In Silico Evaluation of Potential Hit Molecules Against Multiple Serotypes of Dengue Virus Envelope Glycoprotein
Abstract
1. Introduction
2. Results and Discussion
2.1. Docking of the Controls and Pharmacophore Screening
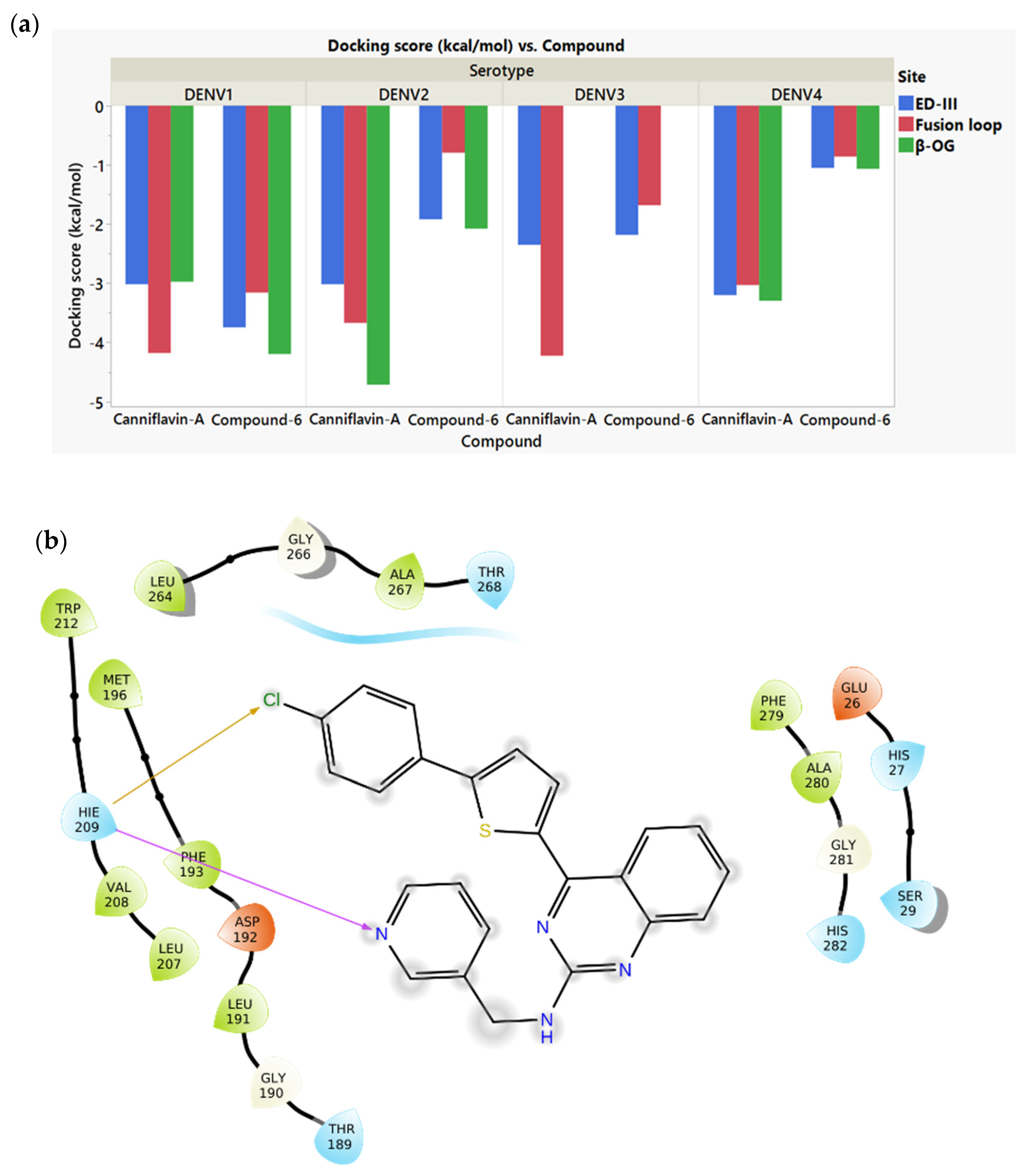
2.2. Docking of the Screened Compounds
2.3. ADMET Properties
2.4. Molecular Dynamics Simulation of the Selected Molecules
2.5. Binding Kinetics of CAP1 and Paclitaxel
3. Materials and Methods
3.1. Protein and Ligand Structures
3.2. Molecular Docking
3.3. Pharmacophore Screening
3.4. Molecular Dynamics (MD)
3.5. Prime MM-GBSA Binding Energy Calculations
3.6. Assessment of Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) Properties
3.7. Viral E Proteins and Chemicals
3.8. Bio-Layer Interferometry (BLI)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dengue and Severe Dengue. 2024. Available online: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue (accessed on 12 February 2024).
