Affinity Ultrafiltration Mass Spectrometry for Screening Active Ingredients in Traditional Chinese Medicine: A Review of the Past Decade (2014–2024)
Abstract
1. Introduction
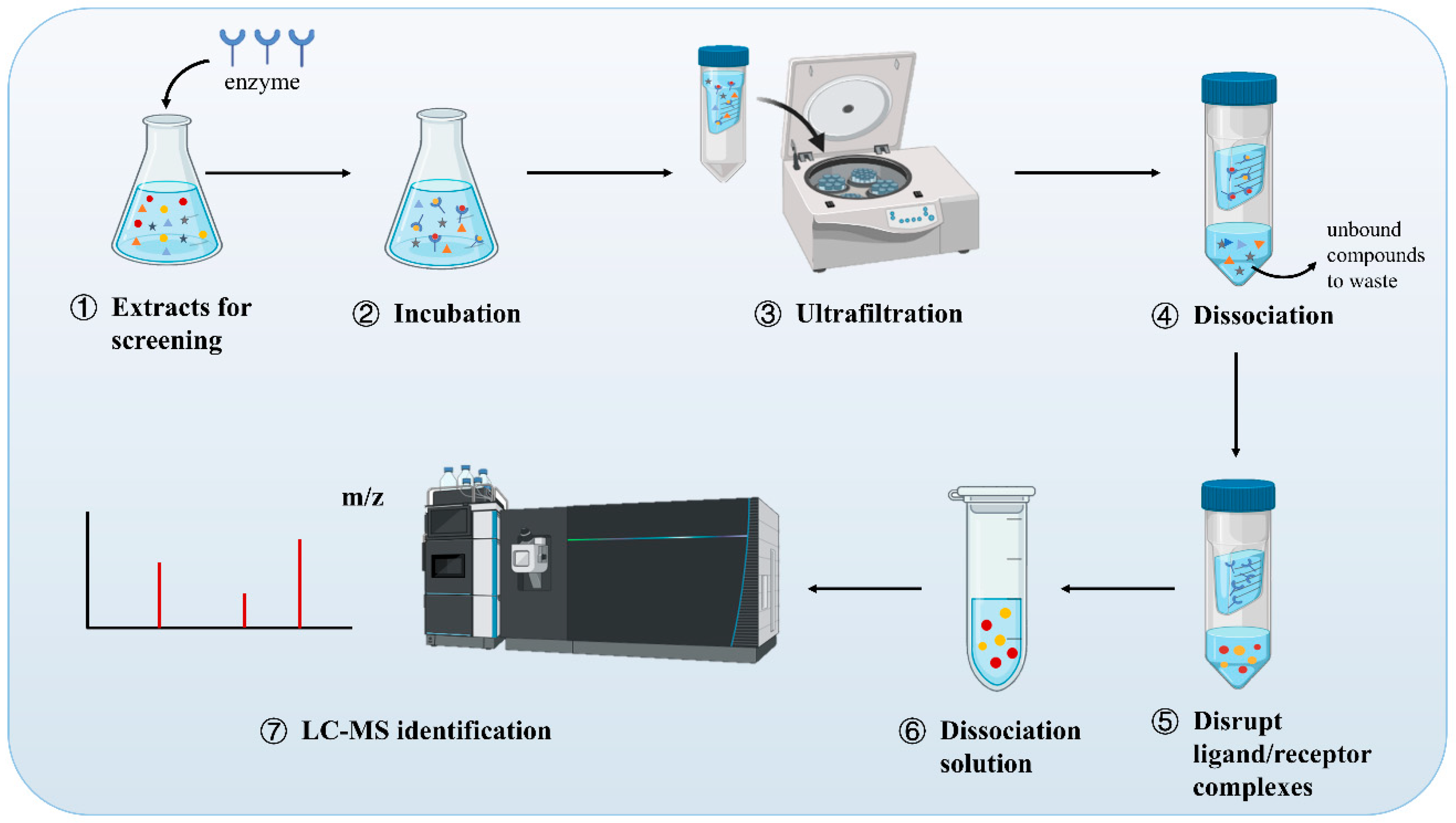
2. AUF-MS: An Overview
3. Advantages and Characteristics of AUF-LC-MS
4. Factors Affecting AUF Screening
4.1. Concentration of the Target and the Screened Substances
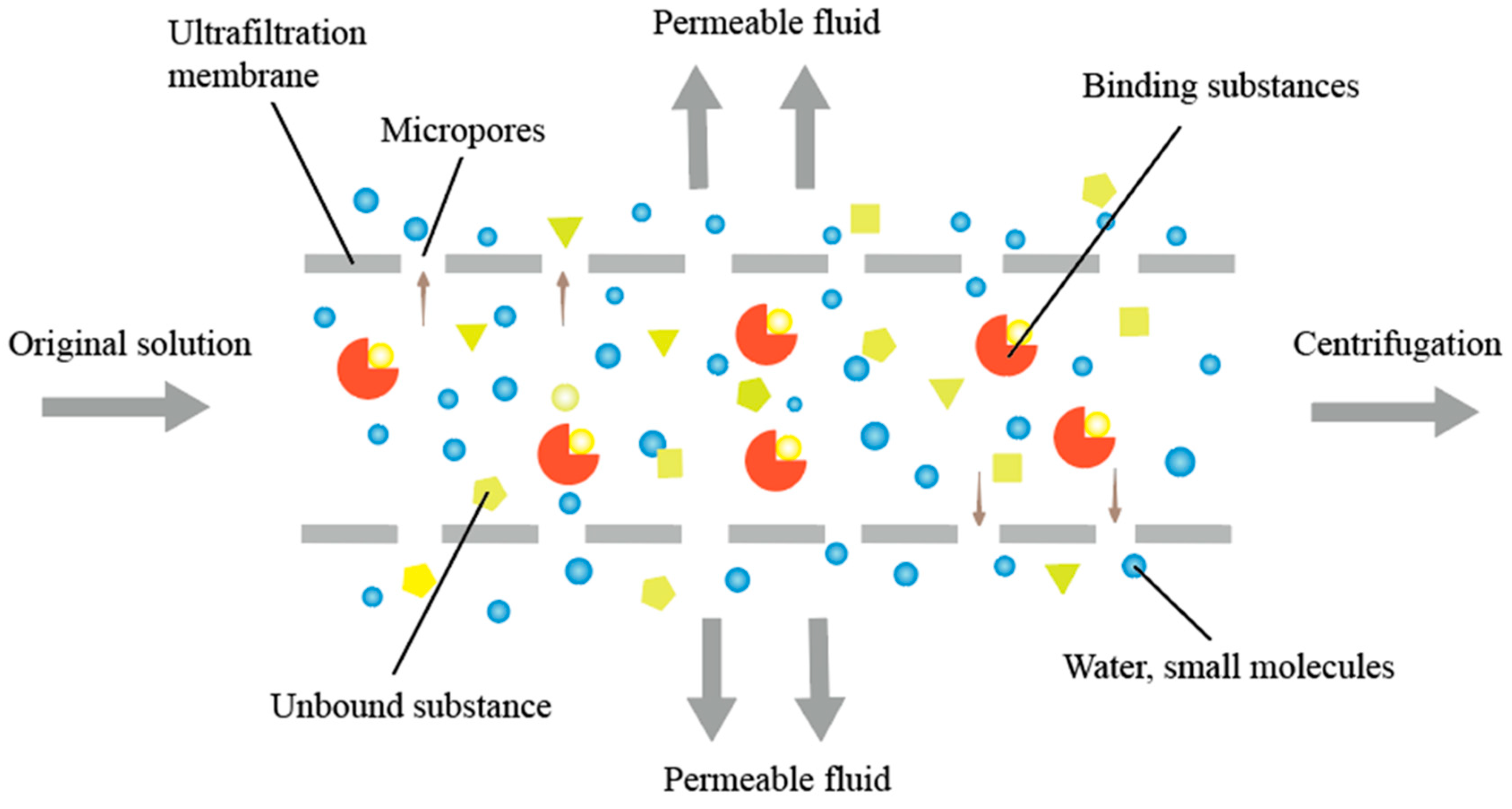
4.2. Ultrafiltration Membrane Material
4.3. Choice of Dissociation Solvent
5. Screening Technology and Application
5.1. High-Throughput Screening (HTS) of Active Ingredients of TCM
| Target Protein | Natural Products | Active Ingredients | Ref. |
|---|---|---|---|
| α-Glucosidase | Panax Ginseng | Twenty-four compounds | [8] |
| Rhizoma Coptidis | Jatrorrhizine, epiberberine, coptisine, palmatine, berberine | [10] | |
| Moringa oleifera leaves | Fourteen compounds | [21] | |
| Perilla frutescens | Nine compounds | [49] | |
| Cichorium glandulosum | Esculetin, chlorogenic acid, isochlorogenic acid B, isochlorogenic acid A | [59] | |
| Cichorium glandulosum | Quercetin, lactucin, 3-O-methylquercetin, hyperoside, lactucopicrin, isochlorogenic acid B | [60] | |
| Rubus suavissimus leaves | Twenty-six compounds | [61] | |
| Inonotus obliquus | (E)-4-(3,4-dihydroxyphenyl) but-3-en-2-one | [62] | |
| Siraitia grosvenorii Roots | Seventeen compounds | [63] | |
| Cichorium glundulosum root | Baicalin, lactupicrin | [66] | |
| Trifolium pratense | Daidzin, ononin, daidzein, genistein, fomononetin, biochanin A | [67] | |
| Cyclocarya paliurus leaves | Mainly damarane-type triterpenoid saponins | [68] | |
| Radix Astragali | Thirteen prototype isoflavonoids and one monohydroxylated metabolic isoflavonoid | [69] | |
| Scutellaria baicalensis Georgi | Baicalin, wogonoside, 5,7,3,2′,6′-pentahydroxy flflavanone, chrysin-6-C-arabinosyl-8-C-glucoside, chrysin-6-C-glucosyl-8-C-arabinoside, wogonin | [70] | |
| Polygonatum odoratum | Five phenethyl cinnamides and four homoisoflavanones | [71] | |
| Scutellaria baicalensis Georgi | Baicalein, baicalein, wogonin, chrysin, oroxylin A | [72] | |
| Ginkgo biloba | Eleven compounds | [73] | |
| Buddleja Flos | Thirteen phenylethanoid glycosides and twenty flavonoids | [74] | |
| Cercis chinensis | Twelve compounds | [75] | |
| Cyclooxygenase 2 | Dysosma versipellis | Nine compounds | [22] |
| Anemarrhenae rhizoma | Timosaponin A-II, timosaponin A-III, timosaponin B-II, timosaponin B-III, anemarrhenasaponin I | [51] | |
| Andrographis paniculata | Andrographolide, 14-deoxy-11,12-didehydroandrographiside, andrographidine E, andrographidine D, deoxyandrographolide | [64] | |
| Andrographis paniculata | Eleven compounds | [65] | |
| Kadsura coccinea | Twenty-one compounds | [76] | |
| Rhamnus davurica | Vitexin, Taxifolin, Aromadendrin, Kaempferol 7-O-glucoside, berberine III, apigenin, kaempferol, rhamnocitrin, sakuranetin, questin, physcion | [77] | |
| Trifolium pratense L. | Rothindin, ononin, daidzein, trifoside, pseudobaptigenin, formononetin, biochanin A | [78] | |
| Paris polyphylla | Polyphyllin I, II, VI, VII | [79] | |
| Curcuma longa | Thirteen compounds | [80] | |
| Sinopodophyllum hexandrum | Rutin, quercetin 3-O-glucoside, kaempferol 3-O-glucoside, β-Apopicropodophyllin, quercetin, isorhamnetin, kaempferol, podophyllotoxin | [81] | |
| Saussurea obvallata | Coniferin, syringin, roseoside, grasshopper ketone | [81] | |
| Moutan cortex | Gallic acid, methyl gallate, galloylpaeoniflflorin, 1,2,3,6-Tetra-O-galloyl-β-D-glucose, 1,2,3,4,6-Penta-O-galloyl-β-D-glucopyranose | [82] | |
| Xanthine oxidase | Perilla frutescens | Kaempferol-3-O-rutinoside, rosmarinic acid, methyl-rosmarinic acid, apigenin, 4′,5,7-trimethoxyflflavone were identifified, from total eleven compounds | [47] |
| Trifolium pratense | Daidzin, ononin, daidzein, genistein, fomononetin, biochanin A | [67] | |
| Panax japlcus var. | 24(R)-majoroside R1, chikusetsusaponin IVa, oleanolic acid-28-O-β-D-glucopyranoside, notoginsenoside Fe, ginsenoside Rb2, ginsenoside Rd | [83] | |
| Flos Chrysanthemum | Luteolin-7-O-glucoside, apigenin-7-O-glucoside, luteolin, apigenin | [84] | |
| Selaginella tamariscina | Amentoflavone, robustaflavone | [85] | |
| Celery seeds (Apium graveolens L.) | Luteolin-7-O-apinosyl glucoside, luteolin-7-O-glucoside, luteolin-7-O-malonyl apinoside, luteolin-7-O-6′-malonyl glucoside, luteolin, apigenin, chrysoeriol | [86] | |
| the roots of Lindera reflexa Hemsl | Pinosylvin, pinocembrin, methoxy-5-hydroxy-trans-stilbene | [87] | |
| Azadirachta indica | Carnosic acid | [88] | |
| Ligusticum chuanxiong | Isochlorogenic acid C, senkyunolide I | [89] | |
| Curcumae Rhizoma | Fifteen compounds | [89] | |
| Curcuma phaeocaulis Valeton | Fifteen compounds | [90] | |
| Polygonum Amplexicaule | Gallic acid, procyanidin B2-3″O-gallate, 11-O-galloylbergenin, (−)-epicatechin gallate, di-galloyl-O-bergenin | [91] | |
| Salvia miltiorrhiza Bge. | Seventeen compounds | [92] | |
| Acetylcholinesterase | Panax ginseng | Ginsenoside Ro, Rb2, Rg1, Re, Rf, Rb1, Rc, Rb3, Rd, Rs1, Ra6, chikusetsusaponin IVa, gypenoside XVl, compound O, pseudoginseoside Rc1, zingibroside R1 | [9] |
| Terminalia chebula fruits | Mainly gallotannins and ellagitannins | [68] | |
| Fibraurea recisa Pierre. | Twelve compounds | [93] | |
| Coptis chinensis Franch | Columbamine, jatrorrhizine, coptisine, palmatine, berberine | [94] | |
| Zanthoxylum nitidum | Jatrorrhizine, columbamine, skimmianine, palmatine, epiberberine | [94] | |
| Azadirachta indica | D-(+)-catechin, (−)-epicatechin, carnosol | [95] | |
| Hedyotis diffusa | Quercetin-3-O-sophoroside, quercetin-3-O-[2-O-(6-O-E-sinapoyl)-β-D-glucopyranosyl]-β-D-glucopyanoside, quercetin-3-O-[2-O-(6-O-E-feruloyl)-β-D-glucopy-ranosyl]-β-D-glucopyranoside, (E)-6-O-p-coumaroyl scandoside methyl ester | [96] | |
| Topoisomerase I | Dysosma versipellis | Twelve compounds | [22] |
| Lycoris radiate | Hippeastrine, camptothecin | [44] | |
