Inhibition Profiling of Rhodiola crenulata on Lactate Dehydrogenase Isoenzymes
Abstract
1. Introduction
2. Results and Discussion
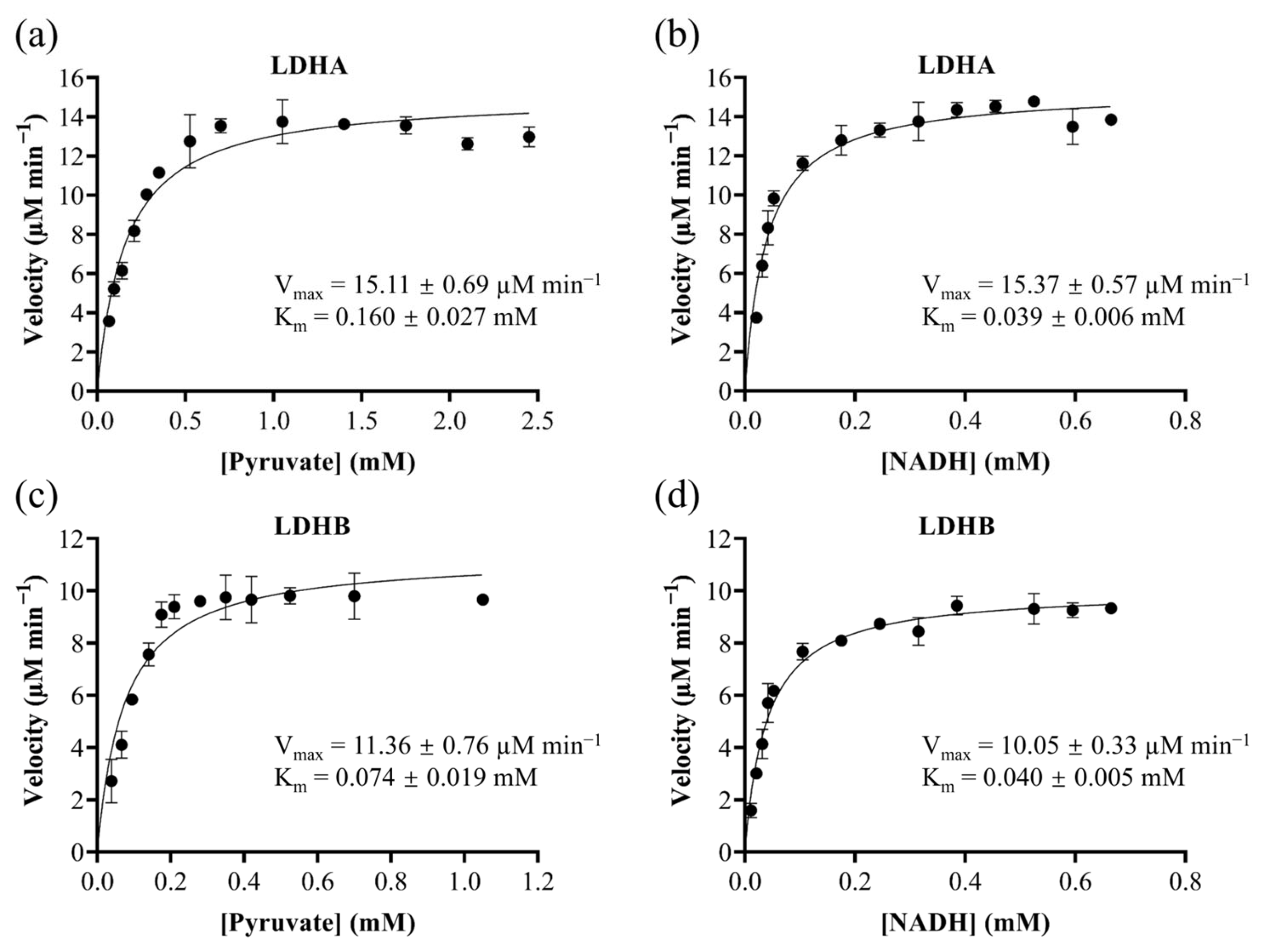
