Predicting the Toxicity of Drug Molecules with Selecting Effective Descriptors Using a Binary Ant Colony Optimization (BACO) Feature Selection Approach
Abstract
1. Introduction
2. Results and Discussion
2.1. Comparison Between Initially Screened Features and High-Frequency Features
2.2. Comparison Between BACO and Several Benchmark Feature Selection Approaches
2.3. Impact of the Parameter on the Performance of BACO
2.4. Impact of Adopting Different Basic Classifiers in BACO
2.5. Detecting the Generalizability and Applicability of BACO
2.6. Discussion and Further Suggestions
3. Materials and Methods
3.1. Datasets and Their Representations
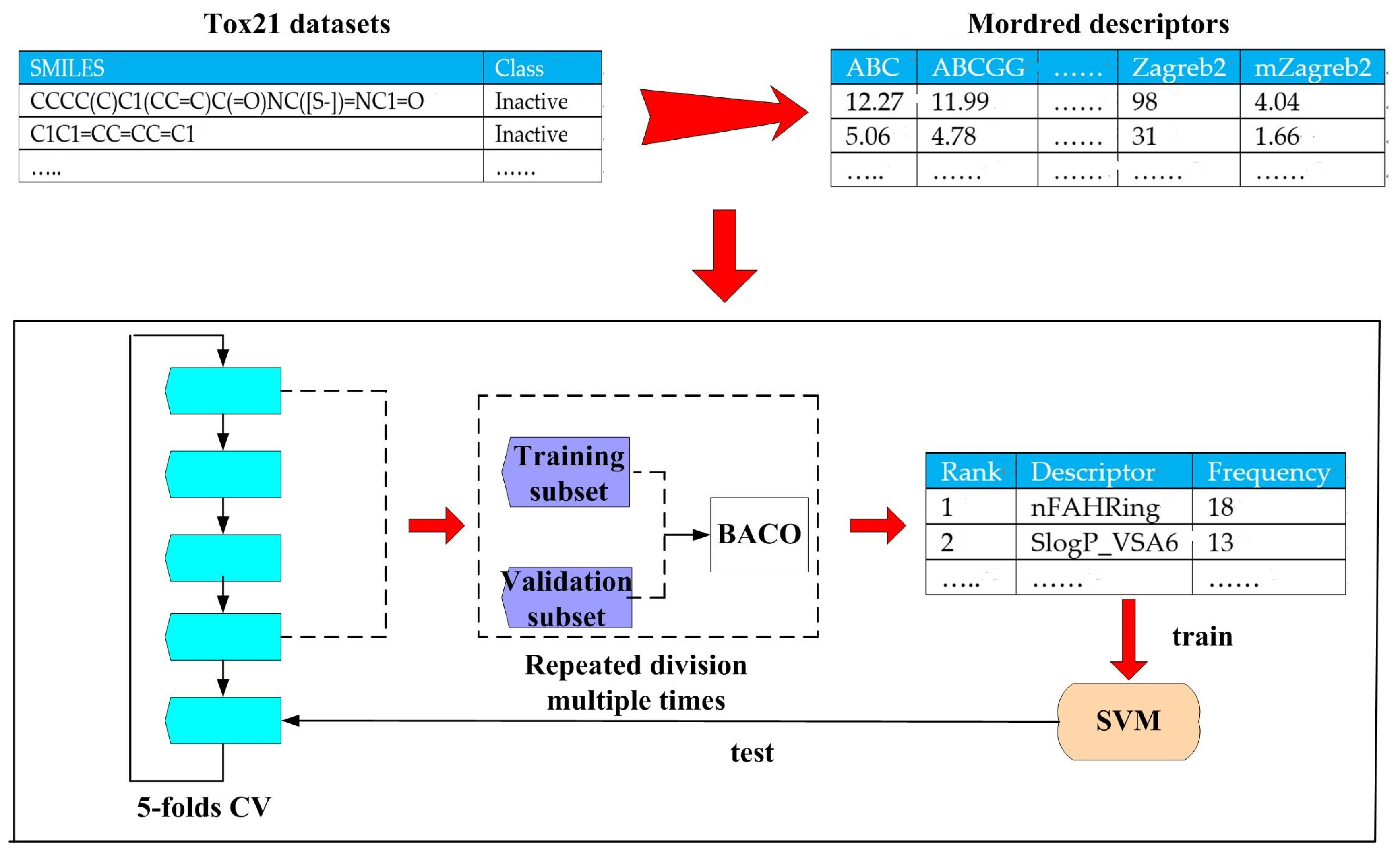
3.1.1. Tox21 Datasets
3.1.2. Modred Descriptor Calculator
3.2. Binary Ant Colony Optimization (BACO) Feature (Descriptor) Selection Algorithm
3.2.1. Optimal Feature Group Search Using BACO
3.2.2. Filter Feature Selection Based on Frequency Statistics Acquired by BACO
3.2.3. Default Parameter Settings and Time Complexity Analysis
3.2.4. Applicability and Limitations of BACO
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gupta, R.; Polaka, S.; Rajpoot, K.; Tekade, M.; Sharma, M.C.; Tekade, R.K. Importance of toxicity testing in drug discovery and research. In Pharmacokinetics and Toxicokinetic Considerations; Academic Press: Cambridge, MA, USA, 2022; pp. 117–144. [Google Scholar]
- Kelleci Çelik, F.; Karaduman, G. In silico QSAR modeling to predict the safe use of antibiotics during pregnancy. Drug Chem. Toxicol. 2023, 46, 962–971. [Google Scholar] [PubMed]
- Krewski, D.; Andersen, M.E.; Tyshenko, M.G.; Krishnan, K.; Hartung, T.; Boekelheide, K.; Wambaugh, J.F.; Jones, D.; Whelan, M.; Thomas, R.; et al. Toxicity testing in the 21st century: Progress in the past decade and future perspectives. Arch. Toxicol. 2020, 94, 1–58. [Google Scholar] [PubMed]
- De, P.; Kar, S.; Ambure, P.; Roy, K. Prediction reliability of QSAR models: An overview of various validation tools. Arch. Toxicol. 2022, 96, 1279–1295. [Google Scholar] [CrossRef]
- Tran, T.T.V.; Surya Wibowo, A.; Tayara, H.; Chong, K.T. Artificial intelligence in drug toxicity prediction: Recent advances, challenges, and future perspectives. J. Chem. Inf. Model. 2023, 63, 2628–2643. [Google Scholar] [PubMed]
- Achar, J.; Firman, J.W.; Tran, C.; Kim, D.; Cronin, M.T.; Öberg, G. Analysis of implicit and explicit uncertainties in QSAR prediction of chemical toxicity: A case study of neurotoxicity. Regul. Toxicol. Pharmacol. 2024, 154, 105716. [Google Scholar]
- Tropsha, A. Best practices for QSAR model development, validation, and exploitation. Mol. Inform. 2010, 29, 476–488. [Google Scholar] [CrossRef]
- Keyvanpour, M.R.; Shirzad, M.B. An analysis of QSAR research based on machine learning concepts. Curr. Drug Discov. Technol. 2021, 18, 17–30. [Google Scholar]
- Zhang, F.; Wang, Z.; Peijnenburg, W.J.; Vijver, M.G. Machine learning-driven QSAR models for predicting the mixture toxicity of nanoparticles. Environ. Int. 2023, 177, 108025. [Google Scholar]
- Cerruela García, G.; Pérez-Parras Toledano, J.; de Haro García, A.; García-Pedrajas, N. Filter feature selectors in the development of binary QSAR models. SAR QSAR Environ. Res. 2019, 30, 313–345. [Google Scholar]