| Rhamnus davurica Pall. | Eleven compounds | [77] | |
| Paris polyphylla | Polyphyllin I, II, VI, VII | [79] | |
| Sinopodophyllum hexandrum | Isocorydine, rutin, quercetin 3-O-glucoside, kaempferol 3-O-glucoside, β-apopicropodophyllin, quercetin, isorhamnetin, kaempferol, podophyllotoxin | [80] | |
| Rhamnus davurica | Aromadendrin, naringeninb, apigenin, quercetina, rhamnocitrinb, sakuranetin, questinb, physcionb | [97] | |
| Arachidonate 5-lipoxygenase | Saposhnikovia divaricata (Turcz.) Schischk | Prim-O-glucosylcimifugin, 4′-O-β-D-glucosyl-5-O-methylvisamminol, cimifugin, sec-O-glucosylhamaudol | [98] |
| Smilax glabra Roxb. | Astilbin, isoastilbin, engelitin, isoengelitin, resveratrol | [98] | |
| Pueraria lobata | puerarin, daidzin, 3′-methoxy-puerarin, 3′-hydroxy-puerarin, daidzein | [98] | |
| Carthamus tinctorius | Hydroxyl safflower yellow A, anhydrosafflor yellow B | [98] | |
| Radix Saposhnikoviae via | Prim-O-glucosylcimifugin, cimifugin, 5-O-methylvisamminol, sec-O-glucosylhamaudol, hamaudol | [99] | |
| Topoisomerase II | Dysosma versipellis | Twelve compounds | [22] |
| Paris polyphylla | Polyphyllin I, II, VI, VII | [79] | |
| Sinopodophyllum hexandrum | Isocorydine, rutin, quercetin 3-O-glucoside, quercetin, isorhamnetin, kaempferol | [80] | |
| Augmented reality | Lysimachia christinae | 1,5-di-hydroxy-1,5-di-[(E)-3-(4-hydroxyphenyl)-2-propenoic]-3-pentanonyl | [100] |
| Peruvian tea plant infusions | Chlorogenic acid, 3,5-di-O-caffeoylquinic acid, 1,3,5-tri-O-caffeoylquinic acid | [101] | |
| Hypericum laricifolium Juss. | Protocatechuic acid, chlorogenic acid, caffeic acid, kaempferol 3-O-glucuronide, quercetin, kaempferol | [102] | |
| Neuraminidase | Polygonum cuspidatum | Trans-polydatin, cis-polydatin, emodin-1-O-β-D-glucoside, emodin-8-O-β-D-glucoside, emodin | [103] |
| Baphicacanthus cusia | 2,4(1H,3H)-quinazolinedione, 4(3H)-quinazolinone, 2(3H)-benzoxazolone, tryptanthrin, indirubin | [104] | |
| Angelica pubescens | Thirteen compounds | [105] | |
| Angiotensin-converting enzyme 2 | Dysosma versipellis | Twelve compounds | [22] |
| Andrographis paniculata | Eleven compounds | [65] | |
| Sinopodophyllum hexandrum | Isocorydine, rutin, quercetin 3-O-glucoside, kaempferol 3-O-glucoside, β-apopicropodophyllin, isorhamnetin, kaempferol, podophyllotoxin | [83] | |
| Pancreatic lipase | Moringa oleifera leaves | Eleven compounds | [21] |
| Dendrobium officinale | Vicenin II, isoschaftoside, schaftoside, vitexin 2″-O-glucoside, vitexin 2″-O-rhamnoside, rutin, isoquercetrin, kaempferol 3-O-β-D-glucopyranoside, naringenine, linolenic acid, palmitic acid | [106] | |
| Protein Tyrosine Phosphatase-1B | Puerariae Lobatae Radix | Daidzin, Puerarin | [107] |
| Black tea | (−)-epicatechin-3-O-gallate, epigallocatechin gallate, positive control | [107] | |
| Estrogen receptor | Arnebia euchroma | Twenty-one compounds | [108] |
| G-quadruplex DNA | Macleaya cordata | Protopine, allocryptopine, sanguinarne, chelerythrine | [109] |
| Superoxide dismutase | Azadirachta indica | Gallic acid, protocatechuic acid, (−)-epicatechin | [88] |
| Lactate dehydrogenase | Trifolium pratense | Biochanin A, genistein, fomononetin, ononin | [67] |
| Monoamine oxidase type-B | Panax ginseng | Ginsenoside Rg1, Re, Rb1, Rc, Ro, Rb2, Rd | [9] |
| N-Methyl-D-aspartic acid | Panax ginseng | Ginsenoside Rg1, Re, Rf | [9] |
| Matrix Metallopeptidase 2 | Smilax glabra Roxb., Smilax china L., Saposhnikovia divaricate (Turcz.) Schischk | Resveratrol, engelitin, asibinn, 4′-O-β-D-glucosyl-5-O-methylvisamminol, cimifugin, prim-O-glucosylcimifugin, sec-O-glucosylhamaudol | [110] |
| UDP-glucuronosyltransferase 1A1 | Polygonum multiflorum root | Cis-2,3,5,4′-tetrahydroxystilbene-2-O-β-glucoside, trans-2,3,5,4′-tetrahydroxystilbene-2-O-β-d-glucoside, emodin-8-O-β-d-glucoside, emodin | [111] |
| Interleukin-6 | Andrographis paniculata | Eleven compounds | [65] |
| Tyrosinase | Dryopteris crassirhizoma rhizome | Twenty-two compounds | [112] |
| Lactate dehydrogenases | Azadirachta Indica | Carnosol | [95] |
| Epidermal growth factor receptor erbB1 | Psoralea Fructus | Psorachalcone A, psoralen, bakuchalcone | [113] |
5.2. Screening of Active Ingredients in TCM Compound Preparations
6. Fingerprint Analysis of the Active Components of TCM
7. Analysis of Metabolites of Small-Molecule Drugs
8. Summary and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Calixto, J. The role of natural products in modern drug discovery. An. Acad. Bras. Cienc. 2019, 91, e20190105. [Google Scholar] [CrossRef]
- Song, H.; Yang, H.; Gao, W.; Chen, J.; Li, P. A progress on the key technologies for discovery of bioactive compounds from traditional Chinese medicines. World Sci. Technol. Mod. Tradit. Chin. Med. 2016, 18, 1093–1098. [Google Scholar]
- Li, S.P.; Zhao, J.; Yang, B. Strategies for quality control of Chinese medicines. J. Pharm. Biomed. Anal. 2011, 55, 802–809. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Huang, B.; Guo, M. Current advances in screening for bioactive components from medicinal plants by affinity ultrafiltration mass spectrometry. Phytochem. Anal. 2018, 29, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Zhao, J.; Zhong, F.; Zhu, W.; Jiang, J.; Wu, S.; Yang, D.; Li, D.; Quan, L. Biotransformation of Panax ginseng extract by rat intestinal microflora: Identification and quantification of metabolites using liquid chromatography-tandem mass spectrometry. J. Ginseng Res. 2017, 41, 540–547. [Google Scholar] [CrossRef] [PubMed]
- Jeong, D.; Irfan, M.; Kim, S.; Kim, S.; Oh, J.; Park, C.; Kim, H.; Rhee, M. Ginsenoside Rg3-enriched red ginseng extract inhibits platelet activation and in vivo thrombus formation. J. Ginseng Res. 2017, 41, 548–555. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Jiang, L.; Xu, C.; Luo, D.; Zeng, C.; Liu, P.; Yue, M.; Liu, Y.; Hu, X.; Hu, H. Ginsenoside Rg1 inhibits platelet activation and arterial thrombosis. Thromb. Res. 2014, 133, 57–65. [Google Scholar] [CrossRef]