2.1. Substrate-Specific Kinetic Parameters of LDHA and LDHB
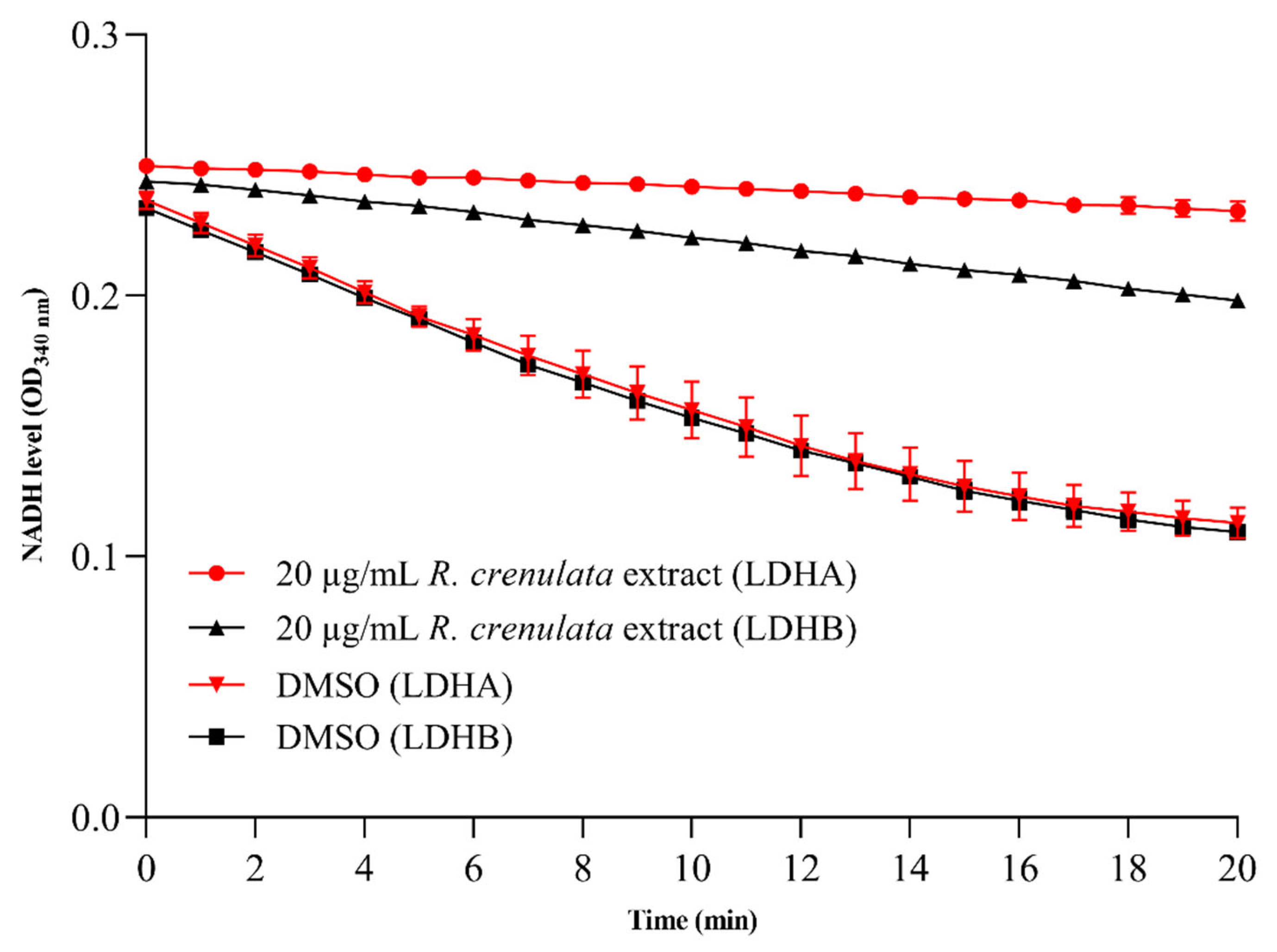
2.2. Validation of the LDHA and LDHB Inhibitor Screening Platforms
