Identification of KKL-35 as a Novel Carnosine Dipeptidase 2 (CNDP2) Inhibitor by In Silico Screening
Abstract
1. Introduction
2. Results
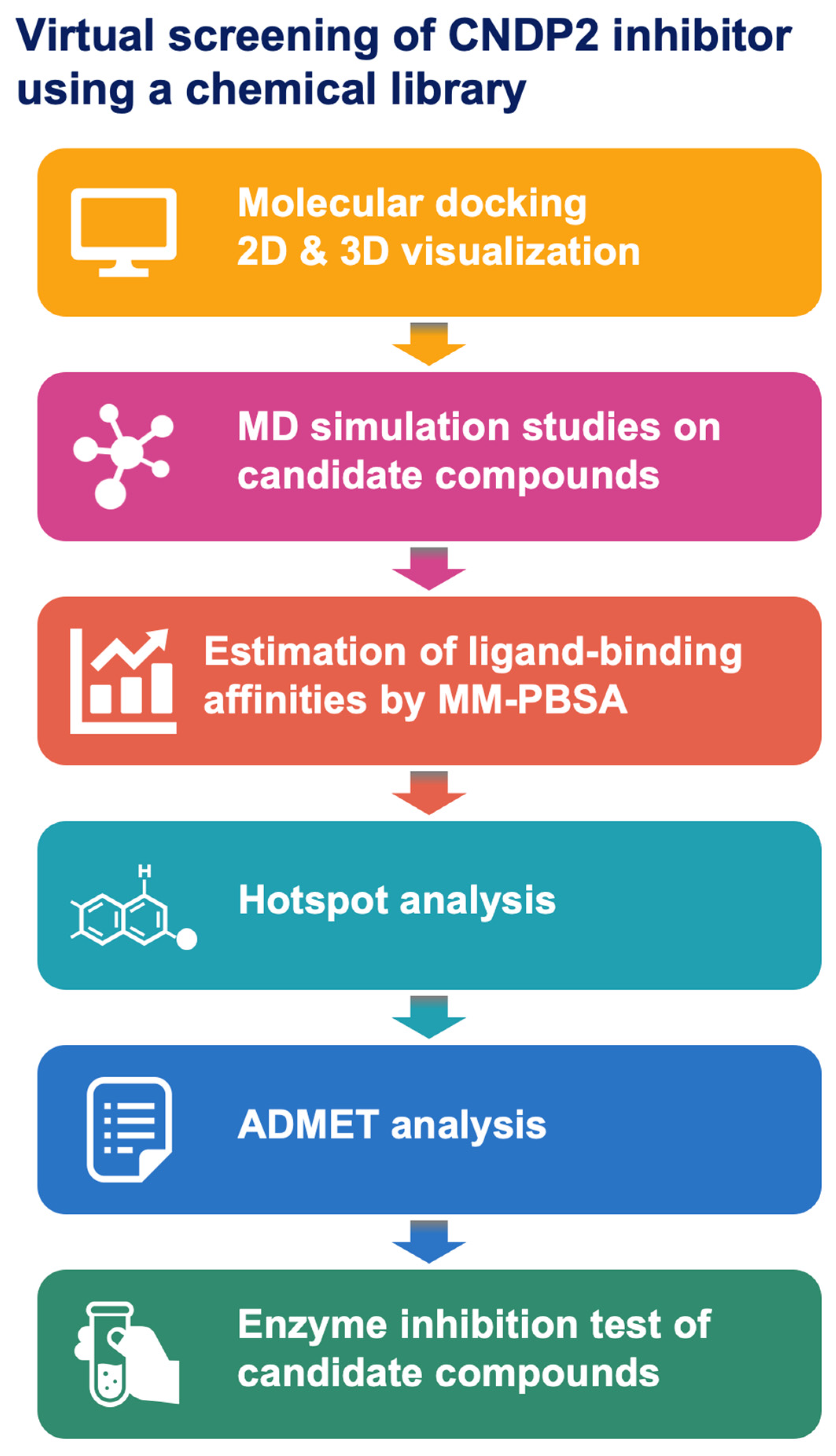
2.1. Overview of the Screening Workflow
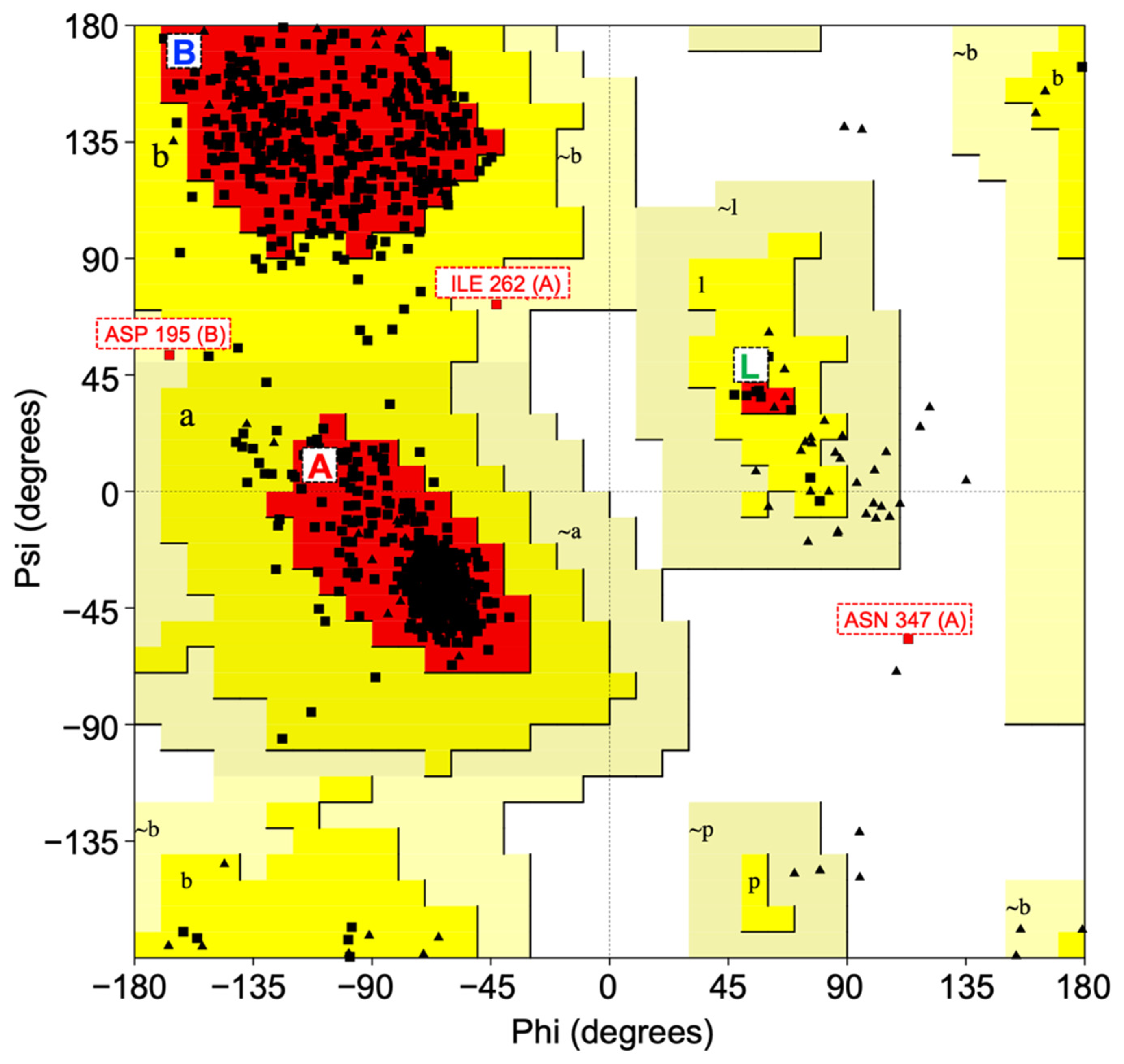
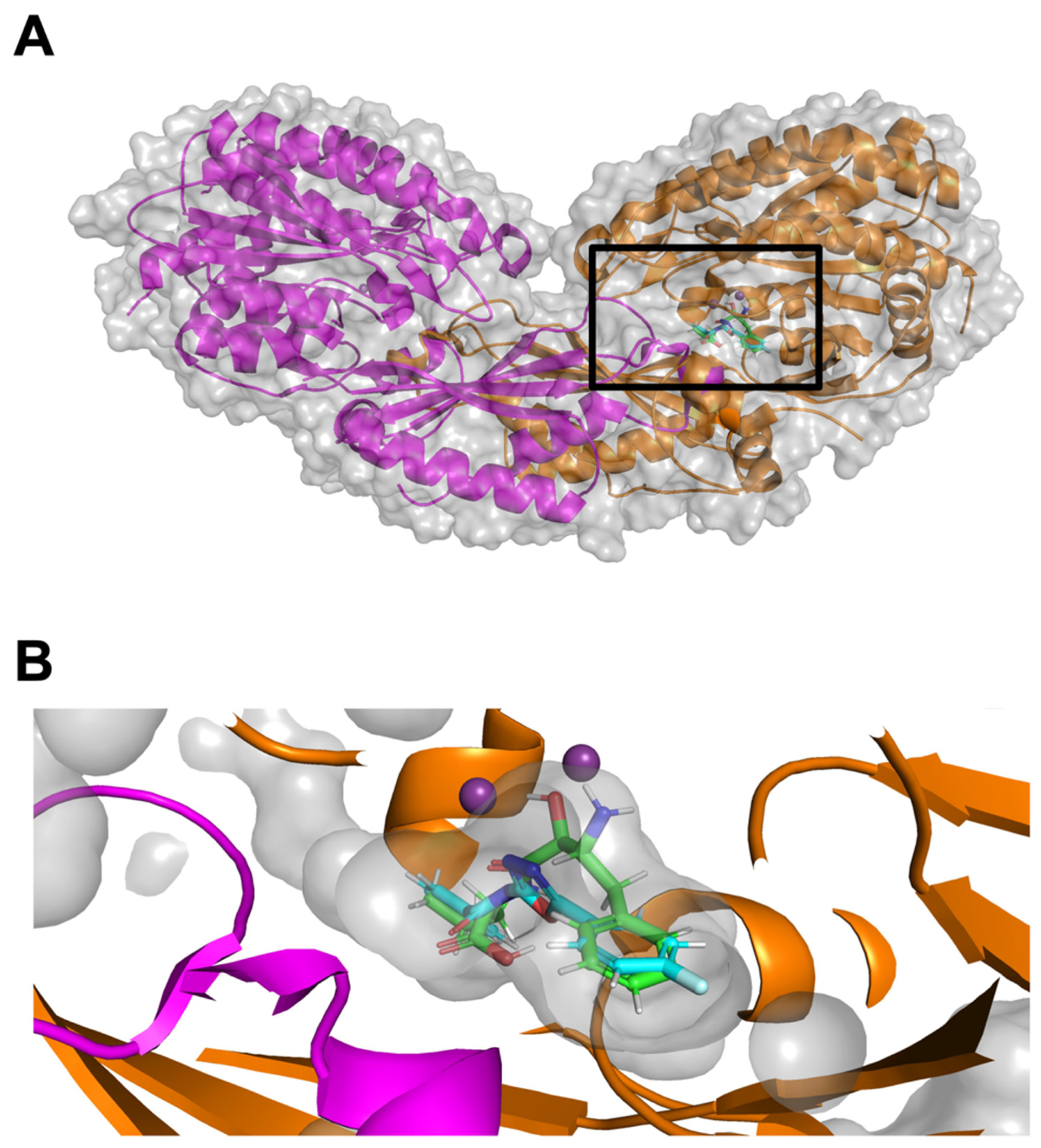
2.2. Structural Validation of CNDP2
2.3. Identification of Potential CNDP2 Inhibitors
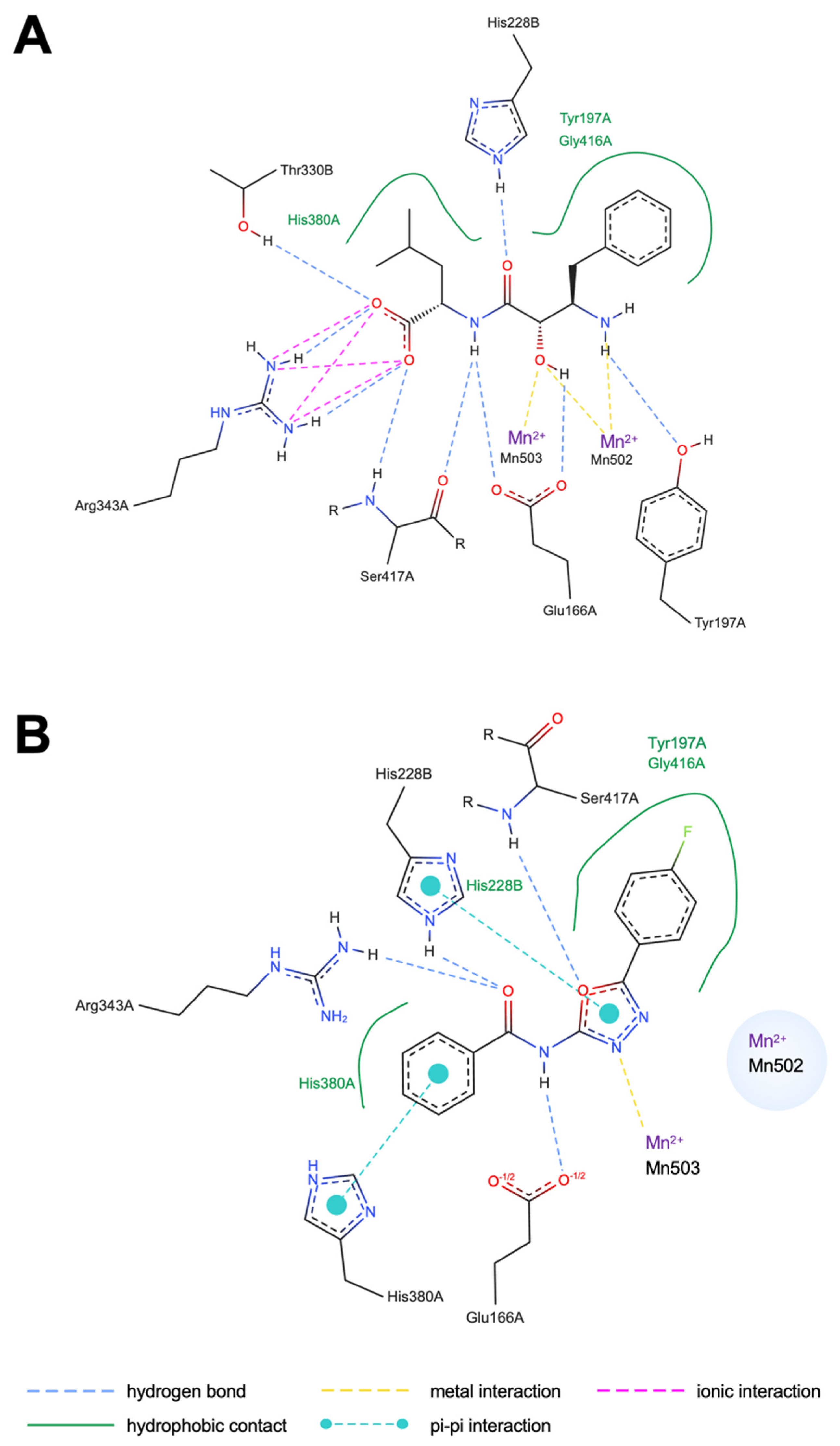
2.4. Binding Interactions and Dynamic Stability of CNDP2–Ligand Complexes
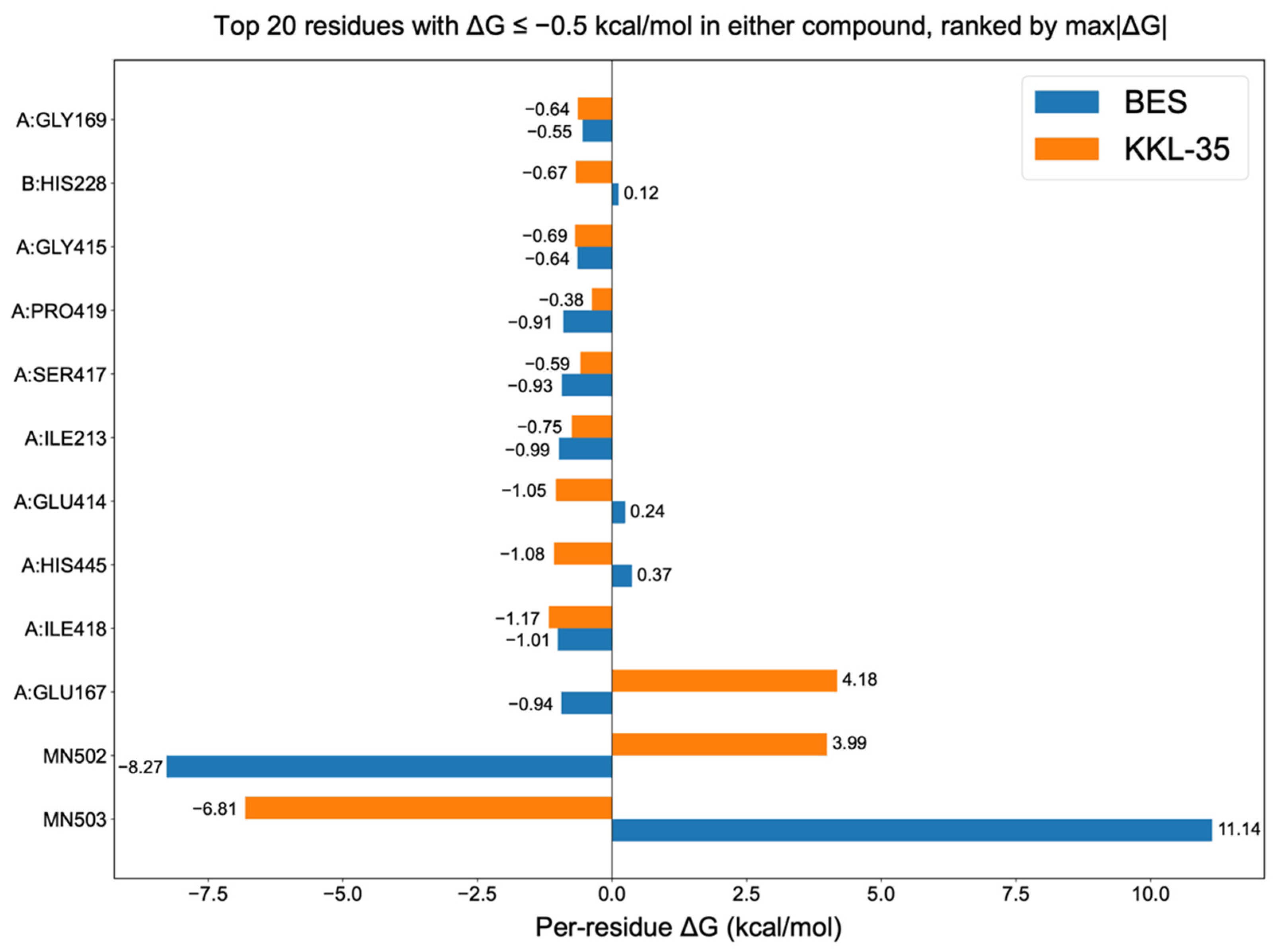
2.5. Binding Free Energy Decomposition and Residue Contributions
2.6. ADMET and Drug-Likeness Analysis
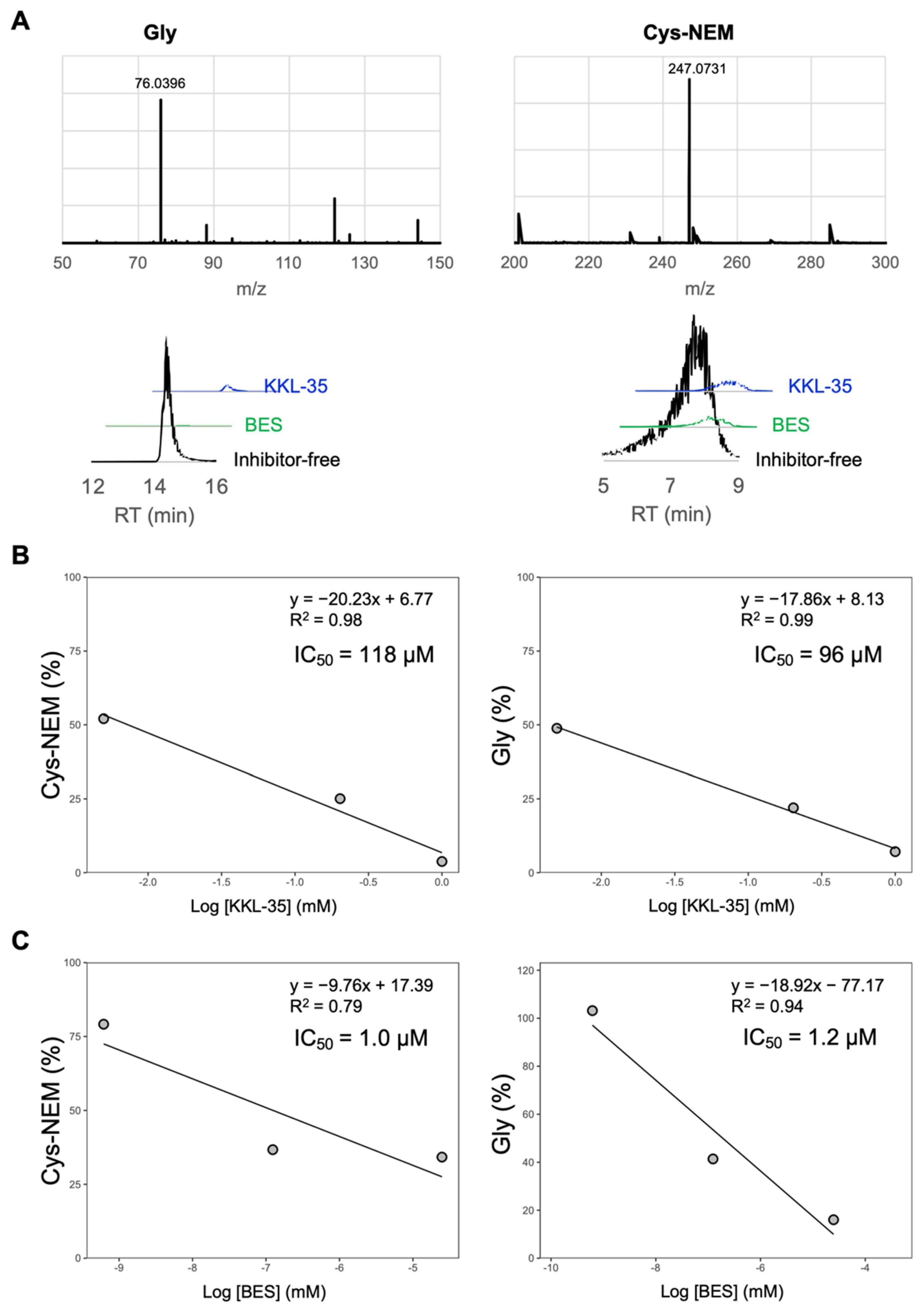
2.7. In Vitro Inhibitory Analysis of Ligands
3. Discussion
4. Materials and Methods
4.1. Protein Preparation for In Silico Screening
4.2. Library Preparation
4.3. In Silico Screening
4.4. MD Simulation and MM-PB/GBSA Free Energy Calculation
4.5. In Silico Pharmacokinetic and Toxicological Analysis
4.6. Target Prediction
4.7. Cell Culture and Chemicals
4.8. Lentiviral Expression and FLAG Affinity Purification of CNDP2
4.9. Analysis of Dipeptidase Activity
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaur, H.; Kumar, C.; Junot, C.; Toledano, M.B.; Bachhawat, A.K. Dug1p Is a Cys-Gly Peptidase of the γ-Glutamyl Cycle of Saccharomyces Cerevisiae and Represents a Novel Family of Cys-Gly Peptidases. J. Biol. Chem. 2009, 284, 14493–14502. [Google Scholar] [CrossRef] [PubMed]
- Meister, A.; Anderson, M.E. Glutathione. Annu. Rev. Biochem. 1983, 52, 711–760. [Google Scholar] [CrossRef]
- Ikeda, Y.; Fujii, J. The Emerging Roles of γ-Glutamyl Peptides Produced by γ-Glutamyltransferase and the Glutathione Synthesis System. Cells 2023, 12, 2831. [Google Scholar] [CrossRef]
- Kobayashi, S.; Homma, T.; Okumura, N.; Han, J.; Nagaoka, K.; Sato, H.; Konno, H.; Yamada, S.; Takao, T.; Fujii, J. Carnosine Dipeptidase II (CNDP2) Protects Cells under Cysteine Insufficiency by Hydrolyzing Glutathione-Related Peptides. Free Radic. Biol. Med. 2021, 174, 12–27. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Yamakawa-Kobayashi, K.; Ohhara, Y.; Kawashima, T.; Ohishi, Y.; Kayashima, Y. Loss of CNDP Causes a Shorter Lifespan and Higher Sensitivity to Oxidative Stress in Drosophila Melanogaster. Biomed. Res. 2020, 41, 131–138. [Google Scholar] [CrossRef]