- Tsheten, T.; Clements, A.C.A.; Gray, D.J.; Adhikary, R.K.; Furuya-Kanamori, L.; Wangdi, K. Clinical predictors of severe dengue: A systematic review and meta-analysis. Infect. Dis. Poverty 2021, 10, 123. [Google Scholar] [CrossRef]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef]
- Malavige, G.N.; Fernando, S.; Fernando, D.J.; Seneviratne, S.L. Dengue viral infections. Postgrad. Med. J. 2004, 80, 588–601. [Google Scholar] [CrossRef]
- Bhatt, P.; Sabeena, S.P.; Varma, M.; Arunkumar, G. Current Understanding of the Pathogenesis of Dengue Virus Infection. Curr. Microbiol. 2021, 78, 17–32. [Google Scholar] [CrossRef]
- Kularatne, S.A.; Dalugama, C. Dengue infection: Global importance, immunopathology and management. Clin. Med. 2022, 22, 9–13. [Google Scholar] [CrossRef]
- Lee, M.F.; Wu, Y.S.; Poh, C.L. Molecular Mechanisms of Antiviral Agents against Dengue Virus. Viruses 2023, 15, 705. [Google Scholar] [CrossRef]
- Rashmi, S.H.; Disha, K.S.; Sudheesh, N.; Karunakaran, J.; Joseph, A.; Jagadesh, A.; Mudgal, P.P. Repurposing of approved antivirals against dengue virus serotypes: An in silico and in vitro mechanistic study. Mol. Divers. 2023, 28, 2831–2844. [Google Scholar] [CrossRef]
- Troost, B.; Smit, J.M. Recent advances in antiviral drug development towards dengue virus. Curr. Opin. Virol. 2020, 43, 9–21. [Google Scholar] [CrossRef]
- Obi, J.O.; Gutiérrez-Barbosa, H.; Chua, J.V.; Deredge, D.J. Current Trends and Limitations in Dengue Antiviral Research. Trop. Med. Infect. Dis. 2021, 6, 180. [Google Scholar] [CrossRef]
- Fredeking, T.M.; Zavala-Castro, J.E.; González-Martínez, P.; Moguel-Rodríguez, W.; Sanchez, E.C.; Foster, M.J.; Diaz-Quijano, F.A. Dengue Patients Treated with Doxycycline Showed Lower Mortality Associated to a Reduction in IL-6 and TNF Levels. Recent Pat. Anti-Infect. Drug Discov. 2015, 10, 51–58. [Google Scholar] [CrossRef]
- Low, J.G.; Sung, C.; Wijaya, L.; Wei, Y.; Rathore, A.P.S.; Watanabe, S.; Tan, B.H.; Toh, L.; Chua, L.T.; Hou, Y.a.; et al. Efficacy and safety of celgosivir in patients with dengue fever (CELADEN): A phase 1b, randomised, double-blind, placebo-controlled, proof-of-concept trial. Lancet Infect. Dis. 2014, 14, 706–715. [Google Scholar] [CrossRef] [PubMed]
- Whitehorn, J.; Nguyen, C.V.V.; Khanh, L.P.; Kien, D.T.H.; Quyen, N.T.H.; Tran, N.T.T.; Hang, N.T.; Truong, N.T.; Hue Tai, L.T.; Cam Huong, N.T.; et al. Lovastatin for the Treatment of Adult Patients With Dengue: A Randomized, Double-Blind, Placebo-Controlled Trial. Clin. Infect. Dis. 2016, 62, 468–476. [Google Scholar] [CrossRef]
- Palanichamy Kala, M.; St John, A.L.; Rathore, A.P.S. Dengue: Update on Clinically Relevant Therapeutic Strategies and Vaccines. Curr. Treat. Options Infect. Dis. 2023, 15, 27–52. [Google Scholar] [CrossRef] [PubMed]