- Wang, H.; Fan, C.; Lin, Z.; Yin, Q.; Zhao, C.; Peng, P.; Zhang, R.; Wang, Z.; Du, J.; Wang, Z. Screening of potential α-Glucosidase inhibitors from the roots and rhizomes of Panax Ginseng by affinity ultrafiltration screening coupled with UPLC-ESI-Orbitrap-MS method. Molecules 2023, 28, 2069. [Google Scholar] [CrossRef]
- Yu, L.; Wei, F.; Liang, J.; Ren, G.; Liu, X.; Wang, C.; Yuan, J.; Zeng, J.; Luo, Y.; Bi, Y.; et al. Target molecular-based neuroactivity screening and analysis of Panax ginseng by affinity ultrafiltration, UPLC-QTOF-MS and molecular docking. Am. J. Chin. Med. 2019, 47, 1345–1363. [Google Scholar] [CrossRef]
- Tang, Y.; Li, S.; Li, S.; Yang, X.; Qin, Y.; Liu, C.; Zhang, Y. Screening and isolating potential α-glucosidase inhibitors from Rhizoma Coptidis by ultrafiltration LC-PDA-ESI/MS combined with high-speed countercurrent chromatography and reverse-phase medium-pressure liquid chromatography. Med. Chem. Res. 2017, 26, 3384–3394. [Google Scholar] [CrossRef]
- Lan, Z.; Yang, R.; Wang, H.; Xue, X.; Sun, Y.; Wang, S.; Zhang, Y.; Meng, J. Rapid identifying of COX-2 inhibitors from turmeric (Curcuma longa) by bioaffinity ultrafiltration coupled with UPLC-Q Exactive-Orbitrap-MS and zebrafish-based in vivo validation. Bioorg. Chem. 2024, 147, 107357. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Yang, H.; Li, P. Application of the affinity ultrafiltration coupled with LC-MS technology in screening active components of traditional Chinese medicines. Acta Pharm. Sin. 2016, 51, 1060–1067. [Google Scholar]
- Whitlam, J.B.; Brown, K.F. Ultrafiltration in serum protein binding determinations. J. Pharm. Sci. 1981, 70, 146–150. [Google Scholar] [CrossRef]
- Babine, R.E.; Bender, S.L. Molecular recognition of protein-ligand complexes: Applications to drug design. Chem. Rev. 1997, 97, 1359–1472. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, R.; Freudenberg, U.; Schweiss, R.; Küttner, D.; Werner, C. Hydroxide and hydronium ion adsorption—A survey. Curr. Opin. Colloid Interface Sci. 2010, 15, 196–202. [Google Scholar] [CrossRef]
- Yang, X.; Hsia, T.; Merenda, A.; AL-Attabi, R.; Dumee, L.; Thang, S.; Kong, L. Constructing novel nanofibrous polyacrylonitrile (PAN)-based anion exchange membrane adsorber for protein separation. Sep. Purif. Technol. 2022, 285, 120364. [Google Scholar] [CrossRef]
- Ye, J.; Wang, X.; Chu, J.; Yao, D.; Zhang, Y.; Meng, J. Electrospun poly(styrene-co-maleic anhydride) nanofibrous membrane: A versatile platform for mixed mode membrane adsorbers. Appl. Surf. Sci. 2019, 484, 62–71. [Google Scholar] [CrossRef]
- Li, J.; Ge, J.; Yin, Y.; Zhong, W. Multiplexed affinity-based protein complex purification. Anal. Chem. 2008, 80, 7068–7074. [Google Scholar] [CrossRef]
- Tran, T.; Gustavsson, R.; Martinsson, E.; Bergqvist, F.; Axen, A.; Lundström, I.; Mandenius, C.; Aili, D. In-line fiber optical sensor for detection of IgG aggregates in affinity chromatography. J. Chromatogr. A 2024, 1730, 465129. [Google Scholar] [CrossRef]
- Cloutier, T.E.; Comess, K.M. Library Screening Using Ultrafiltration and Mass Spectrometry. In Mass Spectrometry in Medicinal Chemistry: Applications in Drug Discovery, Methods and Principles in Medicinal Chemistry; Wanner, K.T., Höfner, G., Eds.; Wiley-VCH: Weinheim/Berlin, Germany, 2007; pp. 157–183. [Google Scholar] [CrossRef]
- Chen, G.; Xu, Y.; Wu, J.; Li, N.; Guo, M. Hypoglycemic and hypolipidemic effects of Moringa oleifera leaves and their functional chemical constituents. Food Chem. 2020, 333, 127478. [Google Scholar] [CrossRef]
- Feng, H.; Chen, G.; Zhang, Y.; Guo, M. Potential multifunctional bioactive compounds from Dysosma versipellis explored by bioaffinity ultrafiltration-HPLC/MS with Topo I, Topo II, COX-2 and ACE2. J. Inflamm. Res. 2022, 15, 4677–4692. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-J.; Chen, S.; Woodbury, C.P.; Venton, D.L. Pulsed ultrafiltration characterization of binding. Anal. Biochem. 1998, 261, 164–182. [Google Scholar] [CrossRef]
- Van Breemen, R.B.; Huang, C.R.; Nikolic, D.; Woodbury, C.P.; Zhao, Y.; Venton, D.L. Pulsed ultrafiltration mass spectrometry: A new method for screening combinatorial libraries. Anal. Chem. 1997, 69, 2159–2164. [Google Scholar] [CrossRef] [PubMed]
- Beverly, M.B.; West, P.; Julian, R.K. Evaluation of a micro volume pulsed ultrafiltration cell for screening ligands in non-covalent complexes. Comb. Chem. High Throughput Screen. 2002, 5, 65–73. [Google Scholar] [CrossRef]
- Yates, J.R., III. A century of mass spectrometry: From atoms to proteomes. Nat. Methods 2011, 8, 633–637. [Google Scholar] [CrossRef]
- Bennett, J.L.; Nguyen, G.T.H.; Donald, W.A. Protein-Small Molecule Interactions in Native Mass Spectrometry. Chem. Rev. 2022, 122, 7327–7385. [Google Scholar] [CrossRef]
- Wieboldt, R.; Zweigenbaum, J.; Henion, J. Immunoaffinity ultrafiltration with ion spray HPLC/MS for screening small-molecule libraries. Anal. Chem. 1997, 690, 1683–1691. [Google Scholar] [CrossRef]
- Jian, J.; Chen, H.; Hong, Q.; Wang, L.; Zhao, Y.; Li, L.; Zhang, T.; Zhou, H.; Jiang, Z. Research progress in screening technology for active pharmaceutical ingredients of natural products based on liquid chromatography separation. Acta Pharm. Sin. 2020, 55, 1504–1510. [Google Scholar]
- Ma, W.; Wang, C.; Liu, R.; Wang, N.; Lv, Y.; Dai, B.; He, L. Advances in cell membrane chromatography. J. Chromatogr. A 2021, 1639, 461916. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shi, S.; Chen, X.; Peng, M. Functionalized magnetic nanoparticles coupled with mass spectrometry for screening and identification of cyclooxygenase-1 inhibitors from natural products. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2014, 960, 126–132. [Google Scholar] [CrossRef]
- Jin, P.; Chen, L.; Zhong, J.; Yuan, T.; Gan, L.; Huang, J.; Wang, L.; Fan, H.; Lin, C. Screening and identification of lipase inhibitors extracted from Dioscorea nipponica Makino by UV-vis and HPLC coupled to UPLC-Q-TOF-MS/MS. Int. J. Biol. Macromol. 2023, 230, 123427. [Google Scholar] [CrossRef] [PubMed]