- Zhang, L.Q.; Yang, H.Q.; Yang, S.Q.; Wang, Y.; Chen, X.J.; Lu, H.S.; Zhao, L.P. CNDP2 Acts as an Activator for Human Ovarian Cancer Growth and Metastasis via the PI3K/AKT Pathway. Technol. Cancer Res. Treat. 2019, 18, 1533033819874773. [Google Scholar] [CrossRef]
- Xue, C.; Zhang, Z.; Yu, H.; Yu, M.; Yuan, K.; Yang, T.; Miao, M.; Shi, H. Up-Regulation of CNDP2 Facilitates the Proliferation of Colon Cancer. BMC Gastroenterol. 2014, 14, 96. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Z.; Wang, C.; Miao, L.; Zhang, J.; Wang, J.; Jiao, B.; Zhao, S. Quantitative Proteomics Approach to Screening of Potential Diagnostic and Therapeutic Targets for Laryngeal Carcinoma. PLoS ONE 2014, 9, e90181. [Google Scholar] [CrossRef]
- Vu, H.M.; Mohammad, H.B.; Nguyen, T.N.; Lee, J.H.; Do, Y.; Sung, J.Y.; Lee, S.H.; Kim, M.S. Quantitative Proteomic Analysis of Bronchoalveolar Lavage Fluids from Patients with Small Cell Lung Cancers. Proteom. Clin. Appl. 2023, 17, 2300011. [Google Scholar] [CrossRef]
- Guzelsoy, G.; Elorza, S.D.; Ros, M.; Schachtner, L.T.; Hayashi, M.; Hobson-Gutierrez, S.; Rundstrom, P.; Brunner, J.S.; Pillai, R.; Walkowicz, W.E.; et al. Cooperative Nutrient Scavenging Is an Evolutionary Advantage in Cancer. Nature 2025, 640, 534–542. [Google Scholar] [CrossRef]
- Zhang, J.; Pavlova, N.N.; Thompson, C.B. Cancer Cell Metabolism: The Essential Role of the Nonessential Amino Acid, Glutamine. EMBO J. 2017, 36, 1302–1315. [Google Scholar] [CrossRef]
- Mathé, G. Bestatin, an Aminopeptidase Inhibitor with a Multi-Pharmacological Function. Biomed. Pharmacother. 1991, 45, 49–54. [Google Scholar] [CrossRef]
- Orning, L.; Krivi, G.; Fitzpatrick, F.A. Leukotriene A4 Hydrolase. Inhibition by Bestatin and Intrinsic Aminopeptidase Activity Establish Its Functional Resemblance to Metallohydrolase Enzymes. J. Biol. Chem. 1991, 266, 1375–1378. [Google Scholar] [CrossRef]
- Tieku, S.; Hooper, N.M. Inhibition of Aminopeptidases N, A and W. A Re-Evaluation of the Actions of Bestatin and Inhibitors of Angiotensin Converting Enzyme. Biochem. Pharmacol. 1992, 44, 1725–1730. [Google Scholar] [CrossRef] [PubMed]
