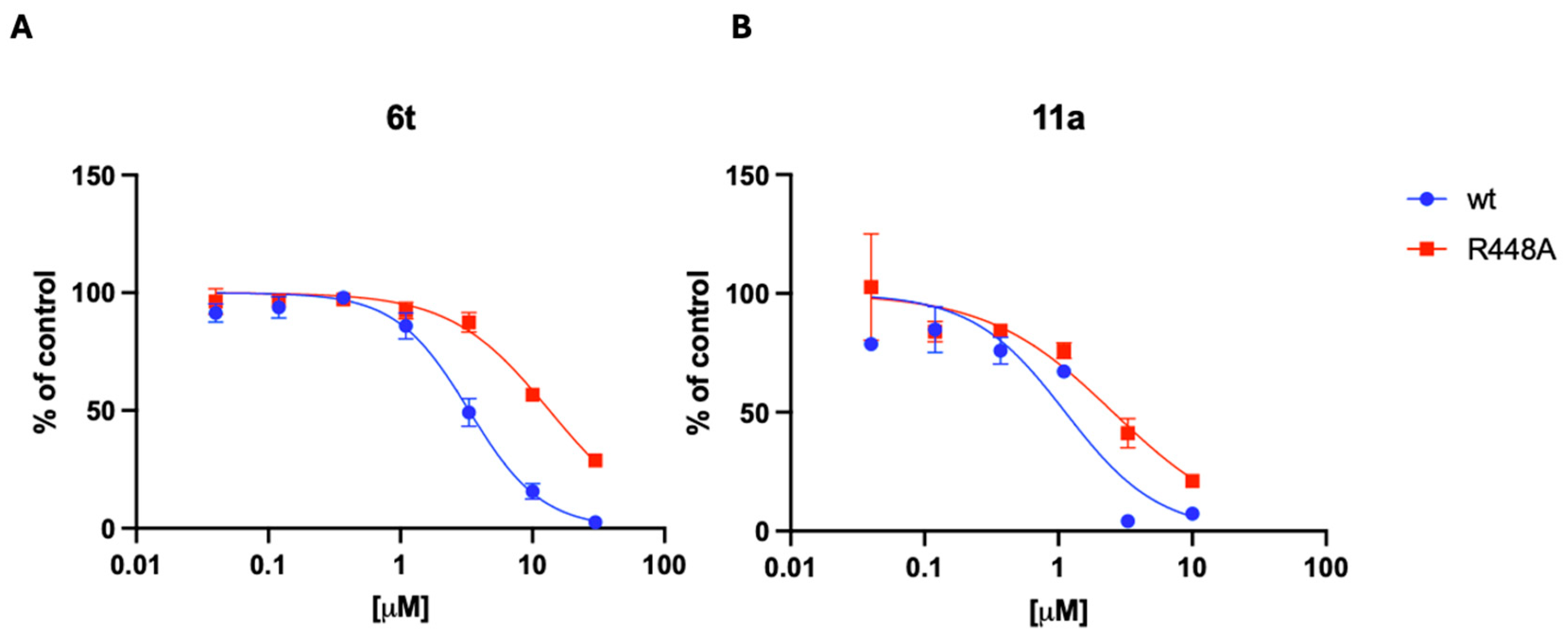
2.5. Evaluation of the Mode of Action
3. Conclusions
4. Experiment
4.1. Chemistry
4.1.1. General Procedure for Preparation of Compound 3
4.1.2. General Procedure for Preparation of Compound 4
4.1.3. General Procedure for Preparation of Compound 5
4.1.4. General Procedure for Preparation of Compounds 6a–6v and 11a–11b
4.1.5. General Procedure for Preparation of Compound 8
4.1.6. General Procedure for Preparation of Compound 9
4.1.7. General Procedure for Preparation of Compound 10
4.2. Biology
4.2.1. Expression and Purification of Recombinant HIV-1 RT
4.2.2. Determination of HIV-1 RNase H Activity
4.2.3. In Vitro Anti-HIV Assay
4.2.4. Determination of Inhibitors Kinetic Parameters
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Broder, S.; Gallo, R.C. A Pathogenic Retrovirus (HTLV-III) Linked to AIDS. N. Engl. J. Med. 1984, 311, 1292–1297. [Google Scholar] [CrossRef] [PubMed]
- Barré-Sinoussi, F.; Chermann, J.C.; Rey, F.; Nugeyre, M.T.; Chamaret, S.; Gruest, J.; Dauguet, C.; Axler-Blin, C.; Vézinet-Brun, F.; Rouzioux, C.; et al. Isolation of a T-Lymphotropic Retrovirus from a Patient at Risk for Acquired Immune Deficiency Syndrome (AIDS). Science 1983, 220, 868–871. [Google Scholar] [CrossRef] [PubMed]
- WHO. HIV and AIDS. Available online: https://www.who.int/news-room/fact-sheets/detail/hiv-aids (accessed on 12 December 2024).