- Eklund, M.; Norinder, U.; Boyer, S.; Carlsson, L. Choosing feature selection and learning algorithms in QSAR. J. Chem. Inf. Model. 2014, 54, 837–843. [Google Scholar]
- MotieGhader, H.; Gharaghani, S.; Masoudi-Sobhanzadeh, Y.; Masoudi-Nejad, A. Sequential and mixed genetic algorithm and learning automata (SGALA, MGALA) for feature selection in QSAR. Iran. J. Pharm. Res. IJPR 2017, 16, 533. [Google Scholar] [PubMed]
- Wang, Y.; Wang, B.; Jiang, J.; Guo, J.; Lai, J.; Lian, X.Y.; Wu, J. Multitask CapsNet: An imbalanced data deep learning method for predicting toxicants. ACS Omega 2021, 6, 26545–26555. [Google Scholar] [PubMed]
- Idakwo, G.; Thangapandian, S.; Luttrell, J.; Li, Y.; Wang, N.; Zhou, Z.; Hong, H.; Yang, B.; Zhang, C.; Gong, P. Structure–activity relationship-based chemical classification of highly imbalanced Tox21 datasets. J. Cheminform. 2020, 12, 1–19. [Google Scholar]
- Kim, C.; Jeong, J.; Choi, J. Effects of Class Imbalance and Data Scarcity on the Performance of Binary Classification Machine Learning Models Developed Based on ToxCast/Tox21 Assay Data. Chem. Res. Toxicol. 2022, 35, 2219–2226. [Google Scholar] [CrossRef]
- Guyon, I.; Weston, J.; Barnhill, S.; Vapnik, V. Gene selection for cancer classification using support vector machines. Mach. Learn. 2002, 46, 389–422. [Google Scholar] [CrossRef]
- Dorigo, M.; Birattari, M.; Stutzle, T. Ant colony optimization. IEEE Comput. Intell. Mag. 2006, 1, 28–39. [Google Scholar]
- Sun, L.; Chen, Y.; Ding, W.; Xu, J. LEFSA: Label enhancement-based feature selection with adaptive neighborhood via ant colony optimization for multilabel learning. Int. J. Mach. Learn. Cybern. 2024, 15, 533–558. [Google Scholar]
- Yu, H.; Ni, J.; Zhao, J. ACOSampling: An ant colony optimization-based undersampling method for classifying imbalanced DNA microarray data. Neurocomputing 2013, 101, 309–318. [Google Scholar]
- Drucker, H.; Wu, D.; Vapnik, V.N. Support vector machines for spam categorization. IEEE Trans. Neural Netw. 1999, 10, 1048–1054. [Google Scholar]
- Boczar, D.; Michalska, K. A review of machine learning and QSAR/QSPR Predictions for complexes of organic molecules with cyclodextrins. Molecules 2024, 29, 3159. [Google Scholar] [CrossRef]
- Rodríguez-Pérez, R.; Bajorath, J. Evolution of support vector machine and regression modeling in chemoinformatics and drug discovery. J. Comput.-Aided Mol. Des. 2022, 36, 355–362. [Google Scholar] [PubMed]
- Czermiński, R.; Yasri, A.; Hartsough, D. Use of support vector machine in pattern classification: Application to QSAR studies. Quant. Struct.-Act. Relatsh. 2001, 20, 227–240. [Google Scholar]
- Du, Z.; Wang, D.; Li, Y. Comprehensive evaluation and comparison of machine learning methods in QSAR modeling of antioxidant tripeptides. ACS Omega 2022, 7, 25760–25771. [Google Scholar]
- Antelo-Collado, A.; Carrasco-Velar, R.; García-Pedrajas, N.; Cerruela-García, G. Effective feature selection method for class-imbalance datasets applied to chemical toxicity prediction. J. Chem. Inf. Model. 2020, 61, 76–94. [Google Scholar] [PubMed]
- Moriwaki, H.; Tian, Y.S.; Kawashita, N.; Takagi, T. Mordred: A molecular descriptor calculator. J. Cheminform. 2018, 10, 4. [Google Scholar] [PubMed]
- Rupapara, V.; Rustam, F.; Ishaq, A.; Lee, E.; Ashraf, I. Chi-square and PCA based feature selection for diabetes detection with ensemble classifier. Intell. Autom. Soft Comput. 2023, 36, 1931–1949. [Google Scholar]