- Almihyawi, R.A.; Naman, Z.T.; Al-Hasani, H.M.; Muhseen, Z.T.; Zhang, S.; Chen, G. Integrated Computer-Aided Drug Design and Biophysical Simulation Approaches to Determine Natural Anti-Bacterial Compounds for Acinetobacter baumannii. Sci. Rep. 2022, 12, 6590. [Google Scholar] [CrossRef] [PubMed]
- Alabbas, A.B. Identification of Promising Methionine Aminopeptidase Enzyme Inhibitors: A Combine Study of Comprehensive Virtual Screening and Dynamics Simulation Study. Saudi Pharm. J. 2023, 31, 101745. [Google Scholar] [CrossRef]
- Ischak, N.I.; Aman, L.O.; Hasan, H.; La Kilo, A.; Asnawi, A. In Silico Screening of Andrographis Paniculata Secondary Metabolites as Anti-Diabetes Mellitus through PDE9 Inhibition. Res. Pharm. Sci. 2023, 18, 100–111. [Google Scholar] [CrossRef]
- Charbe, N.B.; Zacconi, F.C.; Kowthavarapu, V.K.; Gupta, C.; Palakurthi, S.S.; Satheeshkumar, R.; Lokwani, D.K.; Tambuwala, M.M.; Palakurthi, S. Targeting Allosteric Site of PCSK9 Enzyme for the Identification of Small Molecule Inhibitors: An in Silico Drug Repurposing Study. Biomedicines 2024, 12, 286. [Google Scholar] [CrossRef] [PubMed]
- Hameed, A.R.; Ali, S.F.; Alsallameh, S.M.; Muhseen, Z.T.; Almansour, N.M.; ALSuhaymi, N.; Alsugoor, M.H.; Allemailem, K.S. Structural Dynamics of P-Rex1 Complexed with Natural Leads Establishes the Protein as an Attractive Target for Therapeutics to Suppress Cancer Metastasis. BioMed Res. Int. 2023, 2023, 3882081. [Google Scholar] [CrossRef]
- Muhseen, Z.T.; Ali, M.H.; Jaber, N.R.; Mashrea, D.S.; Alfalki, A.M.; Li, G. Determination of Novel Anti-Cancer Agents by Targeting OGG1 Enzyme Using Integrated Bioinformatics Methods. Int. J. Environ. Res. Public Health 2021, 18, 13290. [Google Scholar] [CrossRef]
- Lovell, S.C.; Davis, I.W.; Arendall, W.B., 3rd; De Bakker, P.I.; Word, J.M.; Prisant, M.G.; Richardson, J.S.; Richardson, D.C. Structure Validation by Calpha Geometry: Phi, Psi and Cbeta Deviation. Proteins 2003, 50, 437–450. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Ramadoss, N.S.; Alumasa, J.N.; Cheng, L.; Wang, Y.; Li, S.; Chambers, B.S.; Chang, H.; Chatterjee, A.K.; Brinker, A.; Engels, I.H.; et al. Small Molecule Inhibitors of Trans-Translation Have Broad-Spectrum Antibiotic Activity. Proc. Natl. Acad. Sci. USA 2013, 110, 10282–10287. [Google Scholar] [CrossRef]