- Carr, A.; Cooper, D.A. Adverse effects of antiretroviral therapy. Lancet 2000, 356, 1423–1430. [Google Scholar] [CrossRef] [PubMed]
- Chun, T.W.; Stuyver, L.; Mizell, S.B.; Ehler, L.A.; Mican, J.A.M.; Baseler, M.; Lloyd, A.L.; Nowak, M.A.; Fauci, A.S. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviraltherapy. Proc. Natl. Acad. Sci. USA 1997, 94, 13193–13197. [Google Scholar] [CrossRef]
- Wang, X.; Gao, P.; Menendez-Arias, L.; Liu, X.; Zhan, P. Update on Recent Developments in Small Molecular HIV-1 RNase H Inhibitors (2013–2016): Opportunities and Challenges. Curr. Med. Chem. 2018, 25, 1682–1702. [Google Scholar] [CrossRef]
- Tian, L.; Kim, M.S.; Li, H.; Wang, J.; Yang, W. Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid. Proc. Natl. Acad. Sci. USA 2018, 115, 507–512. [Google Scholar] [CrossRef]
- Hyjek, M.; Figiel, M.; Nowotny, M. RNases H: Structure and mechanism. DNA Repair 2019, 84, 102672. [Google Scholar] [CrossRef]
- Sarafianos, S.G.; Marchand, B.; Das, K.; Himmel, D.M.; Parniak, M.A.; Hughes, S.H.; Arnold, E. Structure and Function of HIV-1 Reverse Transcriptase: Molecular Mechanisms of Polymerization and Inhibition. J. Mol. Biol. 2009, 385, 693–713. [Google Scholar] [CrossRef]
- Kankanala, J.; Kirby, K.A.; Liu, F.; Miller, L.; Nagy, E.; Wilson, D.J.; Parniak, M.A.; Sarafianos, S.G.; Wang, Z. Design, Synthesis, and Biological Evaluations of Hydroxypyridonecarboxylic Acids as Inhibitors of HIV Reverse Transcriptase Associated RNase, H. J. Med. Chem. 2016, 59, 5051–5062. [Google Scholar] [CrossRef]
- Paredes, R.; Lalama, C.M.; Ribaudo, H.J.; Schackman, B.R.; Shikuma, C.; Giduel, F.; Meyer, W.A.; Johnson, V.A.; Fiscus, S.A.; Aqulia, R.T.D.; et al. Pre-existing Minority Drug-Resistant HIV-1 Variants, Adherence, and Risk of Antiretroviral Treatment Failure. J. Infect. Dis. 2010, 201, 662–671. [Google Scholar] [CrossRef]
- Wang, L.; Tang, J.; Huber, A.D.; Casey, M.C.; Kirby, K.A.; Wilson, D.J.; Kankanala, J.; Xie, J.; Parniak, M.A.; Sarafianos, S.G.; et al. 6-Arylthio-3-hydroxypyrimidine-2,4-diones potently inhibited HIV reverse transcriptase-associated RNase H with antiviral activity. Eur. J. Med. Chem. 2018, 156, 652–665. [Google Scholar] [CrossRef] [PubMed]
- Tramontano, E.; Corona, A.; Menéndez-Arias, L. Ribonuclease H, an unexploited target for antiviral intervention against HIV and hepatitis B virus. Antivir. Res. 2019, 171, 104613. [Google Scholar] [CrossRef] [PubMed]
- Di Santo, R. Inhibiting the HIV Integration Process: Past, Present, and the Future. J. Med. Chem. 2014, 57, 539–566. [Google Scholar] [CrossRef]
- Nowotny, M.; Gaidamakov, S.A.; Ghirlando, R.; Cerritelli, S.M.; Crouch, R.J.; Yang, W. Structure of Human RNase H1 Complexed with an RNA/DNA Hybrid: Insight into HIV Reverse Transcription. Mol. Cell 2007, 28, 264–276. [Google Scholar] [CrossRef]