- Menze, B.H.; Kelm, B.M.; Masuch, R.; Himmelreich, U.; Bachert, P.; Petrich, W.; Hamprecht, F.A. A comparison of random forest and its Gini importance with standard chemometric methods for the feature selection and classification of spectral data. BMC Bioinform. 2009, 10, 213. [Google Scholar]
- Peng, H.; Long, F.; Ding, C. Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [PubMed]
- Estévez, P.A.; Tesmer, M.; Perez, C.A.; Zurada, J.M. Normalized mutual information feature selection. IEEE Trans. Neural Netw. 2009, 20, 189–201. [Google Scholar]
- Robnik-Šikonja, M.; Kononenko, I. Theoretical and empirical analysis of ReliefF and RReliefF. Mach. Learn. 2003, 53, 23–69. [Google Scholar]
- Demsar, J. Statistical comparisons of classifiers over multiple data sets. J. Mach. Learn. Res. 2006, 7, 1–30. [Google Scholar]
- Garcia, S.; Fernandez, A.; Luengo, J.; Herrera, F. Advanced nonpara-metric tests for multiple comparisons in the design of experiments in computational intelligence and data mining: Experimental analysis of power. Inf. Sci. 2010, 180, 2044–2064. [Google Scholar]
- Makarov, D.M.; Ksenofontov, A.A.; Budkov, Y.A. Consensus Modeling for Predicting Chemical Binding to Transthyretin as the Winning Solution of the Tox24 Challenge. Chem. Res. Toxicol. 2025, 38, 392–399. [Google Scholar] [PubMed]
- Loh, W.Y. Classification and regression trees. Wiley Data Min. Knowl. Discov. 2011, 1, 14–23. [Google Scholar]
- Nick, T.G.; Campbell, K.M. Logistic regression. Top. Biostat. 2007, 273–301. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Petricoin, E.F.; Ardekani, A.M.; Hitt, B.A.; Levine, P.J.; Fusaro, V.A.; Steinberg, S.M.; Mills, G.B.; Simone, C.; Fishman, D.A.; Kohn, E.C.; et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet 2002, 359, 572–577. [Google Scholar]
- Alon, U.; Barkai, N.; Notterman, D.A.; Gish, K.; Ybarra, S.; Mack, D.; Levine, A.J. Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 1999, 96, 6745–6750. [Google Scholar]
- Srisongkram, T. Ensemble quantitative read-across structure–activity relationship algorithm for predicting skin cytotoxicity. Chem. Res. Toxicol. 2023, 36, 1961–1972. [Google Scholar]
- Krishnan, S.R.; Roy, A.; Gromiha, M.M. Reliable method for predicting the binding affinity of RNA-small molecule interactions using machine learning. Brief. Bioinform. 2024, 25, bbae002. [Google Scholar]
- Clare, B.W. QSAR of aromatic substances: Toxicity of polychlorodibenzofurans. J. Mol. Struct. Theochem 2006, 763, 205–213. [Google Scholar]
- Podder, T.; Kumar, A.; Bhattacharjee, A.; Ojha, P.K. Exploring regression-based QSTR and i-QSTR modeling for ecotoxicity prediction of diverse pesticides on multiple avian species. Environ. Sci. Adv. 2023, 2, 1399–1422. [Google Scholar]
- Kumar, A.; Ojha, P.K.; Roy, K. QSAR modeling of chronic rat toxicity of diverse organic chemicals. Comput. Toxicol. 2023, 26, 100270. [Google Scholar]
- Edros, R.; Feng, T.W.; Dong, R.H. Utilizing machine learning techniques to predict the blood-brain barrier permeability of compounds detected using LCQTOF-MS in Malaysian Kelulut honey. SAR QSAR Environ. Res. 2023, 34, 475–500. [Google Scholar]