- Shishodia, S.; Nuñez, R.; Strohmier, B.P.; Bursch, K.L.; Goetz, C.J.; Olp, M.D.; Jensen, D.R.; Fenske, T.G.; Ordonez-Rubiano, S.C.; Blau, M.E.; et al. Selective and cell-active PBRM1 bromodomain inhibitors discovered through NMR fragment screening. J. Med. Chem. 2022, 65, 13714–13735. [Google Scholar] [CrossRef]
- Yan, F.; He, S.; Han, X.; Wang, J.; Tian, X.; Wang, C.; James, T.D.; Cui, J.; Ma, X.; Feng, L. High-throughput fluorescent screening of β-lactamase inhibitors to improve antibiotic treatment strategies for tuberculosis. Biosens. Bioelectron. 2022, 216, 114606. [Google Scholar] [CrossRef]
- Liang, Q.; Huang, Y.; Wang, M.; Kuang, D.; Yang, J.; Yi, Y.; Shi, H.; Li, J.; Yang, J.; Li, G. An electrochemical biosensor for SARS-CoV-2 detection via its papain-like cysteine protease and the protease inhibitor screening. Chem. Eng. J. 2023, 452, 139646. [Google Scholar] [CrossRef] [PubMed]
- Mulabaqal, V.; Calderón, A.I. Development of an ultrafiltration-liquid chromatography/mass spectrometry (UF-LC/MS) based ligand-binding assay and an LC/MS based functional assay for Mycobacterium tuberculosis Shikimate Kinase. Anal. Chem. 2010, 82, 3616–3621. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, X.; Wang, X.; Fan, X.; Liu, K.; Sa, Y.; Wilson, G.; Ma, X.; Chen, G. Establishment and application of a screening method for α-glucosidase inhibitors based on dual sensing and affinity chromatography. J. Chromatogr. A 2024, 1720, 464822. [Google Scholar] [CrossRef]
- Liu, Y.; Hu, J.; Lin, L.; Yang, B.; Huang, M.; Chang, M.; Huang, X.; Dai, Z.; Sun, S.; Ren, L.; et al. Overcoming the fluorescent interference during Raman spectroscopy detection of microplastics. Sci. Total Environ. 2023, 897, 165333. [Google Scholar] [CrossRef]
- Leong, I.; Kishimoto, S.; Tsutsui, M.; Taniguchi, M. Interference of electrochemical ion diffusion in nanopore sensing. iScience 2022, 25, 105073. [Google Scholar] [CrossRef]
- Johnson, B.; Nikolic, D.; van Breemen, R. Applications of pulsed ultrafiltration-mass spectrometry. Mass Spectrom. Rev. 2002, 21, 76–86. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Wang, S.; Geng, X. Coating and fusing cell membranes onto a silica surface and their chromatographic characteristics. Chromatographia 2001, 54, 71–76. [Google Scholar] [CrossRef]
- Yasuda, M.; Wilson, D.; Fugmann, S.; Moaddel, R. Synthesis and characterization of SIRT6 protein coated magnetic beads: Identification of a novel inhibitor of SIRT6 deacetylase from medicinal plant extracts. Anal. Chem. 2011, 83, 7400–7407. [Google Scholar] [CrossRef]
- Zhang, H.; Yao, J.; Xiao, G.; Xie, J.; Mao, S.; Sun, C.; Yao, J.; Yan, J.; Tu, P. Discovery of drug targets based on traditional Chinese medicine microspheres (TCM-MPs) fishing strategy combined with bio-layer interferometry (BLI) technology. Anal. Chim. Acta 2024, 1305, 342542. [Google Scholar] [CrossRef]
- Zhao, H.; Zhang, Y.; Xiao, C. Advances in the study of affinity selection-ultrafiltration/HPLC-MS. Acta Pharm. Sin. 2009, 44, 1084–1088. [Google Scholar]
- Lohmann, W.; Karst, U. Generation and identification of reactive metabolites by electrochemistry and immobilized enzymes coupled on-line to liquid chromatography/mass spectrometry. Anal. Chem. 2007, 79, 6831–6839. [Google Scholar] [CrossRef]
- Wang, S.; Sun, M.; Zhang, Y.; Du, H.; He, L. A new A431/cell membrane chromatography and online high performance liquid chromatography/mass spectrometry method for screening epidermal growth factor receptor antagonists from Radix sophorae flavescentis. J. Chromatogr. A 2010, 1217, 5246–5252. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Kwon, S.H.; Hwang, S.H.; Kang, Y.-H.; Lee, J.-Y.; Lim, S.S. Competitive binding experiments can reduce the false positive results of affinity-based ultrafiltration-HPLC: A case study for identification of potent xanthine oxidase inhibitors from Perilla frutescens extract. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2017, 1048, 30–37. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, Y.; Sun, L.; Wang, Y.; Gao, X.; Cheng, Y. An ultrafiltration high-performance liquid chromatography coupled with diode array detector and mass spectrometry approach for screening and characterizing tyrosinase inhibitors from mulberry leaves. Anal. Chim. Acta 2012, 719, 87–95. [Google Scholar] [CrossRef]
- Wang, Z.; Zuo, G.; Hwang, S.H.; Kwon, S.H.; Kang, Y.-H.; Lee, J.-Y.; Lim, S.S. Affinity measurement of ligands in Perilla frutescens extract towards α-glucosidase using affinity-based ultrafiltration-high-performance liquid chromatography. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2019, 1125, 121725. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhao, B.; Li, C.; Wang, K.; Zhang, T.; Wang, S.; Liu, R.; He, J. Research progress on screening active components from medicinal plants based on affinity ultrafiltration coupled with LC-MS technology. Chin. J. Exp. Tradit. Med. Formulae 2021, 27, 196–208. [Google Scholar]
- Xie, L.; Lee, D.Y.-W.; Shang, Y.; Cao, X.; Wang, S.; Liao, J.; Zhang, T.; Dai, R. Characterization of spirostanol glycosides and furostanol glycosides from anemarrhenae rhizoma as dual targeted inhibitors of 5-lipoxygenase and Cyclooxygenase-2 by employing a combination of affinity ultrafiltration and HPLC/MS. Phytomedicine 2020, 77, 153284. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Liu, S.; Li, X.; Song, F.; Liu, Z.; Liu, S. Bioactivity fingerprint analysis of cyclooxygenase-2 ligands from radix Aconiti by ultrafiltration-UPLC-MSn. Anal. Bioanal. Chem. 2013, 405, 7437–7445. [Google Scholar] [CrossRef] [PubMed]
- Mulabagal, V.; Calderón, A.I. Development of binding assays to screen ligands for Plasmodium falciparum thioredoxin and glutathione reductases by ultrafiltration and liquid chromatography/mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2010, 878, 987–993. [Google Scholar] [CrossRef]