- Hadinegoro, S.R.; Arredondo-García, J.L.; Capeding, M.R.; Deseda, C.; Chotpitayasunondh, T.; Dietze, R.; Ismail, H.I.H.M.; Reynales, H.; Limkittikul, K.; Rivera-Medina, D.M.; et al. Efficacy and Long-Term Safety of a Dengue Vaccine in Regions of Endemic Disease. N. Engl. J. Med. 2015, 373, 1195–1206. [Google Scholar] [CrossRef]
- Capeding, M.R.; Tran, N.H.; Hadinegoro, S.R.S.; Ismail, H.I.H.J.M.; Chotpitayasunondh, T.; Chua, M.N.; Luong, C.Q.; Rusmil, K.; Wirawan, D.N.; Nallusamy, R.; et al. Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: A phase 3, randomised, observer-masked, placebo-controlled trial. Lancet 2014, 384, 1358–1365. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, S.; Luedtke, A.; Langevin, E.; Zhu, M.; Bonaparte, M.; Machabert, T.; Savarino, S.; Zambrano, B.; Moureau, A.; Khromava, A.; et al. Effect of Dengue Serostatus on Dengue Vaccine Safety and Efficacy. N. Engl. J. Med. 2018, 379, 327–340. [Google Scholar] [CrossRef]
- Kaptein, S.J.F.; Goethals, O.; Kiemel, D.; Marchand, A.; Kesteleyn, B.; Bonfanti, J.-F.; Bardiot, D.; Stoops, B.; Jonckers, T.H.M.; Dallmeier, K.; et al. A pan-serotype dengue virus inhibitor targeting the NS3–NS4B interaction. Nature 2021, 598, 504–509. [Google Scholar] [CrossRef]
- Hou, J.; Ye, W.; Chen, J. Current Development and Challenges of Tetravalent Live-Attenuated Dengue Vaccines. Front. Immunol. 2022, 13, 840104. [Google Scholar] [CrossRef]
- Crill, W.D.; Roehrig, J.T. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J. Virol. 2001, 75, 7769–7773. [Google Scholar] [CrossRef]
- Lee, M.F.; Anasir, M.I.; Poh, C.L. Development of novel antiviral peptides against dengue serotypes 1–4. Virology 2023, 580, 10–27. [Google Scholar] [CrossRef]
- Kesteleyn, B.; Bonfanti, J.F.; Bardiot, D.; De Boeck, B.; Goethals, O.; Kaptein, S.J.F.; Stoops, B.; Coesemans, E.; Fortin, J.; Muller, P.; et al. Discovery of JNJ-1802, a First-in-Class Pan-Serotype Dengue Virus NS4B Inhibitor. J. Med. Chem. 2024, 67, 4063–4082. [Google Scholar] [CrossRef] [PubMed]
- Timiri, A.K.; Sinha, B.N.; Jayaprakash, V. Progress and prospects on DENV protease inhibitors. Eur. J. Med. Chem. 2016, 117, 125–143. [Google Scholar] [CrossRef]
- Lee, J.-C.; Chang, F.-R.; Chen, S.-R.; Wu, Y.-H.; Hu, H.-C.; Wu, Y.-C.; Backlund, A.; Cheng, Y.-B. Anti-Dengue Virus Constituents from Formosan Zoanthid Palythoa mutuki. Mar. Drugs 2016, 14, 151. [Google Scholar] [CrossRef]
- Lin, C.-S.; Lu, C.-H.; Lin, T.-H.; Kiu, Y.-T.; Kan, J.-Y.; Chang, Y.-J.; Hung, P.-Y.; Koval’skaya, A.V.; Tsypyshev, D.O.; Tsypysheva, I.P.; et al. Inhibition of dengue viruses by N-methylcytisine thio derivatives through targeting viral envelope protein and NS2B-NS3 protease. Bioorganic Med. Chem. Lett. 2024, 99, 129623. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.-Y.; Patel, S.J.; Vangrevelinghe, E.; Xu, H.Y.; Rao, R.; Jaber, D.; Schul, W.; Gu, F.; Heudi, O.; Ma, N.L.; et al. A Small-Molecule Dengue Virus Entry Inhibitor. Antimicrob. Agents Chemother. 2009, 53, 1823–1831. [Google Scholar] [CrossRef]