- Ramírez, D.; Caballero, J. Is It Reliable to Take the Molecular Docking Top Scoring Position as the Best Solution without Considering Available Structural Data? Molecules 2018, 23, 1038. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, S.A.; Dror, R.O. Molecular Dynamics Simulation for All. Neuron 2018, 99, 1129–1143. [Google Scholar] [CrossRef]
- Karplus, M.; McCammon, J.A. Molecular Dynamics Simulations of Biomolecules. Nat. Struct. Biol. 2002, 9, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Lobanov, M.Y.; Bogatyreva, N.S.; Galzitskaya, O.V. Radius of Gyration as an Indicator of Protein Structure Compactness. Mol. Biol. 2008, 42, 623–628. [Google Scholar] [CrossRef]
- Luzar, A.; Chandler, D. Hydrogen-Bond Kinetics in Liquid Water. Nature 1996, 379, 55–57. [Google Scholar] [CrossRef]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W.; et al. Calculating Structures and Free Energies of Complex Molecules: Combining Molecular Mechanics and Continuum Models. Acc. Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. Gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef]
- Hou, T.; Wang, J.; Li, Y.; Wang, W. Assessing the Performance of the Molecular Mechanics/Poisson Boltzmann Surface Area and Molecular Mechanics/Generalized Born Surface Area Methods. II. The Accuracy of Ranking Poses Generated from Docking. J. Comput. Chem. 2011, 32, 866–877. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 2012, 64, 4–17. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A Knowledge-Based Approach in Designing Combinatorial or Medicinal Chemistry Libraries for Drug Discovery. 1. A Qualitative and Quantitative Characterization of Known Drug Databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Egan, W.J.; Merz, K.M., Jr.; Baldwin, J.J. Prediction of Drug Absorption Using Multivariate Statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef]
- Muegge, I.; Heald, S.L.; Brittelli, D. Simple Selection Criteria for Drug-like Chemical Matter. J. Med. Chem. 2001, 44, 1841–1846. [Google Scholar] [CrossRef]
- Baell, J.B.; Holloway, G.A. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef]