- McDougall, G.J.; Stewart, D. The inhibitory effects of berry polyphenols on digestive enzymes. BioFactors 2005, 23, 189–195. [Google Scholar] [CrossRef]
- Mohan, S.; Eskandari, R.; Pinto, B.M. Naturally occurring sulfonium-ion glucosidase inhibitors and their derivatives: A promising class of potential antidiabetic agents. Acc. Chem. Res. 2014, 47, 211–225. [Google Scholar] [CrossRef]
- Kihara, Y.; Ogami, Y.; Tabaru, A.; Unoki, H.; Otsuki, M. Safe and effective treatment of diabetes mellitus associated with chronic liver diseases with an alphaglucosidase inhibitor, acarbose. J. Gastroenterol. 1997, 32, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.; Wu, H.; Lv, Q.; Aisa, H.A. Hypoglycemic Effect of Extracts from Cichorium glandulosum. Nat. Prod. Res. Dev. 2012, 24, 234–238. [Google Scholar]
- Dalar, A.; Konczak, I. Cichorium intybus from Eastern Anatolia: Phenolic composition, antioxidant and enzyme inhibitory activities. Ind. Crops Prod. 2014, 60, 79–85. [Google Scholar] [CrossRef]
- Chen, H.; Qin, H.; Long, F.; Yu, W.; Wang, Y.; Chen, L.; Li, Q.; Chen, W.; Qin, D.; Han, B. Screening of high-affinity α-glucosidase inhibitors from cichorium glandulosum boiss. et Hout seed based on ultrafiltration liquid chromatography-mass spectrometry and molecular docking. Anal. Chem. 2017, 45, 889–897. [Google Scholar]
- Abudurexiti, A.; Abdurahman, A.; Zhang, R.; Zhong, Y.; Lei, Y.; Qi, S.; Hou, W.; Ma, X. Screening of α-Glucosidase inhibitors in Cichorium glandulosum Boiss. et Huet extracts and study of interaction mechanisms. ACS Omega 2024, 9, 19401–19417. [Google Scholar] [CrossRef]
- Liu, M.; Li, X.; Liu, Q.; Xie, S.; Chen, M.; Wang, L.; Feng, Y.; Chen, X. Comprehensive profiling of α-glucosidase inhibitors from the leaves of Rubus suavissimus using an off-line hyphenation of HSCCC, ultrafiltration HPLC-UV-MS and prep-HPLC. J. Food Compos. Anal. 2020, 85, 103336. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, L.; Liu, C.; Zhang, Y.; Li, S. Isolation of potential α-glucosidase inhibitor from Inonotus obliquus by combining ultrafiltration-liquid chromatography and consecutive high-speed countercurrent chromatography. Anal. Methods 2021, 13, 918–924. [Google Scholar] [CrossRef]
- Lu, F.; Sun, J.; Jiang, X.; Song, J.; Yan, X.; Teng, Q.; Li, D. Identification and isolation of α-Glucosidase inhibitors from Siraitia grosvenorii roots using bio-affinity ultrafiltration and comprehensive chromatography. Int. J. Mol. Sci. 2023, 24, 10178. [Google Scholar] [CrossRef]
- Jiao, J.; Yang, Y.; Wu, Z.; Li, B.; Zheng, Q.; Wei, S.; Wang, Y.; Yang, M. Screening cyclooxygenase-2 inhibitors from Andrographis paniculata to treat inflammation based on bio-affinity ultrafiltration coupled with UPLC-Q-TOF-MS. Fitoterapia 2019, 137, 104259. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Chen, G.; Guo, M. Exploring multifunctional components from Andrographis paniculata by affinity ultrafiltration with three molecular targets. Food Chem. 2023, 404, 134515. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Ma, S.; Jiang, M.; Wang, Q.; Zang, J.; Qin, H.; Chen, W.; Han, B. Screening of α-glucosidase inhibitors from roots of Cichorium glundulosum by UF-LC-MS and molecular docking. Chin. Tradit. Herb. Drugs 2019, 50, 344–351. [Google Scholar]
- Hao, Y.; Liu, C.; Li, S.; Wang, Y.; Hou, W.; Wu, T. Screening of bioactive ligands in Trifolium pratense byaffinity ultrafiltration mass spectrometry. North Hortic. 2019, 17, 102–107. [Google Scholar]
- Li, Y. Studies on affinity screening acetylcholinesterase and α-glucosidase inhibitors from traditional Chinese medicines and interactions with enzymes. PhD Thesis, Lanzhou University, Lanzhou, Gansu, China, 2023. [Google Scholar]
- Zhao, H.; Zhang, Y.; Guo, Y.; Shi, S. Identification of major α-glucosidase inhibitors in Radix Astragali and its human microsomal metabolites using ultrafiltration HPLC-DAD-MSn. J. Pharm. Biomed. Anal. 2015, 104, 31–37. [Google Scholar] [CrossRef]
- Wang, J.; Liu, S.; Li, S.; Song, F.; Zhang, Y.; Liu, Z.; Liu, C. Ultrafiltration LC-PDA-ESI/MS combined with reverse phase-medium pressure liquid chromatography for screening and isolation potential α-glucosidase inhibitors from Scutellaria baicalensis Georgi. Anal. Methods 2014, 6, 5918–5924. [Google Scholar] [CrossRef]
- Zhou, X.; Liang, J.; Zhang, Y.; Zhao, H.; Guo, Y.; Shi, S. Separation and purification of α-glucosidase inhibitors from Polygonatum odoratum by stepwise high-speed counter-current chromatography combined with Sephadex LH-20 chromatography target-guided by ultrafiltration-HPLC screening. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2015, 985, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Luo, J.; Kong, L. Determination of α-glucosidase inhibitors from Scutellaria baicalensis using liquid chromatography with quadrupole time of flight tandem mass spectrometry coupled with centrifugal ultrafiltration. Chin. J. Nat. Med. 2015, 13, 208–214. [Google Scholar] [CrossRef]
- Wu, B.; Song, H.; Zhou, X.; Liu, X.; Gao, W.; Dong, X.; Li, H.; Li, P.; Yang, H. Screening of minor bioactive compounds from herbal medicines by in silico docking and the trace peak exposure methods. J. Chromatogr. A 2016, 1436, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Fu, Q.; Shi, S.; Li, J.; Zhou, X. Rapid and comprehensive profiling of α-glucosidase inhibitors in Buddleja flos by ultrafiltration HPLC-QTOF-MS/MS with diagnostic ions filtering strategy. Food Chem. 2021, 344, 128651. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Liao, X.; Chen, Z. Screening and characterization of potential α-glucosidase inhibitors from Cercis chinensis Bunge fruits using ultrafiltration coupled with HPLC-ESI-MS/MS. Food Chem. 2022, 372, 131316. [Google Scholar] [CrossRef]
- Sun, X. Study on cyclooxygenase-2 inhibitors in Kadsura coccinea based on affinity ultrafiltration coupled with high-performance liquid chromatography and quadruple time-of-flight mass spectrometry. PhD Thesis, Zhengzhou University, Zhengzhou, Henan, China, 2020. [Google Scholar]