- Powers, C.N.; Setzer, W.N. An In-Silico Investigation of Phytochemicals as Antiviral Agents Against Dengue Fever. Comb. Chem. High Throughput Screen. 2016, 19, 516–536. [Google Scholar] [CrossRef]
- Fleith, R.C.; Lobo, F.P.; dos Santos, P.F.; Rocha, M.M.; Bordignon, J.; Strottmann, D.M.; Patricio, D.O.; Pavanelli, W.R.; Lo Sarzi, M.; Santos, C.N.D.; et al. Genome-wide analyses reveal a highly conserved Dengue virus envelope peptide which is critical for virus viability and antigenic in humans. Sci. Rep. 2016, 6, 36339. [Google Scholar] [CrossRef] [PubMed]
- Rathore, A.S.; Sarker, A.; Gupta, R.D. Designing antibody against highly conserved region of dengue envelope protein by in silico screening of scFv mutant library. PLoS ONE 2019, 14, e0209576. [Google Scholar] [CrossRef]
- Cui, X.; Wu, Y.; Fan, D.; Gao, N.; Ming, Y.; Wang, P.; An, J. Peptides P4 and P7 derived from E protein inhibit entry of dengue virus serotype 2 via interacting with β3 integrin. Antivir. Res. 2018, 155, 20–27. [Google Scholar] [CrossRef]
- Liao, M.; Sánchez-San Martín, C.; Zheng, A.; Kielian, M. In vitro reconstitution reveals key intermediate states of trimer formation by the dengue virus membrane fusion protein. J. Virol. 2010, 84, 5730–5740. [Google Scholar] [CrossRef]
- Wang, H.; Mulgaonkar, N.; Pérez, L.M.; Fernando, S. ELIXIR-A: An Interactive Visualization Tool for Multi-Target Pharmacophore Refinement. ACS Omega 2022, 7, 12707–12715. [Google Scholar] [CrossRef] [PubMed]
- Sunseri, J.; Koes, D.R. Pharmit: Interactive exploration of chemical space. Nucleic Acids Res. 2016, 44, W442–W448. [Google Scholar] [CrossRef]
- Ragavan, R.M.; Purushothaman, I.; Swaminathan, R.; Almutairi, S.M.; Hussein, D.S.; Rasheed, R.A.; Narasingam, A. Malacitanolide, reissantin E and paclitaxel compounds as inhibitors of envelope, NS5 and NS2B/NS3 target proteins of dengue virus: Computational docking and molecular dynamics simulations studies. J. King Saud Univ.-Sci. 2023, 35, 102868. [Google Scholar] [CrossRef]
- Yenamandra, S.P.; Koo, C.; Chiang, S.; Lim, H.S.J.; Yeo, Z.Y.; Ng, L.C.; Hapuarachchi, H.C. Evolution, heterogeneity and global dispersal of cosmopolitan genotype of Dengue virus type 2. Sci. Rep. 2021, 11, 13496. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.-J.; Chang, K.; Chen, C.-H.; Liao, C.-L.; Chen, L.-J.; Tsai, Y.-Y.; Tsai, C.-Y.; Lin, P.-C.; Hsu, M.-C.; Liu, L.-T. Dengue virus serotype did not contribute to clinical severity or mortality in Taiwan’s largest dengue outbreak in 2015. Eur. J. Med. Res. 2023, 28, 482. [Google Scholar] [CrossRef]
- Dalpiaz, A.; Pavan, B. Nose-to-Brain Delivery of Antiviral Drugs: A Way to Overcome Their Active Efflux? Pharmaceutics 2018, 10, 39. [Google Scholar] [CrossRef]