- Fujimoto, T.; Gotoh, H. Prediction and chemical interpretation of singlet-oxygen-scavenging activity of small molecule compounds by using machine learning. Antioxidants 2021, 10, 1751. [Google Scholar] [CrossRef]
- Galvez-Llompart, M.; Hierrezuelo, J.; Blasco, M.; Zanni, R.; Galvez, J.; de Vicente, A.; Pérez-García, A.; Romero, D. Targeting bacterial growth in biofilm conditions: Rational design of novel inhibitors to mitigate clinical and food contamination using QSAR. J. Enzym. Inhib. Med. Chem. 2024, 39, 2330907. [Google Scholar]
- Castillo-Garit, J.A.; Barigye, S.J.; Pham-The, H.; Pérez-Doñate, V.; Torrens, F.; Pérez-Giménez, F. Computational identification of chemical compounds with potential anti-Chagas activity using a classification tree. SAR QSAR Environ. Res. 2021, 32, 71–83. [Google Scholar] [PubMed]
- Wang, G.; Wong, K.W.; Lu, J. AUC-based extreme learning machines for supervised and semi-supervised imbalanced classification. IEEE Trans. Syst. Man Cybern. Syst. 2020, 51, 7919–7930. [Google Scholar]
- Saito, T.; Rehmsmeier, M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS ONE 2015, 10, e0118432. [Google Scholar]
- Efron, B. The bootstrap and modern statistics. J. Am. Stat. Assoc. 2000, 95, 1293–1296. [Google Scholar]
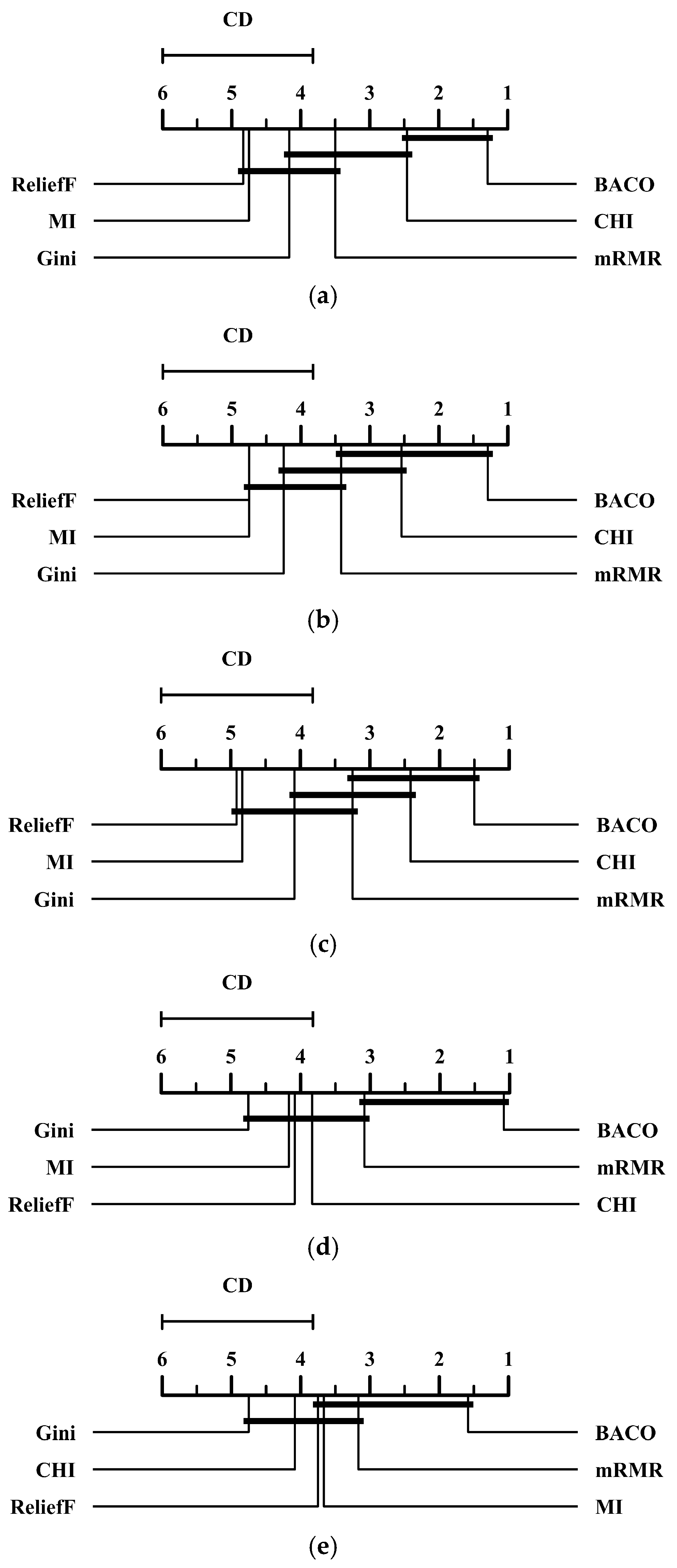
| Dataset | Initially Screened Features | High-Frequency Features | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # Descriptors | F-Measure | G-Mean | MCC | AUC | PR-AUC | # Descriptors | F-Measure | G-Mean | MCC | AUC | PR-AUC | |
| DS1 | 672 | 0.5519 | 0.6467 | 0.5727 | 0.7128 | 0.0897 | 20 | 0.6029 | 0.6866 | 0.6170 | 0.7657 | 0.1616 |
| DS2 | 669 | 0.5732 | 0.6790 | 0.5819 | 0.7931 | 0.0732 | 20 | 0.6168 | 0.7286 | 0.6142 | 0.8336 | 0.1033 |
| DS3 | 672 | 0.0898 | 0.2173 | 0.1809 | 0.6659 | 0.1758 | 20 | 0.2334 | 0.3779 | 0.2568 | 0.7529 | 0.2595 |
| DS4 | 671 | 0.0000 | 0.0000 | 0.0000 | 0.6178 | 0.0597 | 20 | 0.0570 | 0.1509 | 0.1365 | 0.6856 | 0.1191 |
| DS5 | 670 | 0.0437 | 0.1484 | 0.1389 | 0.5993 | 0.1571 | 20 | 0.1997 | 0.3367 | 0.2722 | 0.7198 | 0.2948 |
| DS6 | 670 | 0.0000 | 0.0000 | 0.0000 | 0.6037 | 0.0614 | 20 | 0.1465 | 0.2801 | 0.2488 | 0.6854 | 0.1732 |
| DS7 | 670 | 0.0000 | 0.0000 | 0.0000 | 0.6227 | 0.0391 | 20 | 0.0000 | 0.0000 | 0.0000 | 0.6496 | 0.0547 |
| DS8 | 629 | 0.0000 | 0.0000 | 0.0000 | 0.5949 | 0.1732 | 20 | 0.0884 | 0.2174 | 0.1345 | 0.8159 | 0.2193 |
| DS9 | 671 | 0.0000 | 0.0000 | 0.0000 | 0.5558 | 0.0588 | 20 | 0.0236 | 0.0848 | 0.0833 | 0.6434 | 0.1490 |
| DS10 | 629 | 0.0000 | 0.0000 | 0.0000 | 0.6337 | 0.0742 | 20 | 0.0311 | 0.0970 | 0.0947 | 0.7225 | 0.1229 |
| DS11 | 672 | 0.1302 | 0.2579 | 0.2018 | 0.7907 | 0.2335 | 20 | 0.2722 | 0.4110 | 0.2816 | 0.8471 | 0.3845 |
| DS12 | 670 | 0.0000 | 0.0000 | 0.0000 | 0.5892 | 0.0698 | 20 | 0.0620 | 0.1778 | 0.1142 | 0.6778 | 0.1871 |
| Dataset | CHI | Gini | mRMR | MI | ReliefF | BACO |
|---|---|---|---|---|---|---|
| DS1 | 0.5959 | 0.5545 | 0.5366 | 0.5570 | 0.4379 | 0.6029 |
| DS2 | 0.6060 | 0.5591 | 0.5543 | 0.5774 | 0.5586 | 0.6168 |
| DS3 | 0.1717 | 0.0776 | 0.1738 | 0.0028 | 0.0287 | 0.2334 |
| DS4 | 0.0394 | 0.0131 | 0.0384 | 0.0000 | 0.0000 | 0.0570 |
| DS5 | 0.1014 | 0.0000 | 0.0639 | 0.0000 | 0.0000 | 0.1997 |
| DS6 | 0.1247 | 0.0395 | 0.1148 | 0.0000 | 0.0000 | 0.1465 |
| DS7 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| DS8 | 0.0756 | 0.0000 | 0.0000 | 0.0000 | 0.0068 | 0.0884 |