- Chen, G.; Wu, J.; Li, N.; Guo, M. Screening for anti-proliferative and anti-inflammatory components from Rhamnus davurica Pall. using bio-affinity ultrafiltration with multiple drug targets. Anal. Bioanal. Chem. 2018, 410, 3587–3595. [Google Scholar] [CrossRef]
- Hou, W.; Li, S.; Li, S.; Shi, D.; Liu, C. Screening and isolation of cyclooxygenase-2 inhibitors from Trifolium pratense L. via ultrafiltration, enzyme-immobilized magnetic beads, semi-preparative high-performance liquid chromatography and high-speed counter-current chromatography. J. Sep. Sci. 2019, 42, 1133–1143. [Google Scholar] [CrossRef]
- Chen, G.; Guo, M. Rapid re-evaluation of bioactive saponins from Paris polyphylla using affinity ultrafiltration-LC/MS with multiple drug targets. Int. J. Mass Spectrom. 2018, 434, 87–92. [Google Scholar] [CrossRef]
- Feng, H.; Chen, G.; Zhang, Y.; Guo, M. Potential multiple bioactive components from Sinopodophyllum hexandrum explored by affinity ultrafiltration with four drug targets. Phytomed. Plus 2022, 2, 100219. [Google Scholar] [CrossRef]
- Wang, W.; Mei, L.; Yue, H.; Tao, Y.; Liu, Z. Targeted isolation of cyclooxygenase-2 inhibitors from Saussurea obvallata using affinity ultrafiltration combined with preparative liquid chromatography. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2023, 1217, 123620. [Google Scholar] [CrossRef]
- Zou, C.; Chen, Q.; Li, J.; Lin, X.; Xue, X.; Cai, X.; Chen, Y.; Sun, Y.; Wang, S.; Zhang, Y.; et al. Identification of potential anti-inflammatory components in Moutan Cortex by bio-affinity ultrafiltration coupled with ultra-performance liquid chromatography mass spectrometry. Front. Pharmacol. 2024, 15, 1358640. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Tang, Y.; Liu, C.; Li, J.; Guo, L.; Zhang, Y. Development of a method to screen and isolate potential xanthine oxidase inhibitors from Panax japlcus var via ultrafiltration liquid chromatography combined with counter-current chromatography. Talanta 2015, 134, 665–673. [Google Scholar] [CrossRef]
- Song, H.; Zhang, H.; Fu, Y.; Mo, H.; Zhang, M.; Chen, J.; Li, P. Screening for selective inhibitors of xanthine oxidase from Flos Chrysanthemum using ultrafiltration LC-MS combined with enzyme channel blocking. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2014, 961, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, S.; Ma, B.; Chen, L.; Song, F.; Liu, Z.; Liu, C. Rapid screening and detection of XOD inhibitors from S. tamariscina by ultrafiltration LC-PDA-ESI-MS combined with HPCCC. Anal. Bioanal. Chem. 2014, 406, 7379–7387. [Google Scholar] [CrossRef] [PubMed]
- Gan, X.; Peng, B.; Chen, L.; Jiang, Y.; Li, T.; Li, B.; Liu, X. Identification of xanthine oxidase inhibitors from celery seeds using affinity ultrafiltration-liquid chromatography-mass spectrometry. Molecules 2023, 28, 6048. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Yang, J.; Chen, S.; Sun, X.; Zhao, P.; Xie, Z. Screening, and identification of the binding position, of xanthine oxidase inhibitors in the roots of Lindera reflexa Hemsl using ultrafiltration LC-MS combined with enzyme blocking. Biomed. Chromatogr. 2019, 33, e4577. [Google Scholar] [CrossRef]
- Fan, M.; Chen, G.; Guo, M. Potential antioxidative components in Azadirachta indica revealed by bio-affinity ultrafiltration with SOD and XOD. Antioxidants 2022, 11, 658. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yang, Y.; Li, S.; Wang, Y.; Yang, F.; Chen, H.; Xia, Z. An ultrafiltration and high performance liquid chromatography coupled with diode array detector and mass spectrometry approach for screening and characterizing thrombin inhibitors from Rhizoma Chuanxiong. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2017, 1061–1062, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Lan, Z.; Zhang, Y.; Sun, Y.; Wang, L.; Huang, Y.; Cao, H.; Wang, S.; Meng, J. Identifying of anti-thrombin active components from Curcumae rhizoma by affinity-ultrafiltration coupled with UPLC-Q-Exactive Orbitrap/MS. Front. Pharmacol. 2021, 10, 769021. [Google Scholar] [CrossRef]
- Huang, C.; Yan, S.; Zhao, Y.; He, X.; Huang, S.; Zhao, H.; Yuan, Z.; Xu, G. Determination of anti-thrombin active components in Polygonum amplexicaule radix by affinity-ultrafiltration (AUF) and high-performance-tandem mass spectrometry (HPLC-MS/MS). Anal. Lett. 2024, 57, 1632–1645. [Google Scholar] [CrossRef]
- Hu, H.; Lai, L.; Wang, S.; Xie, Y. Ultra-filtration affinity mass spectrometry coupled with molecular docking to explore effective form of blood-activating and stasis-resolving active ingredient group in Salvia miltiorrhiza. Chin. Tradit. Herb. Drugs 2024, 55, 7217–7229. [Google Scholar]
- He, Z.; Lyu, N.; Nan, M.; Zhao, Y.; He, Y.; Meng, L.; Sun, J.; Zhang, L. Sereening and structure characterization of acetylcholinesterase inhibitorsfrom total alkaloids of Fibraurea recisa Pierre. by target molecule affinity-liquid chromatography-tandem mass spectrometry. Chin. J. Anal. Chem. 2017, 45, 211–216. [Google Scholar]
- Liu, M.; Liu, Q.; Chen, M.; Huang, X.; Chen, X. Large-scale separation of acetylcholinesterase inhibitors from Zanthoxylum nitidum by pH-zone-refining counter-current chromatography target-guided by ultrafiltration high-performance liquid chromatography with ultraviolet and mass spectrometry screening. J. Sep. Sci. 2019, 42, 1194–1201. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Rakotondrabe, T.F.; Chen, G.; Guo, M. Antiparasiticactivity and potential active compounds from Azadirachta indica revealed by affinity ultrafiltration chromatography-mass spectrometry with acetylcholinesterase and lactate dehydrogenases. J. Anal. Test. 2024, 8, 403–414. [Google Scholar] [CrossRef]
- Li, J.; Yang, G.; Shi, W.; Fang, X.; Han, L.; Cao, Y. Anti-Alzheimer’s disease active components screened out and identified from Hedyotis diffusa combining bioaffinity ultrafiltration LC-MS with acetylcholinesterase. J. Ethnopharmacol. 2022, 296, 115460. [Google Scholar] [CrossRef]