- Calderón-Peláez, M.-A.; Velandia-Romero, M.L.; Bastidas-Legarda, L.Y.; Beltrán, E.O.; Camacho-Ortega, S.J.; Castellanos, J.E. Dengue Virus Infection of Blood–Brain Barrier Cells: Consequences of Severe Disease. Front. Microbiol. 2019, 10, 1435. [Google Scholar] [CrossRef]
- Bar-Hai, A.; Domb, A.J.; Hoffman, A. Strategies for enhancing the oral bioavailability of cannabinoids. Expert Opin. Drug Metab. Toxicol. 2022, 18, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Thilakarathna, S.H.; Rupasinghe, H.P.V. Flavonoid Bioavailability and Attempts for Bioavailability Enhancement. Nutrients 2013, 5, 3367–3387. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Kakhar Umar, A.; Zothantluanga, J.H.; Luckanagul, J.A.; Limpikirati, P.; Sriwidodo, S. Structure-based computational screening of 470 natural quercetin derivatives for identification of SARS-CoV-2 M(pro) inhibitor. PeerJ 2023, 11, e14915. [Google Scholar] [CrossRef] [PubMed]
- Mulgaonkar, N.; Wang, H.; Mallawarachchi, S.; Růžek, D.; Martina, B.; Fernando, S. In silico and in vitro evaluation of imatinib as an inhibitor for SARS-CoV-2. J. Biomol. Struct. Dyn. 2023, 41, 3052–3061. [Google Scholar] [CrossRef]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Wang, E.; Sun, H.; Wang, J.; Wang, Z.; Liu, H.; Zhang, J.Z.H.; Hou, T. End-Point Binding Free Energy Calculation with MM/PBSA and MM/GBSA: Strategies and Applications in Drug Design. Chem. Rev. 2019, 119, 9478–9508. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Mulgaonkar, N.; Mallawarachchi, S.; Ramasamy, M.; Padilla, C.S.; Irigoyen, S.; Coaker, G.; Mandadi, K.K.; Fernando, S. Evaluation of Candidatus Liberibacter Asiaticus Efflux Pump Inhibition by Antimicrobial Peptides. Molecules 2022, 27, 8729. [Google Scholar] [CrossRef]
- Sharma, A.; Zhang, X.; Dejnirattisai, W.; Dai, X.; Gong, D.; Wongwiwat, W.; Duquerroy, S.; Rouvinski, A.; Vaney, M.-C.; Guardado-Calvo, P.; et al. The epitope arrangement on flavivirus particles contributes to Mab C10’s extraordinary neutralization breadth across Zika and dengue viruses. Cell 2021, 184, 6052–6066. [Google Scholar] [CrossRef]
- Kudlacek, S.T.; Metz, S.; Thiono, D.; Payne, A.M.; Phan, T.T.N.; Tian, S.; Forsberg, L.J.; Maguire, J.; Seim, I.; Zhang, S.; et al. Designed, highly expressing, thermostable dengue virus 2 envelope protein dimers elicit quaternary epitope antibodies. Sci. Adv. 2021, 7, eabg4084. [Google Scholar] [CrossRef] [PubMed]
- Modis, Y.; Ogata, S.; Clements, D.; Harrison, S.C. Variable Surface Epitopes in the Crystal Structure of Dengue Virus Type 3 Envelope Glycoprotein. J. Virol. 2005, 79, 1223–1231. [Google Scholar] [CrossRef]
- Giordano, D.; Biancaniello, C.; Argenio, M.A.; Facchiano, A. Drug Design by Pharmacophore and Virtual Screening Approach. Pharmaceuticals 2022, 15, 646. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Cheng, F.; Li, W.; Zhou, Y.; Shen, J.; Wu, Z.; Liu, G.; Lee, P.W.; Tang, Y. admetSAR: A comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inf. Model. 2012, 52, 3099–3105. [Google Scholar] [CrossRef]