- Nowotny, M.; Gaidamakov, S.A.; Crouch, R.J.; Yang, W. Crystal Structures of RNase H Bound to an RNA/DNA Hybrid: Substrate Specificity and Metal-Dependent Catalysis. Cell 2005, 121, 1005–1016. [Google Scholar] [CrossRef]
- Beilhartz, G.L.; Ngure, M.; Johns, B.A.; DeAnda, F.; Gerondelis, P.; Götte, M. Inhibition of the Ribonuclease H Activity of HIV-1 Reverse Transcriptase by GSK5750 Correlates with Slow Enzyme-Inhibitor Dissociation. J. Biol. Chem. 2014, 289, 16270–16277. [Google Scholar] [CrossRef]
- Vernekar, S.K.V.; Tang, J.; Wu, B.; Huber, A.D.; Casey, M.C.; Myshakina, N.; Wilson, D.J.; Kankanala, J.; Kirby, K.A.; Parniak, M.A.; et al. Double-Winged 3-Hydroxypyrimidine-2,4-diones: Potent and Selective Inhibition against HIV-1 RNase H with Significant Antiviral Activity. J. Med. Chem. 2017, 60, 5045–5056. [Google Scholar] [CrossRef]
- Himmel, D.M.; Sarafianos, S.G.; Dharmasena, S.; Hossain, M.M.; McCoy-Simandle, K.; Ilina, T.; Clark, D., Jr.; Knight, J.L.; Julias, J.G.; Clark, P.K.; et al. HIV-1 Reverse Transcriptase Structure with RNase H Inhibitor Dihydroxy Benzoyl Naphthyl Hydrazone Bound at a Novel Site. ACS Chem. Biol. 2006, 1, 702–712. [Google Scholar] [CrossRef]
- Corona, A.; Meleddu, R.; Esposito, F.; Distinto, S.; Bianco, G.; Masaoka, T.; Maccioni, E.; Menéndez-Arias, L.; Alcaro, S.; Le Grice, S.F.J.; et al. Ribonuclease H/DNA Polymerase HIV-1 Reverse Transcriptase Dual Inhibitor: Mechanistic Studies on the Allosteric Mode of Action of Isatin-Based Compound RMNC6. PLoS ONE 2016, 11, e0147225. [Google Scholar] [CrossRef]
- White, J.D.; Hansen, J.D. Asymmetric Synthesis of Epicylindrospermopsin via Intramolecular Nitrone Cycloaddition. Assignment of Absolute Configuration. J. Am. Chem. Soc. 2002, 124, 4950–4951. [Google Scholar] [CrossRef]
- Zhao, K.X.; Zhang, Y.Y.; Wang, J.S.; Wang, S.; Corona, A.; Maloccu, S.; Tramontano, E.; Pannecouque, C.; Clercq, E.D.; Meng, G.; et al. Design, synthesis and biological evaluation of Thiazolo [3,2-a]Pyrimidine derivatives as novel RNase H inhibitors. Bioorganic Chem. 2024, 148, 107495. [Google Scholar] [CrossRef] [PubMed]
- Felts, A.K.; LaBarge, K.; Bauman, J.D.; Patel, D.V.; Himmel, D.M.; Arnold, E.; Parniak, M.A.; Levy, R.M. Identification of Alternative Binding Sites for Inhibitors of HIV-1 Ribonuclease H Through Comparative Analysis of Virtual Enrichment Studies. J. Chem. Inf. Model 2011, 51, 1986–1998. [Google Scholar] [CrossRef] [PubMed]
- Sarafianos, S.G.; Das, K.; Tantillo, C.; Clark, A.D., Jr.; Ding, J.; Whitcomb, J.M.; Boyer, P.L.; Hughes, S.H.; Arnold, E. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 2001, 20, 1449–1461. [Google Scholar] [CrossRef] [PubMed]