| DS9 | 0.0236 | 0.0000 | 0.0236 | 0.0000 | 0.0000 | 0.0236 |
| DS10 | 0.0098 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0311 |
| DS11 | 0.1877 | 0.2264 | 0.2115 | 0.0000 | 0.0421 | 0.2722 |
| DS12 | 0.0346 | 0.0000 | 0.0541 | 0.0000 | 0.0000 | 0.0620 |
| Dataset | CHI | Gini | mRMR | MI | ReliefF | BACO |
|---|---|---|---|---|---|---|
| DS1 | 0.6827 | 0.6434 | 0.6375 | 0.6478 | 0.5273 | 0.6866 |
| DS2 | 0.7107 | 0.6753 | 0.6688 | 0.6858 | 0.6804 | 0.7286 |
| DS3 | 0.3133 | 0.2003 | 0.3144 | 0.0167 | 0.1187 | 0.3779 |
| DS4 | 0.1085 | 0.0517 | 0.1221 | 0.0000 | 0.0000 | 0.1509 |
| DS5 | 0.2306 | 0.0000 | 0.1803 | 0.0000 | 0.0000 | 0.3367 |
| DS6 | 0.2546 | 0.0896 | 0.2447 | 0.0000 | 0.0000 | 0.2801 |
| DS7 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| DS8 | 0.1978 | 0.0000 | 0.0000 | 0.0000 | 0.0451 | 0.2174 |
| DS9 | 0.0848 | 0.0000 | 0.0848 | 0.0000 | 0.0000 | 0.0848 |
| DS10 | 0.0446 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0970 |
| DS11 | 0.3274 | 0.3652 | 0.3469 | 0.0000 | 0.1370 | 0.4110 |
| DS12 | 0.1032 | 0.0000 | 0.1636 | 0.0000 | 0.0000 | 0.1778 |
| Dataset | CHI | Gini | mRMR | MI | ReliefF | BACO |
|---|---|---|---|---|---|---|
| DS1 | 0.6106 | 0.5803 | 0.5541 | 0.5790 | 0.4451 | 0.6170 |
| DS2 | 0.6090 | 0.5653 | 0.5630 | 0.5849 | 0.5603 | 0.6142 |
| DS3 | 0.2166 | 0.1606 | 0.2254 | 0.0157 | 0.1056 | 0.2568 |
| DS4 | 0.1057 | 0.0503 | 0.1189 | 0.0000 | 0.0000 | 0.1365 |
| DS5 | 0.2064 | 0.0000 | 0.1653 | 0.0000 | 0.0000 | 0.2722 |
| DS6 | 0.2488 | 0.0875 | 0.2391 | 0.0000 | 0.0000 | 0.2488 |
| DS7 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| DS8 | 0.1356 | 0.0000 | 0.0000 | 0.0000 | 0.0389 | 0.1345 |
| DS9 | 0.0833 | 0.0000 | 0.0833 | 0.0000 | 0.0000 | 0.0833 |
| DS10 | 0.0418 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0947 |
| DS11 | 0.2372 | 0.2694 | 0.2512 | 0.0000 | 0.0901 | 0.2816 |
| DS12 | 0.0875 | 0.0000 | 0.1408 | 0.0000 | 0.0000 | 0.1142 |
| Dataset | CHI | Gini | mRMR | MI | ReliefF | BACO |
|---|---|---|---|---|---|---|
| DS1 | 0.7436 | 0.7099 | 0.7528 | 0.7244 | 0.7497 | 0.7657 |
| DS2 | 0.7887 | 0.7792 | 0.8429 | 0.8007 | 0.7861 | 0.8336 |
| DS3 | 0.6730 | 0.6701 | 0.6733 | 0.6728 | 0.7125 | 0.7529 |
| DS4 | 0.6766 | 0.6429 | 0.6407 | 0.6541 | 0.6250 | 0.6856 |
| DS5 | 0.7046 | 0.6288 | 0.6773 | 0.6332 | 0.6590 | 0.7198 |
| DS6 | 0.6468 | 0.6577 | 0.6680 | 0.6145 | 0.6399 | 0.6854 |
| DS7 | 0.6081 | 0.6346 | 0.6229 | 0.6458 | 0.6335 | 0.6496 |
| DS8 | 0.7032 | 0.6758 | 0.7327 | 0.7565 | 0.7021 | 0.8159 |
| DS9 | 0.6218 | 0.5421 | 0.6036 | 0.5976 | 0.5808 | 0.6434 |
| DS10 | 0.5979 | 0.6544 | 0.6770 | 0.6231 | 0.6080 | 0.7225 |
| DS11 | 0.7499 | 0.8006 | 0.8138 | 0.7999 | 0.8302 | 0.8471 |
| DS12 | 0.6656 | 0.6247 | 0.6242 | 0.6108 | 0.6334 | 0.6778 |
| Dataset | CHI | Gini | mRMR | MI | ReliefF | BACO |
|---|---|---|---|---|---|---|
| DS1 | 0.1412 | 0.1009 | 0.1458 | 0.0950 | 0.1132 | 0.1616 |
| DS2 | 0.1330 | 0.1217 | 0.1448 | 0.0979 | 0.0856 | 0.1033 |
| DS3 | 0.1810 | 0.2009 | 0.2032 | 0.2406 | 0.2129 | 0.2595 |
| DS4 | 0.0228 | 0.0240 | 0.0775 | 0.0957 | 0.0656 | 0.1191 |
| DS5 | 0.2369 | 0.1952 | 0.2244 | 0.2667 | 0.2324 | 0.2948 |
| DS6 | 0.0838 | 0.0992 | 0.1348 | 0.1252 | 0.1387 | 0.1732 |
| DS7 | 0.0556 | 0.0622 | 0.0617 | 0.0438 | 0.0610 | 0.0547 |