- Yang, H.; Lou, C.; Sun, L.; Li, J.; Cai, Y.; Wang, Z.; Li, W.; Liu, G.; Tang, Y. admetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 2019, 35, 1067–1069. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Kemmler, E.; Dunkel, M.; Preissner, R. ProTox 3.0: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2024, 52, W513–W520. [Google Scholar] [CrossRef] [PubMed]
- Dubrow, A.; Zuniga, B.; Topo, E.; Cho, J.-H. Suppressing Nonspecific Binding in Biolayer Interferometry Experiments for Weak Ligand–Analyte Interactions. ACS Omega 2022, 7, 9206–9211. [Google Scholar] [CrossRef] [PubMed]
- Brand, Y.M.; Roa-Linares, V.; Santiago-Dugarte, C.; Olmo, E.d.; López-Pérez, J.L.; Betancur-Galvis, L.; Gallego-Gómez, J.C.; Feliciano, A.S. A new host-targeted antiviral cyclolignan (SAU-22.107) for Dengue Virus infection in cell cultures. Potential action mechanisms based on cell imaging. Virus Res. 2022, 323, 198995. [Google Scholar] [CrossRef]
- Orthwein, T.; Huergo, L.F.; Forchhammer, K.; Selim, K.A. Kinetic Analysis of a Protein-protein Complex to Determine its Dissociation Constant (KD) and the Effective Concentration (EC50) of an Interplaying Effector Molecule Using Bio-layer Interferometry. Bio-Protocol 2021, 11, e4152. [Google Scholar] [CrossRef]





| DENV1 | DENV2 | DENV3 | DENV4 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β-OG | ED-III | Fusion Loop | β-OG | ED-III | Fusion Loop | β-OG | ED-III | Fusion Loop | β-OG | ED-III | Fusion Loop | |
| Paclitaxel | - | −4.04 | - | - | - | - | - | - | 3.73 | - | - | −2.41 |
| Malacitanolide | −3.06 | −3.38 | −3.17 | −3.28 | - | −4.18 | −1.70 | −3.71 | −3.03 | - | −2.97 | −3.32 |
| Canniflavin A | −2.97 | −3.02 | −4.18 | −4.72 | −3.02 | −3.67 | - | −2.35 | −4.23 | −3.30 | −3.20 | −3.03 |
| CAP1 | −2.85 | −2.66 | −3.03 | −4.44 | −4.44 | −2.48 | −2.86 | −4.64 | −3.52 | −3.26 | −3.62 | −4.16 |
| CAP2 | −3.50 | −3.09 | −3.77 | −4.15 | −3.81 | −3.23 | −3.02 | −3.76 | −3.09 | −2.00 | −3.22 | −4.13 |
| CAP3 | −3.41 | −3.39 | −4.07 | −3.41 | −2.77 | −2.97 | −1.59 | −4.56 | −2.79 | −3.01 | −2.66 | −4.01 |
| Compound 6 | −4.20 | −3.75 | −3.16 | −2.08 | −1.92 | −0.79 | - | −2.18 | −1.68 | −1.07 | −1.05 | −0.86 |
| C6P1 | - | −2.76 | −3.34 | −4.12 | −2.13 | −2.52 | - | −3.07 | −3.16 | −3.05 | −2.77 | −3.32 |
| C6P2 | −2.97 | −3.64 | −3.55 | −4.00 | −3.05 | −3.89 | −3.94 | −3.31 | −3.36 | - | −2.18 | −3.15 |
| C6P3 | −4.08 | −3.57 | −3.75 | −4.03 | −2.87 | −2.73 | −2.78 | −4.52 | −2.66 | −4.05 | −2.21 | −2.74 |
| Name | BBB | HIA | Caco2 Permeability | AMES Mutagenicity | Carcinogenesis | Hepatotoxicity | Biodegradation | Aqueous Solubility LogS | LD50 (mg/kg) |
|---|---|---|---|---|---|---|---|---|---|
| Paclitaxel | - | + | - | - | - | - | - | −3.8727 | 134 |
| Malacitanolide | + | + | - | - | - | - | - | −3.8155 | 1330 |