- Gong, Q.; Menon, L.; Ilina, T.; Miller, L.G.; Ahn, J.; Parniak, M.A.; Ishima, R. Interaction of HIV-1 Reverse Transcriptase Ribonuclease H with an Acylhydrazone Inhibitor. Chem. Biol. Drug Des. 2011, 77, 39–47. [Google Scholar] [CrossRef]
- Christen, M.T.; Menon, L.; Myshakina, N.S.; Ahn, J.; Parniak, M.A.; Ishima, R. Structural Basis of the Allosteric Inhibitor Interaction on the HIV-1 Reverse Transcriptase RNase H Domain. Chem. Biol. Drug Des. 2012, 80, 706–716. [Google Scholar] [CrossRef]
- Lansdon, E.B.; Liu, Q.; Leavitt, S.A.; Balakrishnan, M.; Perry, J.K.; Lancaster-Moyer, C.; Kutty, N.; Liu, X.; Squires, N.H.; Watkins, W.J.; et al. Structural and Binding Analysis of Pyrimidinol Carboxylic Acid and N-Hydroxy Quinazolinedione HIV-1 RNase H Inhibitors. Antimicrob. Agents Chemother. 2011, 55, 2905–2915. [Google Scholar] [CrossRef]
- Sail, B.S.; Naik, V.H.; Kamli, M.R.; Prasanna, B.M. Synthesis, spectral, in vitro microbial and DNA cleavage studies of isatin bishydrozone metal complexes. J. Mol. Struct. 2023, 1277, 134837. [Google Scholar] [CrossRef]
- Rodríguez-Argüelles, M.C.; Ferrari, M.B.; Bisceglie, F.; Pelizzi, C.; Pelosi, G.; Pinelli, S.; Sassi, M. Synthesis, characterization and biological activity of Ni, Cu and Zn complexes of isatin hydrazones. J. Inorg. Biochem. 2004, 98, 313–321. [Google Scholar] [CrossRef]
- Mohanan, K.; Murukan, B. Complexes of Manganese(II), Iron(II), Cobalt(II), Nickel(II), Copper(II), and Zinc(II) with a Bishydrazone. Synth. React. Inorg. Met.-Org. Nano.-Met. Chem. 2005, 35, 837–844. [Google Scholar] [CrossRef]
- Rodríguez-Argüelles, M.C.; Cao, R.; García-Deibe, A.M.; Pelizzi, C.; Sanmartín-Matalobos, J.; Zani, F. Antibacterial and antifungal activity of metal(II) complexes of acylhydrazones of 3-isatin and 3-(N-methyl)isatin. Polyhedron 2009, 28, 2187–2195. [Google Scholar] [CrossRef]
- Zhu, X.D.; Corona, A.; Maloccu, S.; Tramontano, E.; Wang, S.; Pannecouque, C.; Clercq, E.D.; Meng, G.; Chen, F.E. Structure-Based Design of Novel Thiazolone[3,2-a]pyrimidine Derivatives as Potent RNase H Inhibitors for HIV Therapy. Molecules 2024, 29, 738–754. [Google Scholar] [CrossRef] [PubMed]
- Corona, A.; Ballana, E.; Distinto, S.; Rogolino, D.; Del Vecchio, C.; Carcelli, M.; Badia, R.; Riveira-Munoz, E.; Esposito, F.; Parolin, C.; et al. Targeting HIV-1 RNase H: N’-(2-Hydroxy-benzylidene)-3,4,5-Trihydroxybenzoylhydrazone as Selective Inhibitor Active against NNRTIs-Resistant Variants. Viruses 2020, 12, 729. [Google Scholar] [CrossRef] [PubMed]