2.3. Differential Inhibitory Effects of R. crenulata Extract on LDHA and LDHB
2.4. Constituent Profiling of R. crenulata
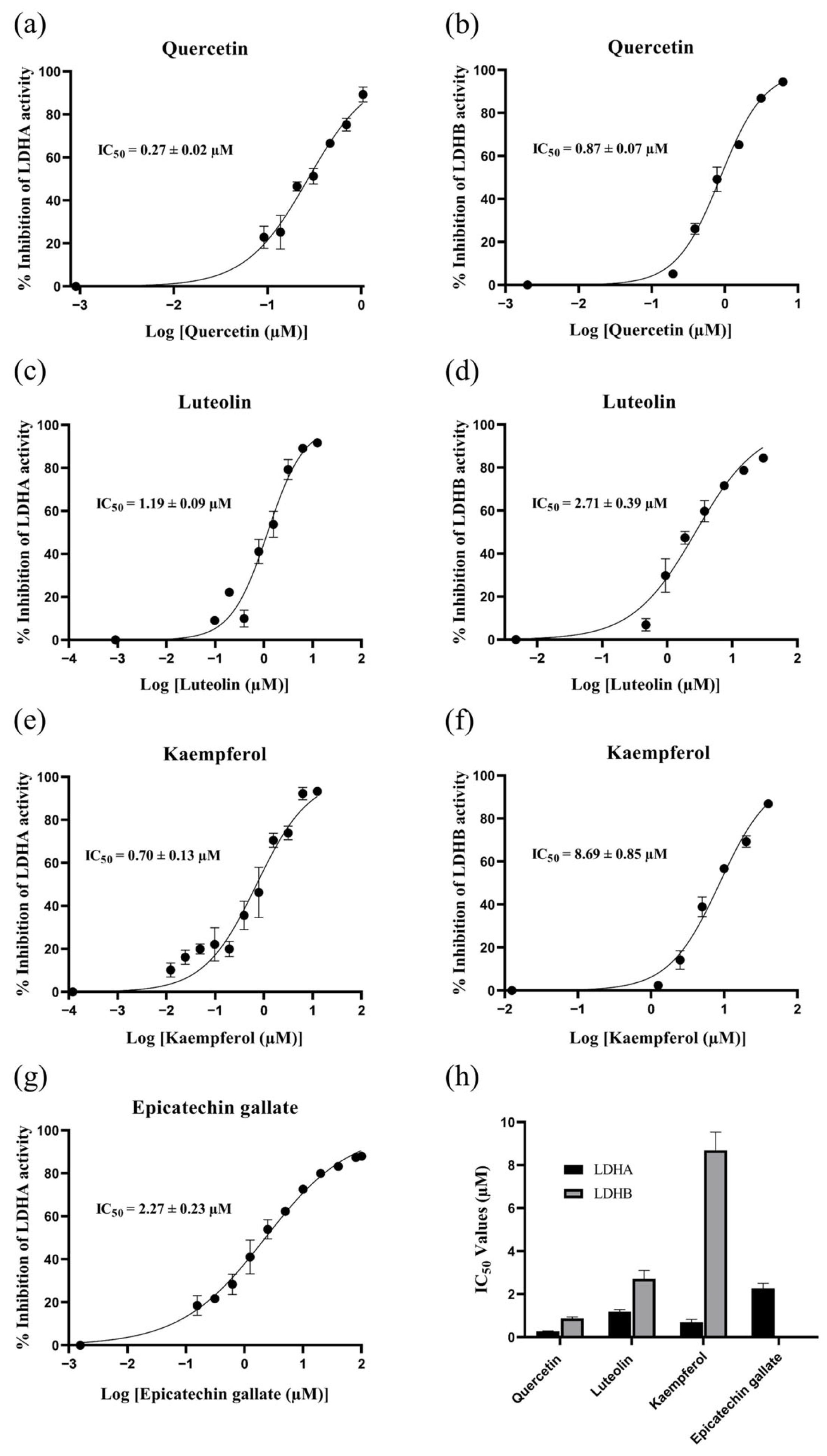
2.5. Identification of LDHA and LDHB