- Brenk, R.; Schipani, A.; James, D.; Krasowski, A.; Gilbert, I.H.; Frearson, J.; Wyatt, P.G. Lessons Learnt from Assembling Screening Libraries for Drug Discovery for Neglected Diseases. ChemMedChem 2008, 3, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Ertl, P.; Rohde, B.; Selzer, P. Fast Calculation of Molecular Polar Surface Area as a Sum of Fragment-Based Contributions and Its Application to the Prediction of Drug Transport Properties. J. Med. Chem. 2000, 43, 3714–3717. [Google Scholar] [CrossRef] [PubMed]
- Zanger, U.M.; Schwab, M. Cytochrome P450 Enzymes in Drug Metabolism: Regulation of Gene Expression, Enzyme Activities, and Impact of Genetic Variation. Pharmacol. Ther. 2013, 138, 103–141. [Google Scholar] [CrossRef]
- Potts, R.O.; Guy, R.H. Predicting Skin Permeability. Pharm. Res. 1992, 9, 663–669. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Lenney, J.F.; Peppers, S.C.; Kucera-Orallo, C.M.; George, R.P. Characterization of Human Tissue Carnosinase. Biochem. J. 1985, 228, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A. PDBsum: Summaries and Analyses of PDB Structures. Nucleic Acids Res. 2001, 29, 221–222. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, J.; Marsili, M. Iterative Partial Equalization of Orbital Electronegativity—A Rapid Access to Atomic Charges. Tetrahedron 1980, 36, 3219–3228. [Google Scholar] [CrossRef]
- Halgren, T.A. Merck Molecular Force Field. I. Basis, Form, Scope, Parameterization, and Performance of MMFF94. J. Comput. Chem. 1996, 17, 490–519. [Google Scholar] [CrossRef]
- Koes, D.R.; Baumgartner, M.P.; Camacho, C.J. Lessons Learned in Empirical Scoring with Smina from the CSAR 2011 Benchmarking Exercise. J. Chem. Inf. Model. 2013, 53, 1893–1904. [Google Scholar] [CrossRef]
- Diedrich, K.; Krause, B.; Berg, O.; Rarey, M. PoseEdit: Enhanced Ligand Binding Mode Communication by Interactive 2D Diagrams. J. Comput. Aided Mol. Des. 2023, 37, 491–503. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Lemkul, J.A. Introductory Tutorials for Simulating Protein Dynamics with GROMACS. J. Phys. Chem. B 2024, 128, 9418–9435. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. Ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from Ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and Testing of a General Amber Force Field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Sousa da Silva, A.W.; Vranken, W.F. ACPYPE—AnteChamber PYthon Parser InterfacE. BMC Res. Notes 2012, 5, 367. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Li, P.; Roberts, B.P.; Chakravorty, D.K.; Merz, K.M., Jr. Rational Design of Particle Mesh Ewald Compatible Lennard-Jones Parameters for +2 Metal Cations in Explicit Solvent. J. Chem. Theory Comput. 2013, 9, 2733–2748. [Google Scholar] [CrossRef] [PubMed]
- Brünger, A.; Brooks, C.L., III; Karplus, M. Stochastic Boundary Conditions for Molecular Dynamics Simulations of ST2 Water. Chem. Phys. Lett. 1984, 105, 495–500. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Bernetti, M.; Bussi, G. Pressure Control Using Stochastic Cell Rescaling. J. Chem. Phys. 2020, 153, 114107. [Google Scholar] [CrossRef]
- Salentin, S.; Schreiber, S.; Haupt, V.J.; Adasme, M.F.; Schroeder, M. PLIP: Fully Automated Protein–Ligand Interaction Profiler. Nucleic Acids Res. 2015, 43, W443–W447. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.; Blundell, T.L.; Ascher, D.B. PkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated Data and New Features for Efficient Prediction of Protein Targets of Small Molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef] [PubMed]

| Rank | ID | Name | Binding Energy Score (kcal/mol) |
|---|---|---|---|
| 1 | drug2490 | Bestatin (BES) | −11.40 |
| 2 | drug2591 | KKL-35 | −11.20 |
| 3 | drug6171 | WAY-660898 | −11.00 |
| 4 | drug5628 | WAY-656494 | −11.00 |
| 5 | drug5970 | WAY-326270-A | −10.90 |
| 6 | drug4125 | IRAK-4 protein kinase inhibitor 2 | −10.90 |
| 7 | drug4843 | 3,6-Dihydroxyflavone | −10.80 |
| 8 | drug5449 | WAY-233765 | −10.70 |
| 9 | drug384 | A-803467 | −10.70 |
| 10 | drug4196 | SIRT7 inhibitor 97491 | −10.60 |
| Compound | Docking Interaction | Docking Residue | MD Interaction | MD Residue |
|---|---|---|---|---|
| BES | HB | Ser417A | HB | Ser417A |
| BES | HB | Tyr197A | HB | Tyr197A |
| BES | HB | Thr330B | HB | Thr330B |
| BES | Hphobic | Tyr197A | Hphobic | Tyr197A |
| BES | II | Arg343A | II | Arg343A |
| BES | HB | Glu166A | ||
| BES | HB | His228B | ||
| BES | Hphobic | Gly416A | ||
| BES | Hphobic | His380A | ||
| BES | HB | Gln103A | ||
| BES | HB | Asp195A | ||