| DS8 | 0.1991 | 0.1846 | 0.2033 | 0.1996 | 0.2148 | 0.2193 |
| DS9 | 0.1336 | 0.1028 | 0.0793 | 0.0881 | 0.0742 | 0.1490 |
| DS10 | 0.0889 | 0.0795 | 0.1034 | 0.0929 | 0.1036 | 0.1229 |
| DS11 | 0.2827 | 0.2541 | 0.3136 | 0.3376 | 0.3051 | 0.3845 |
| DS12 | 0.1141 | 0.0782 | 0.0914 | 0.1081 | 0.0905 | 0.1871 |
| Number of Selected Descriptors K | F-Measure | G-Mean | MCC | AUC | PR-AUC |
|---|---|---|---|---|---|
| DS1 | |||||
| 5 | 0.5737 | 0.6699 | 0.5867 | 0.6759 | 0.0917 |
| 10 | 0.5968 | 0.6861 | 0.6082 | 0.7421 | 0.1356 |
| 20 | 0.6029 | 0.6866 | 0.6170 | 0.7657 | 0.1616 |
| 30 | 0.6041 | 0.6886 | 0.6177 | 0.7698 | 0.1681 |
| 50 | 0.6087 | 0.6941 | 0.6214 | 0.7736 | 0.1744 |
| 100 | 0.6085 | 0.6941 | 0.6213 | 0.8105 | 0.1752 |
| 200 | 0.6110 | 0.6962 | 0.6235 | 0.8032 | 0.1810 |
| 300 | 0.6153 | 0.6987 | 0.6278 | 0.7764 | 0.1732 |
| DS2 | |||||
| 5 | 0.5722 | 0.7189 | 0.5642 | 0.8059 | 0.0692 |
| 10 | 0.6164 | 0.7295 | 0.6162 | 0.8357 | 0.1421 |
| 20 | 0.6168 | 0.7286 | 0.6142 | 0.8336 | 0.1033 |
| 30 | 0.6090 | 0.7204 | 0.6079 | 0.8311 | 0.1058 |
| 50 | 0.6073 | 0.7203 | 0.6060 | 0.8230 | 0.1011 |
| 100 | 0.6136 | 0.7206 | 0.6138 | 0.8167 | 0.0872 |
| 200 | 0.6156 | 0.7206 | 0.6131 | 0.8123 | 0.0903 |
| 300 | 0.6139 | 0.7205 | 0.6141 | 0.8056 | 0.0887 |
| DS3 | |||||
| 5 | 0.1568 | 0.2985 | 0.2053 | 0.6577 | 0.1830 |
| 10 | 0.2123 | 0.3557 | 0.2395 | 0.7121 | 0.2093 |
| 20 | 0.2334 | 0.3779 | 0.2568 | 0.7529 | 0.2595 |
| 30 | 0.2612 | 0.4030 | 0.2817 | 0.7636 | 0.2546 |
| 50 | 0.2651 | 0.4027 | 0.2997 | 0.7519 | 0.2319 |
| 100 | 0.2200 | 0.3603 | 0.2602 | 0.7253 | 0.2172 |
| 200 | 0.1990 | 0.3385 | 0.2564 | 0.7034 | 0.2005 |
| 300 | 0.1480 | 0.2863 | 0.2134 | 0.6908 | 0.1891 |
| DS4 | |||||
| 5 | 0.0958 | 0.2190 | 0.2082 | 0.7236 | 0.1459 |
| 10 | 0.0843 | 0.2046 | 0.1995 | 0.6959 | 0.1392 |
| 20 | 0.0570 | 0.1509 | 0.1365 | 0.6856 | 0.1191 |
| 30 | 0.0508 | 0.1434 | 0.1322 | 0.6758 | 0.1201 |
| 50 | 0.0515 | 0.1597 | 0.1470 | 0.6813 | 0.1096 |
| 100 | 0.0443 | 0.1325 | 0.1290 | 0.6467 | 0.0989 |
| 200 | 0.0324 | 0.1140 | 0.1110 | 0.6501 | 0.0807 |
| 300 | 0.0253 | 0.0870 | 0.0847 | 0.6288 | 0.0753 |
| Classifier | F-Measure | G-Mean | MCC | AUC | PR-AUC |
|---|---|---|---|---|---|
| DS1 | |||||
| SVM | 0.6029 | 0.6866 | 0.6170 | 0.7657 | 0.1616 |
| CART | 0.5746 | 0.6517 | 0.5842 | 0.7126 | 0.1110 |
| LR | 0.5978 | 0.6429 | 0.6011 | 0.6959 | 0.0979 |
| RF | 0.5898 | 0.6917 | 0.6154 | 0.7452 | 0.1842 |
| XGBoost | 0.6232 | 0.7135 | 0.6127 | 0.7591 | 0.1721 |
| DS2 | |||||
| SVM | 0.6168 | 0.7286 | 0.6142 | 0.8336 | 0.1033 |
| CART | 0.5820 | 0.6899 | 0.5776 | 0.7984 | 0.0811 |
| LR | 0.5491 | 0.6372 | 0.5531 | 0.7521 | 0.0623 |
| RF | 0.6029 | 0.7141 | 0.5982 | 0.8419 | 0.0976 |
| XGBoost | 0.6171 | 0.7188 | 0.6075 | 0.8501 | 0.0928 |
| DS3 | |||||
| SVM | 0.2334 | 0.3779 | 0.2568 | 0.7529 | 0.2595 |
| CART | 0.2258 | 0.3556 | 0.2426 | 0.6887 | 0.1984 |
| LR | 0.2096 | 0.3432 | 0.2581 | 0.6931 | 0.1672 |
| RF | 0.2617 | 0.4165 | 0.2607 | 0.7425 | 0.2571 |
| XGBoost | 0.2528 | 0.3974 | 0.2753 | 0.7377 | 0.2692 |
| DS4 | |||||
| SVM | 0.0570 | 0.1509 | 0.1365 | 0.6856 | 0.1191 |
| CART | 0.0296 | 0.1279 | 0.1128 | 0.6572 | 0.0992 |
| LR | 0.0479 | 0.1601 | 0.1306 | 0.6773 | 0.1143 |
| RF | 0.0511 | 0.1548 | 0.1427 | 0.6654 | 0.1242 |