| Cannflavin A | - | - | - | - | - | - | - | −4.4887 | 3919 |
| CAP 1 | + | + | - | - | - | - | - | −4.9531 | 678 |
| CAP 2 | - | + | - | - | - | - | - | −3.732 | 3120 |
| CAP 3 | - | + | - | - | - | - | - | −3.2751 | 5000 |
| Compound 6 | + | + | - | - | - | - | - | −4.2568 | 1330 |
| C6P 1 | + | + | + | - | - | - | - | −2.8656 | 300 |
| C6P 2 | + | + | - | + | - | - | - | −3.2004 | 1000 |
| C6P3 | + | + | - | - | - | - | - | −3.1335 | 550 |
| Name | Molecular Weight (Da) | AlogP | Num. H-Bond Acceptors | Num. H-Bond Donors | Rule Violations |
|---|---|---|---|---|---|
| Paclitaxel | 972.01 | 3.25 | 18 | 5 | 2 |
| Malacitanolide | 394.42 | −0.1 | 8 | 3 | 0 |
| Canniflavin A | 436.5 | 5.82 | 6 | 3 | 1 |
| CAP1 | 628.1 | 6.98 | 4 | 2 | 2 |
| CAP2 | 490.56 | 3.52 | 6 | 3 | 0 |
| CAP3 | 357.37 | 1.89 | 6 | 3 | 0 |
| Compound 6 | 428.95 | 6.69 | 5 | 1 | 1 |
| C6P1 | 368.4 | 4.29 | 6 | 2 | 0 |
| C6P2 | 413.44 | 4.87 | 8 | 1 | 0 |
| C6P3 | 383.41 | 4.86 | 7 | 1 | 0 |
| Serotype | MM-GBSA Binding Energy (kcal/mol) | |||
|---|---|---|---|---|
| Compound 6 | C6P3 | Canniflavin A | CAP1 | |
| DENV1 | −61.88434 | −42.15755 | −38.46022 | −72.25684 |
| DENV2 | −33.57146 | −33.80145 | −57.20999 | −47.91876 |
| DENV3 | 0.00000 | −51.60349 | 0.00000 | −32.30574 |
| DENV4 | −47.78996 | −59.98014 | −36.47438 | −53.62067 |
| Serotype | Molecule | KD (mM) | ka (1/Ms) | kd (1/s) |
|---|---|---|---|---|
| DENV1 | Paclitaxel | (6.408 ± 1.272) × 10−7 | (792.90 ± 157.90) | 4.883 × 10−7 |
| CAP1 | (7.371 ± 0.697) × 10−4 | (3754.50 ± 1253.50) | (2.680 ± 0.663) × 10−3 | |
| DENV2 | Paclitaxel | (6.635 ± 4.016) × 10−7 | (1161.65 ± 703.35) | 4.883 × 10−7 |
| CAP1 | (1.371 ± 0.958) × 10 | (355.79 ± 304.31) | (1.964 ± 0.765) × 10−1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haq, A.; Mallawarachchi, S.; Anderson, A.; Khaleghi, L.; Manujitha, L.; Fernando, S. In Silico Evaluation of Potential Hit Molecules Against Multiple Serotypes of Dengue Virus Envelope Glycoprotein. Molecules 2025, 30, 1268. https://doi.org/10.3390/molecules30061268
Haq A, Mallawarachchi S, Anderson A, Khaleghi L, Manujitha L, Fernando S. In Silico Evaluation of Potential Hit Molecules Against Multiple Serotypes of Dengue Virus Envelope Glycoprotein. Molecules. 2025; 30(6):1268. https://doi.org/10.3390/molecules30061268
Chicago/Turabian StyleHaq, Aadhil, Samavath Mallawarachchi, Aiden Anderson, Leily Khaleghi, Lasan Manujitha, and Sandun Fernando. 2025. "In Silico Evaluation of Potential Hit Molecules Against Multiple Serotypes of Dengue Virus Envelope Glycoprotein" Molecules 30, no. 6: 1268. https://doi.org/10.3390/molecules30061268
APA StyleHaq, A., Mallawarachchi, S., Anderson, A., Khaleghi, L., Manujitha, L., & Fernando, S. (2025). In Silico Evaluation of Potential Hit Molecules Against Multiple Serotypes of Dengue Virus Envelope Glycoprotein. Molecules, 30(6), 1268. https://doi.org/10.3390/molecules30061268