- Rausch, J.W.; Lener, D.; Miller, J.T.; Julias, J.G.; Hughes, S.H.; Le Grice, S.F.J. Altering the RNase H Primer Grip of Human Immunodeficiency Virus Reverse Transcriptase Modifies Cleavage Specificity. Biochemistry 2002, 41, 4856–4865. [Google Scholar] [CrossRef] [PubMed]
- Chiodi, D.; Ishihara, Y. “Magic Chloro”: Profound Effects of the Chlorine Atom in Drug Discovery. J. Med. Chem. 2023, 66, 5305–5331. [Google Scholar] [CrossRef]
- Youssef, H.M.; Abdulhamed, Y.K.; El-Reash, G.M.A.; Yousef, T.A. Cr(III) and Ni(II) complexes of isatin-hydrazone ligand: Preparation, characterization, DFT studies, biological activity, and ion-flotation separation of Ni(II). Inorg. Chem. Commun. 2022, 138, 109278. [Google Scholar] [CrossRef]



 | |||
|---|---|---|---|
| Compd | IC50 [μM] a | Compd | IC50 [μM] a |
| 6a | 81.23 ± 11.36 | 6m | 43.78 ± 7.92 |
| 6b | >100 | 6n | 25.49 ± 1.37 |
| 6c | 58.76 ± 2.40 | 6o | 24.06 ± 7.02 |
| 6d | >100 | 6p | >100 |
| 6e | >100 | 6q | >100 |
| 6f | 8.94 ± 1.43 | 6r | >100 |
| 6g | 56.84 ± 15.90 | 6s | 64.90 ± 0.77 |
| 6h | >100 | 6t | 7.85 ± 0.55 |
| 6i | 23.59 ± 1.11 | 6u | 11.83 ± 0.81 |
| 6j | 15.28 ± 0.01 | 6v | 23.58 ± 3.89 |
| 6k | >100 | 11a | 1.90 ± 0.01 |
| 6l | 46.65 ± 3.25 | 11b | 2.20 ± 0.20 |
| Compd | HIV-1(IIIB) EC50 (μM) a | CC50 (μM) b | SI c |
|---|---|---|---|
| 6t | 109.71 | 294.37 | 3 |
| 11a | 38.97 | 402.2 | 10 |
| 11b | 44.86 | 348.93 | 8 |
| NVP | 0.19 | >15.02 | >80 |
| EFV | 0.0030 | >4.59 | >1538 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Wang, R.; Bu, Y.; Corona, A.; Dettori, L.; Tramontano, E.; Pannecouque, C.; De Clercq, E.; Wang, S.; Meng, G.; et al. Design, Synthesis and Biological Evaluation of 3-Hydrazonoindolin-2-one Derivatives as Novel HIV-1 RNase H Inhibitors. Molecules 2025, 30, 1868. https://doi.org/10.3390/molecules30091868
Zhang Y, Wang R, Bu Y, Corona A, Dettori L, Tramontano E, Pannecouque C, De Clercq E, Wang S, Meng G, et al. Design, Synthesis and Biological Evaluation of 3-Hydrazonoindolin-2-one Derivatives as Novel HIV-1 RNase H Inhibitors. Molecules. 2025; 30(9):1868. https://doi.org/10.3390/molecules30091868
Chicago/Turabian StyleZhang, Yiying, Rao Wang, Yueyue Bu, Angela Corona, Laura Dettori, Enzo Tramontano, Christophe Pannecouque, Erik De Clercq, Shuai Wang, Ge Meng, and et al. 2025. "Design, Synthesis and Biological Evaluation of 3-Hydrazonoindolin-2-one Derivatives as Novel HIV-1 RNase H Inhibitors" Molecules 30, no. 9: 1868. https://doi.org/10.3390/molecules30091868
APA StyleZhang, Y., Wang, R., Bu, Y., Corona, A., Dettori, L., Tramontano, E., Pannecouque, C., De Clercq, E., Wang, S., Meng, G., & Chen, F.-E. (2025). Design, Synthesis and Biological Evaluation of 3-Hydrazonoindolin-2-one Derivatives as Novel HIV-1 RNase H Inhibitors. Molecules, 30(9), 1868. https://doi.org/10.3390/molecules30091868