| XGBoost | 0.0537 | 0.1532 | 0.1358 | 0.7175 | 0.1103 |
| Metric | CHI | Gini | mRMR | MI | ReliefF | BACO |
|---|---|---|---|---|---|---|
| Ovarian I | ||||||
| F-measure | 0.5795 | 0.5218 | 0.5944 | 0.5650 | 0.5717 | 0.7201 |
| G-mean | 0.6773 | 0.6379 | 0.7286 | 0.6582 | 0.6470 | 0.8197 |
| MCC | 0.5169 | 0.4822 | 0.4935 | 0.4278 | 0.4040 | 0.6337 |
| AUC | 0.9121 | 0.9038 | 0.9455 | 0.9256 | 0.9319 | 0.9720 |
| PR-AUC | 0.4752 | 0.3230 | 0.4438 | 0.4196 | 0.5133 | 0.5872 |
| Ovarian II | ||||||
| F-measure | 0.4916 | 0.4783 | 0.4928 | 0.5048 | 0.4732 | 0.6131 |
| G-mean | 0.5742 | 0.5549 | 0.6276 | 0.5478 | 0.5947 | 0.7526 |
| MCC | 0.4554 | 0.4999 | 0.4783 | 0.4362 | 0.4881 | 0.5206 |
| AUC | 0.9570 | 0.9225 | 0.9598 | 0.9439 | 0.9530 | 0.9497 |
| PR-AUC | 0.3657 | 0.3571 | 0.3762 | 0.3545 | 0.4033 | 0.4210 |
| Colon | ||||||
| F-measure | 0.7146 | 0.6964 | 0.7652 | 0.7277 | 0.7532 | 0.8064 |
| G-mean | 0.7759 | 0.7448 | 0.8118 | 0.7529 | 0.8216 | 0.8688 |
| MCC | 0.7072 | 0.7106 | 0.7529 | 0.6988 | 0.7391 | 0.8179 |
| AUC | 0.8456 | 0.8619 | 0.9098 | 0.8373 | 0.8752 | 0.9137 |
| PR-AUC | 0.6912 | 0.6440 | 0.7829 | 0.6827 | 0.7913 | 0.8442 |
| Descriptor Name | Frequency | Descriptor Definition |
|---|---|---|
| nG12FARing | 18 | Twelve-or-greater-membered aliphatic fused ring count |
| nG12FRing | 12 | Twelve-or-greater-membered fused ring count |
| n6ARing | 11 | Six-membered aliphatic ring count |
| nHRing | 9 | Hetero ring count |
| n5ARing | 9 | Five-membered aromatic ring count |
| SMR_VSA4 | 8 | MOE MR VSA Descriptor 4 (2.24 ≤ x < 2.45) |
| nAHRing | 8 | Aliphatic hetero ring count |
| nFHRing | 8 | Fused hetero ring count |
| SRW07 | 7 | Walk count (leg-7, only self returning walk) |
| nBridgehead | 6 | Number of bridgehead atoms |
| SlogP_VSA6 | 6 | MOE logP VSA Descriptor 6 (0.15 ≤ x < 0.20) |
| JGI9 | 6 | Nine-ordered mean topological charge |
| EState_VSA4 | 6 | EState VSA Descriptor 4 (0.72 ≤ x < 1.17) |
| SRW05 | 6 | Walk count (leg-5, only self returning walk) |
| ATS0are | 6 | Moreau-broto autocorrelation of lag 0 weighted by allred-rocow EN |
| PEOE_VSA7 | 6 | MOE Charge VSA Descriptor 7 (−0.05 ≤ x < 0.00) |
| Xpc-4dv | 5 | Four-ordered Chi path-cluster weighted by valence electrons |
| ZMIC0 | 5 | Zero-ordered Z-modified information content |
| ATS7m | 5 | Moreau-broto autocorrelation of lag 7 weighted by mass |
| NaaO | 4 | number of aaO |
| Dataset | # Molecules | # Inactive | # Active | Ratio of Active Molecules | Molecular Pathway Endpoint |
|---|---|---|---|---|---|
| DS1 | 7044 | 6743 | 301 | 4.3% | Androgen receptor MDA-kb2 AR-luc cell line (NR-AR) |
| DS2 | 6572 | 6349 | 223 | 3.4% | Androgen receptor GeneBLAzer AR-UAS-bla-GripTite Cell line (NR-AR-LBD) |
| DS3 | 6358 | 5601 | 757 | 11.9% | Aryl hydrocarbon receptor (NR-AhR) |
| DS4 | 5661 | 5368 | 293 | 5.2% | Aromatase enzyme (NR-Aromatase) |
| DS5 | 6013 | 5247 | 766 | 12.7% | Estrogen receptor α BG1-Luc-4E2 cell line (NR-ER) |
| DS6 | 6752 | 6426 | 326 | 4.8% | Estrogen receptor α ER-α-UAS-bla GripTiteTM cell line (NR-ER-LBD) |
| DS7 | 6273 | 6110 | 163 | 2.6% | Peroxisome proliferator-activated receptor γ (NR-PPAR-γ) |
| DS8 | 5684 | 4784 | 900 | 15.8% | Nuclear factor (erythroid-derived 2)-like 2/antioxidant responsive element (NR-ARE) (SR-ARE) |