| BES | Hphobic | Ile213A | ||
| BES | Hphobic | His228B | ||
| BES | Hphobic | Val231B | ||
| BES | MI | MN502 | ||
| BES | MI | MN503 | ||
| KKL-35 | HB | Ser417A | HB | Ser417A |
| KKL-35 | HB | Glu166A | ||
| KKL-35 | HB | Arg343A | ||
| KKL-35 | π–π | His228B | ||
| KKL-35 | π–π | His380A | ||
| KKL-35 | Hphobic | His380A | ||
| KKL-35 | Hphobic | His197A | ||
| KKL-35 | Hphobic | Gly416A | ||
| KKL-35 | Hphobic | Ile213A | ||
| KKL-35 | Hphobic | Pro419A | ||
| KKL-35 | Hphobic | Thr330B | ||
| KKL-35 | Hphobic | Leu210A | ||
| KKL-35 | Hphobic | Asp195A | ||
| KKL-35 | Hphobic | Ile418A | ||
| KKL-35 | MI | MN503 |
| Compound | ΔG Binding | ΔG Electrostatic | ΔG Bind van der Waals | ΔG Bind Gas Phase | ΔG Polar Solvation | ΔG Nonpolar Solvation | ΔG Solvation |
|---|---|---|---|---|---|---|---|
| KKL-35 | –10.05 | –29.32 | –43.62 | –72.94 | 66.41 | –3.52 | 62.89 |
| BES | –22.59 | –92.73 | –35.01 | –127.75 | 109.15 | –3.99 | 105.16 |
| Compound | KKL-35 | BES | |
|---|---|---|---|
| Physicochemical properties | Formula | C15H9ClFN3O2 | C16H24N2O4 |
| Molecular weight | 317.7 | 308.37 | |
| #Heavy atoms | 22 | 22 | |
| #Aromatic heavy atoms | 17 | 6 | |
| Fraction Csp3 | 0 | 0.5 | |
| #Rotatable bonds | 4 | 9 | |
| #H-bond acceptors | 5 | 5 | |
| #H-bond donors | 1 | 4 | |
| Molar refractivity | 78.92 | 83.31 | |
| TPSA (Å2) | 68.02 | 112.65 | |
| Lipophilicity | Consensus Log P | 3.44 | 0.79 |
| Water solubility | Moderately soluble | Very soluble | |
| Pharmacokinetics | GI absorption | High | High |
| BBB permeant | Yes | No | |
| P-gp substrate | No | No | |
| CYP1A2 inhibitor | Yes | No | |
| CYP2C19 inhibitor | Yes | No | |
| CYP2C9 inhibitor | No | No | |
| CYP2D6 inhibitor | No | No | |
| CYP3A4 inhibitor | No | No | |
| log Kp (cm/s) | −5.89 | −8.86 | |
| Druglikeness | Lipinski | Yes; 0 violation | Yes; 0 violation |
| Ghose | Yes | Yes | |
| Veber | Yes | Yes | |
| Egan | Yes | Yes | |
| Muegge | Yes | Yes | |
| Bioavailability Score | 0.55 | 0.55 | |
| Medicinal chemistry | PAINS #alerts | 0 | 0 |
| Brenk #alerts | 0 | 0 | |
| Synthetic Accessibility | 2.64 | 3.1 |
| Target | Common Name | Probability |
|---|---|---|
| Aminopeptidase N | ANPEP | 0.95 |
| Aminopeptidase B (by homology) | RNPEP | 0.95 |