- Chen, G.; Guo, M. Screening for natural inhibitors of Topoisomerases I from Rhamnus davurica by affinity ultrafiltration and high-performance liquid chromatography-mass spectrometry. Front. Plant Sci. 2017, 8, 1521. [Google Scholar] [CrossRef]
- Zhao, A.; Li, L.; Li, B.; Zheng, M.; Tsao, R. Ultrafiltration LC-ESI-MSn screening of 5-lipoxygenase inhibitors from selected Chinese medicinal herbs Saposhnikovia divaricata, Smilax glabra, Pueraria lobata and Carthamus tinctorius. J. Funct. Foods 2016, 24, 244–253. [Google Scholar] [CrossRef]
- Huang, Y.; Yu, M.; Wu, T.; Hou, W.; Liu, C.; Li, S. Development of a method to screen and isolate lipoxidase inhibitors from Radix saposhnikoviae via ultrafiltration liquid chromatography combined with metablism in vivo. Phytochem. Anal. 2020, 31, 937–947. [Google Scholar] [CrossRef]
- Wang, Z.; Hwang, S.H.; Lim, S.S. Characterization of DHDP, a novel aldose reductase inhibitor isolated from Lysimachia christinae. J. Funct. Foods 2017, 37, 241–248. [Google Scholar] [CrossRef]
- Wang, Z.; Hwang, S.H.; Quispe, Y.N.G.; Arce, P.H.G.; Lim, S.S. Investigation of the antioxidant and aldose reductase inhibitory activities of extracts from Peruvian tea plant infusions. Food Chem. 2017, 231, 222–230. [Google Scholar] [CrossRef]
- Quispe, Y.N.G.; Hwang, S.H.; Wang, Z.; Zuo, G.; Lim, S.S. Screening in vitro targets related to diabetes in herbal extracts from Peru: Identification of active compounds in Hypericum laricifolium Juss. by offline high-performance liquid chromatography. Int. J. Mol. Sci. 2017, 18, 2512. [Google Scholar] [CrossRef]
- Wang, L.; Chen, M.; Sun, Q.; Yang, Y.; Rong, R. Discovery of the potential neuraminidase inhibitors from Polygonum cuspidatum by ultrafiltration combined with mass spectrometry guided by molecular docking. J. Sep. Sci. 2023, 46, 2200937. [Google Scholar] [CrossRef]
- Fan, X.; Li, Y.; Wu, T.; Cheng, Z. Screening and identification of neuraminidase inhibitors from Baphicacanthus cusia by a combination of affinity ultrafiltration, HPLC-MS/MS, molecular docking, and fluorescent techniques. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2023, 1231, 123924. [Google Scholar] [CrossRef]
- Tian, Z.; Sun, L.; Chi, B.; Du, Z.; Zhang, X.; Liu, Y. Affinity ultrafiltration and UPLC-HR-Orbitrap-MS based screening of neuraminidase inhibitors from Angelica pubescens. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2022, 1208, 123398. [Google Scholar] [CrossRef]
- Tao, Y.; Cai, H.; Li, W.; Cai, B. Ultrafiltration coupled with high-performance liquid chromatography and quadrupole-time-of-flight mass spectrometry for screening lipase binders from different extracts of Dendrobium officinale. Anal. Bioanal. Chem. 2015, 407, 6081–6093. [Google Scholar] [CrossRef] [PubMed]
- Sun, R. Research of active components in natural products based on ultrafiltration-affnity mass spectrometry sereening and the hypoglyeemic effect study in vitro and in vivo. PhD Thesis, Shanghai Jiao Tong University, Shanghai, China, 2018. [Google Scholar]
- Zhu, L.; Ma, S.; Liu, M.; Li, K.; Shuai, E.; Wang, Z.; Li, S.; Zhang, S.; Cai, W. Screening and characterization estrogen receptor ligands from Arnebia euchroma (Royle) Johnst. via affinity ultrafiltration LC-MS and molecular docking. Front. Plant Sci. 2022, 13, 1012553. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Qin, W.; Ge, Y.; Sun, Y.; Yan, Y.; Zeng, Y.; Wang, F. Sereening of G-quadruplex ligands from Macleaya cordata extract bycontrast ultrafiltration with liquid chromatography-mass spectrometryand molecular docking. Chin. J. Chin. Mater. Med. 2020, 45, 3908–3914. [Google Scholar]
- Li, L.; Li, B.; Zhang, H.; Zhao, A.; Han, B.; Liu, C.; Tsao, R. Ultrafiltration LC-ESI-MSn screening of MMP-2 inhibitors from selected Chinese medicinal herbs Smilax glabra Roxb., Smilax china L. and Saposhnikovia divaricata (Turcz.) Schischk as potential functional food ingredients. J. Funct. Foods 2015, 15, 389–395. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, C.; Zheng, X.; Zhao, Z.; Li, H. Simultaneously screening multiple UGT1A1 inhibitors from Polygonum multiflorum root using ultrafiltration LC-MS. Biomed. Chromatogr. 2022, 36, e5300. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, N.; Han, D.; Yan, H. Characterization of tyrosinase inhibitors in Dryopteris crassirhizoma rhizome using a combination of high-speed counter-current chromatography, cffinity-based ultrafiltration, and liquid chromatography-tandem mass spectrometry. Front. Nutr. 2022, 9, 862773. [Google Scholar] [CrossRef]
- Jiang, B.; Chen, S.; Qian, W.; Yan, X.; Li, Y. Screening of effective components of Psoralea Fructus for endometriacancer by affinity ultrafiltration chromatography-mass spectrometry. Chin. Tradit. Herb. Drugs 2024, 55, 4663–4669. [Google Scholar]
- Huai, J.; Zhao, X.; Wang, S.; Xie, L.; Li, Y.; Zhang, T.; Cheng, C.; Dai, R. Characterization and screening of cyclooxygenase-2 inhibitors from Zi-shen pill by affinity ultrafiltration-ultra performance liquid chromatography mass spectrometry. J. Ethnopharmacol. 2019, 241, 111900. [Google Scholar] [CrossRef]
- Wang, S.; Huai, J.; Shang, Y.; Xie, L.; Cao, X.; Liao, J.; Zhang, T.; Dai, R. Screening for natural inhibitors of 5-lipoxygenase from Zi-shen pill extract by affinity ultrafiltration coupled with ultra performance liquid chromatography-mass spectrometry. J. Ethnopharmacol. 2020, 254, 112733. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, X.; Jiang, H.; Xu, C.; Tong, S.; Yan, J. Screening and identification of α-glucosidase inhibitors from Shenqi Jiangtang Granule by ultrafiltration liquid chromatography and mass spectrometry. J. Sep. Sci. 2018, 41, 797–805. [Google Scholar] [CrossRef]
- Song, H.; Chen, J.; Hong, J.; Hao, H.; Qi, L.; Lu, J.; Fu, Y.; Wu, B.; Yang, H.; Li, P. A strategy for screening of high-quality enzyme inhibitors from herbal medicines based on ultrafiltration LC-MS and in silico molecular docking. Chem. Commun. 2015, 51, 1494–1497. [Google Scholar] [CrossRef]