| DS9 | 6880 | 6633 | 247 | 3.6% | ATAD5 receptor (SR-ATAD5) |
| DS10 | 6294 | 5957 | 337 | 5.4% | Heat shock factor response element (SR-HSE) |
| DS11 | 5334 | 4753 | 881 | 15.6% | Mitochondrial membrane potential (SR-MMP) |
| DS12 | 6586 | 6191 | 395 | 6.0% | p53 signaling pathway (SR-p53) |
| Descriptor Name | Number of Descriptors |
|---|---|
| ABCIndex | 2 |
| AcidBase | 2 |
| AdjacencyMatrix | 13 |
| Aromatic | 2 |
| AtomCount | 17 |
| Autocorrelation | 606 |
| BalabanJ | 1 |
| BaryszMatrix | 104 |
| BCUT | 24 |
| BertzCT | 1 |
| BondCount | 9 |
| CarbonTypes | 11 |
| Chi | 56 |
| Constitutional | 16 |
| DetourMatrix | 14 |
| DistanceMatrix | 13 |
| EccentricConnectivityIndex | 1 |
| Estate | 316 |
| ExtendedTopochemicalAtom | 45 |
| FragmentComplexity | 1 |
| Framework | 1 |
| HydrogenBond | 2 |
| InformationContent | 42 |
| KappaShapeIndex | 3 |
| Lipinski | 2 |
| LogS | 1 |
| McGowanVolume | 1 |
| MoeType | 53 |
| MolecularDistanceEdge | 19 |
| MolecularId | 12 |
| PathCount | 21 |
| Polarizability | 2 |
| RingCount | 138 |
| RotatableBond | 2 |
| SLogP | 2 |
| TopologicalCharge | 21 |
| TopologicalIndex | 4 |
| TopoPSA | 2 |
| VdwVolumeABC | 1 |
| VertexAdjacencyInformation | 1 |
| WalkCount | 21 |
| Weight | 2 |
| WienerIndex | 2 |
| ZagrebIndex | 4 |
| Predicted Positive Class | Predicted Negative Class | |
|---|---|---|
| Real positive class | TP (True positive) | FN (False negative) |
| Real negative class | FP (False positive) | TN (True negative) |
| Parameter Name | Default Setting |
|---|---|
| S: number of ants in the ant colony | 50 |
| R: the iteration times of BACO | 10 |
| M: number of random divisions | 40 |
| ρ: evaporation factor | 0.2 |
| K: number of selecting features | 20 |
| phini1: initial pheromone concentration in pathway 1 | 0.4 |
| phini2: initial pheromone concentration in pathway 2 | 1.0 |
| phmin: the lower bound of pheromone concentration | 0.1 |
| phmax: the lower bound of pheromone concentration | 2.0 |
| α, β: the weights for three performance metrics | 1/3 |
| Kernel function type used in SVM | rbf |
| σ: the parameter of kernel function in SVM | 5 |
| C: the penalty factor in SVM | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dan, Y.; Ruan, J.; Zhu, Z.; Yu, H. Predicting the Toxicity of Drug Molecules with Selecting Effective Descriptors Using a Binary Ant Colony Optimization (BACO) Feature Selection Approach. Molecules 2025, 30, 1548. https://doi.org/10.3390/molecules30071548
Dan Y, Ruan J, Zhu Z, Yu H. Predicting the Toxicity of Drug Molecules with Selecting Effective Descriptors Using a Binary Ant Colony Optimization (BACO) Feature Selection Approach. Molecules. 2025; 30(7):1548. https://doi.org/10.3390/molecules30071548
Chicago/Turabian StyleDan, Yuanyuan, Junhao Ruan, Zhenghua Zhu, and Hualong Yu. 2025. "Predicting the Toxicity of Drug Molecules with Selecting Effective Descriptors Using a Binary Ant Colony Optimization (BACO) Feature Selection Approach" Molecules 30, no. 7: 1548. https://doi.org/10.3390/molecules30071548
APA StyleDan, Y., Ruan, J., Zhu, Z., & Yu, H. (2025). Predicting the Toxicity of Drug Molecules with Selecting Effective Descriptors Using a Binary Ant Colony Optimization (BACO) Feature Selection Approach. Molecules, 30(7), 1548. https://doi.org/10.3390/molecules30071548