| Matrix metalloproteinase 2 | MMP2 | 0.95 |
| Leucine aminopeptidase | LAP3 | 0.95 |
| Leukotriene A4 hydrolase | LTA4H | 0.95 |
| Aspartyl aminopeptidase | DNPEP | 0.14 |
| Angiotensin-converting enzyme | ACE | 0.12 |
| Dipeptidyl peptidase IV | DPP4 | 0.12 |
| Xaa-Pro dipeptidase | PEPD | 0.11 |
| Calpain 1 | CAPN1 | 0.11 |
| Neprilysin (by homology) | MME | 0.11 |
| Beta-secretase 1 | BACE1 | 0.11 |
| Dipeptidyl peptidase VIII | DPP8 | 0.11 |
| Dipeptidyl peptidase IX | DPP9 | 0.11 |
| Methionine aminopeptidase 2 | METAP2 | 0.11 |
| Carboxypeptidase B | CPB1 | 0.11 |
| Renin | REN | 0.11 |
| Xaa-Pro aminopeptidase 2 | XPNPEP2 | 0.11 |
| Prolyl endopeptidase | PREP | 0.11 |
| Fibroblast activation protein alpha | FAP | 0.11 |
| Renal dipeptidase | DPEP1 | 0.11 |
| Beta secretase 2 | BACE2 | 0.11 |
| Caspase-1 | CASP1 | 0.11 |
| Target | Common Name | Probability |
|---|---|---|
| Thrombin and coagulation factor X | F10 | 0.11 |
| Caspase-3 | CASP3 | 0.11 |
| Matrix metalloproteinase 13 | MMP13 | 0.11 |
| Gamma-secretase | PSEN2 | 0.11 |
| Cathepsin (V and K) | CTSV | 0.11 |
| Cathepsin L | CTSL | 0.11 |
| Cathepsin G | CTSG | 0.11 |
| Prolyl endopeptidase | PREP | 0.11 |
| Fibroblast activation protein alpha (by homology) | FAP | 0.11 |
| Caspase-7 | CASP7 | 0.11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Homma, T.; Shinbara, K.; Osaki, T. Identification of KKL-35 as a Novel Carnosine Dipeptidase 2 (CNDP2) Inhibitor by In Silico Screening. Molecules 2025, 30, 4370. https://doi.org/10.3390/molecules30224370
Homma T, Shinbara K, Osaki T. Identification of KKL-35 as a Novel Carnosine Dipeptidase 2 (CNDP2) Inhibitor by In Silico Screening. Molecules. 2025; 30(22):4370. https://doi.org/10.3390/molecules30224370
Chicago/Turabian StyleHomma, Takujiro, Koki Shinbara, and Tsukasa Osaki. 2025. "Identification of KKL-35 as a Novel Carnosine Dipeptidase 2 (CNDP2) Inhibitor by In Silico Screening" Molecules 30, no. 22: 4370. https://doi.org/10.3390/molecules30224370
APA StyleHomma, T., Shinbara, K., & Osaki, T. (2025). Identification of KKL-35 as a Novel Carnosine Dipeptidase 2 (CNDP2) Inhibitor by In Silico Screening. Molecules, 30(22), 4370. https://doi.org/10.3390/molecules30224370