- Xiao, S.; Yu, R.; Ai, N.; Fan, X. Rapid screening natural-origin lipase inhibitors from hypolipidemic decoctions by ultrafiltration combined with liquid chromatography-mass spectrometry. J. Pharm. Biomed. Anal. 2015, 104, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xu, C.; Tian, Q.; Zhang, Y.; Zhang, G.; Guan, Y.; Tong, S.; Yan, J. Screening and characterization of aldose reductase inhibitors from Traditional Chinese medicine based on ultrafiltration-liquid chromatography mass spectrometry and in silico molecular docking. J. Ethnopharmacol. 2021, 264, 113282. [Google Scholar] [CrossRef]
- Fu, W.; Shentu, C.; Chen, D.; Qiu, J.; Zong, C.; Yu, H.; Zhang, Y.; Chen, Y.; Liu, X.; Xu, T. Network pharmacology combined with affinity ultrafiltration to elucidate the potential compounds of Shaoyao Gancao Fuzi Decoction for the treatment of rheumatoid arthritis. J. Ethnopharmacol. 2024, 330, 118268. [Google Scholar] [CrossRef]
- Chen, G.; Li, X.; Saleri, F.; Guo, M. Analysis of flavonoids in Rhamnus davurica and its antiproliferative activities. Molecules 2016, 21, 1275. [Google Scholar] [CrossRef]
- Van Breemen, R.B.; Nikolic, D.; Bolton, J.L. Metabolic screening using on-line ultrafiltration mass spectrometry. Drug Metab. Dispos. 1998, 26, 85–90. [Google Scholar] [PubMed]


| Screening Technology | Features | Compare with AUF | Ref. |
|---|---|---|---|
| Traditional chemical separation methods | The traditional strategy for researching active ingredients in TCM involves “chemical extraction and separation, molecular structure identification, and pharmacological activity evaluation”. | This cumbersome operation and long cycle reduce the efficiency of active ingredient discovery. | [2] |
| Cell chromatography | Able to separate target molecules through specific interactions with stationary phases (e.g., resin, silica gel, etc.), suitable for applications requiring high-purity separations. | Its operation is complex and expensive, and it is not suitable for rapid separation of large-volume samples. | [40,41] |
| Magnetic bead adsorption screening | Different affinity ligands can be modified on the surface to improve selectivity for specific targets; separation using a magnetic field is easy to operate and does not require complex equipment. | This operation is complex and costly and is not suitable for large-scale and high-throughput processing. | [42,43] |
| UV-visible spectroscopy | Suitable for fast, non-destructive quantitative analysis, especially for solution samples with absorbing properties. | Its resolution and sensitivity have certain limitations, and it has certain requirements for sample concentration. | [32] |
| Nuclear magnetic resonance (NMR) technology | It can provide detailed information on the molecular structure and reveal the chemical environment, three-dimensional structure, dynamic behavior of molecules, etc. It can also perform quantitative analysis. | The sample concentration is required to be higher, and the instrument cost and operation difficulty are greater. | [33] |
| Fluorescence technology screening | Suitable for occasions requiring high sensitivity, rapid screening, and dynamic monitoring. | The instrument costs more and requires fluorescent labeling. Improper labeling may affect the results. | [34] |
| Electrochemical method | It is very effective for applications requiring high sensitivity, fast response, and real-time monitoring, especially for the detection of low-concentration substances. | Suitable for small-molecule analysis in liquid samples, but interference issues in complex substrates may affect data accuracy. | [35] |
| Target | Compound Preparation | Active Ingredients | Ref. |
|---|---|---|---|
| Cyclooxygenase-2 | Zi-shen Pill | Twenty compounds | [114] |
| 5-lipoxygenase | Zi-shen Pill | Six compounds | [115] |
| α-Glucosidase | Shenqi Jiangtang granule | Ginsenoside Rc, ginsenoside Rh1, notoginsenoside Fe, quinquenoside L10, schisandrin, isoschisandrin, gomisin D, gomisin J, pregomisin, schisantherin D | [116] |
| Xanthine oxidase | Mai-Luo-Ning injection | 3,4-dicaffeoylquinic acid, 3,5-dicaffeoylquinic acid | [117] |
| Lipase | Wu-Ling-San, Ze-Xie decoction, Xiao-Xian-Xiong decoction, Xiao Chai-Hudecoction | Sixteen compounds | [118] |
| Augmented reality | Shenqi Jiangtang granule | Ginsenoside Rg1, Rf, Rb1, Rh1, Rd, Rg6, Rg3, Rh7, Rh2, Calycosin, Astragaloside A, Notoginsenoside Ft1, Tigloylgomisin H, Gomisin J, K3, E, Schisandrin A | [119] |
| Tumor necrosis factor-α | Shaoyao Gancao Fuzi Decoction | Glycyrrhizic acid, paeoniflorin, formononetin, isoliquiritigenin, benzoyl mesaconitine, glycyrrhetinic acid | [120] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; Zhao, X.; Yu, M.; Yang, D.; Chen, L.; Tang, C.; Zhang, Y. Affinity Ultrafiltration Mass Spectrometry for Screening Active Ingredients in Traditional Chinese Medicine: A Review of the Past Decade (2014–2024). Molecules 2025, 30, 608. https://doi.org/10.3390/molecules30030608
He Y, Zhao X, Yu M, Yang D, Chen L, Tang C, Zhang Y. Affinity Ultrafiltration Mass Spectrometry for Screening Active Ingredients in Traditional Chinese Medicine: A Review of the Past Decade (2014–2024). Molecules. 2025; 30(3):608. https://doi.org/10.3390/molecules30030608
Chicago/Turabian StyleHe, Yuqi, Xinyan Zhao, Muze Yu, Di Yang, Lian Chen, Ce Tang, and Yi Zhang. 2025. "Affinity Ultrafiltration Mass Spectrometry for Screening Active Ingredients in Traditional Chinese Medicine: A Review of the Past Decade (2014–2024)" Molecules 30, no. 3: 608. https://doi.org/10.3390/molecules30030608
APA StyleHe, Y., Zhao, X., Yu, M., Yang, D., Chen, L., Tang, C., & Zhang, Y. (2025). Affinity Ultrafiltration Mass Spectrometry for Screening Active Ingredients in Traditional Chinese Medicine: A Review of the Past Decade (2014–2024). Molecules, 30(3), 608. https://doi.org/10.3390/molecules30030608