Inhibitors from R. crenulata Extract
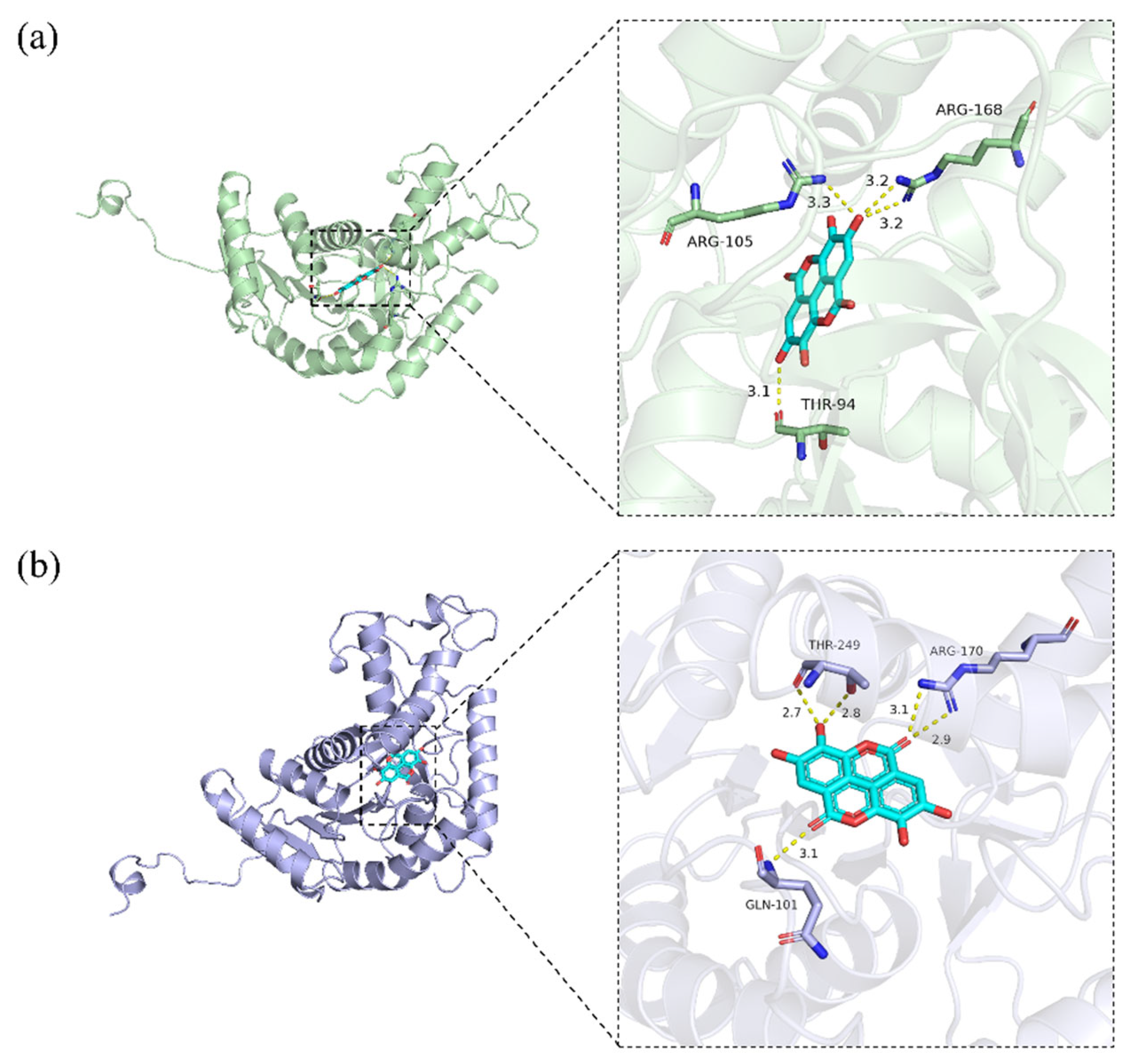
2.6. Molecular Docking of Inhibitory Components to LDHA and LDHB
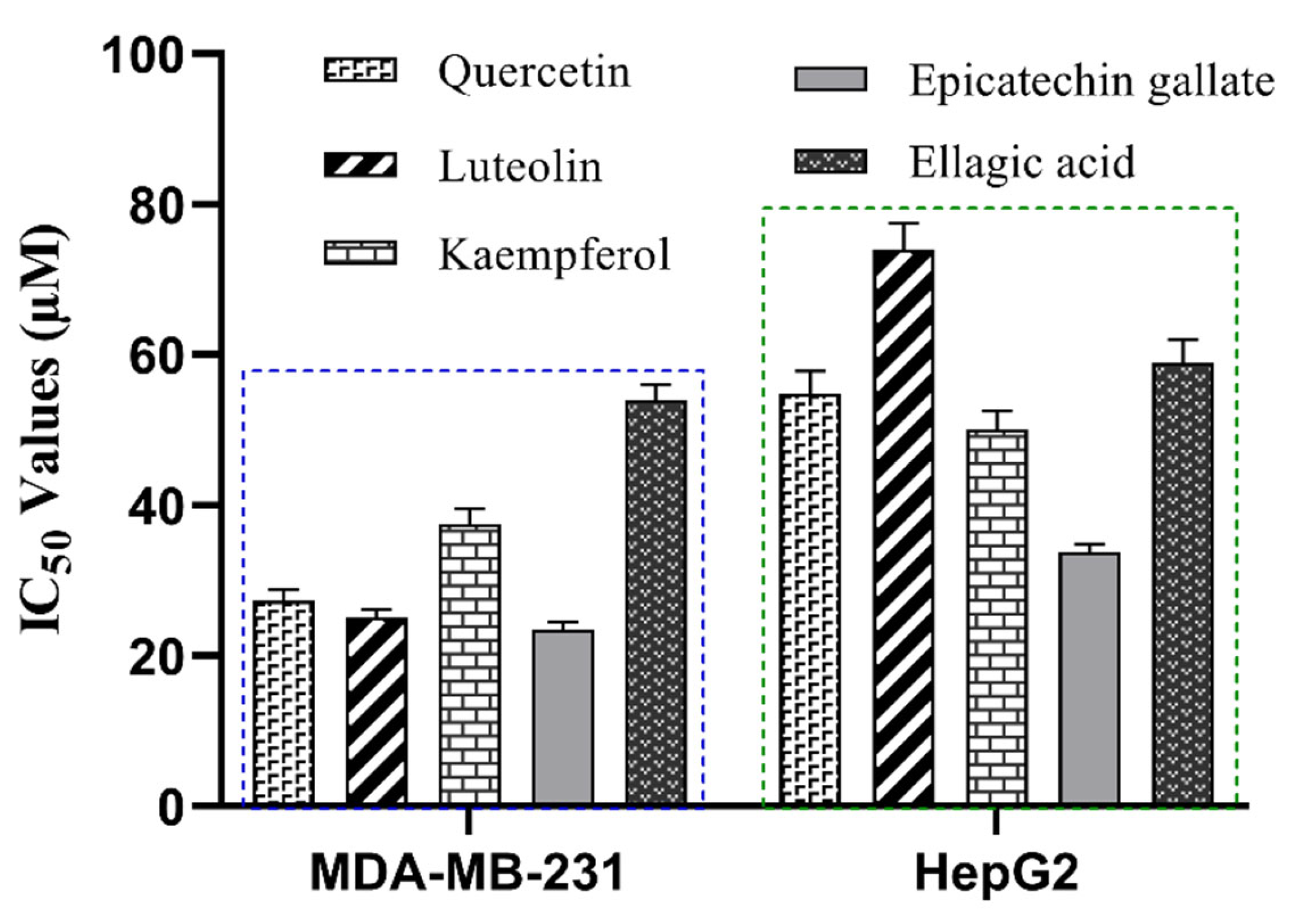
2.7. Anti-Proliferative Effects of R. crenulata Bioactives on MDA-MB-231 and HepG2 Cells
3. Materials and Methods
3.1. Reagents and Materials
3.2. Instruments and Software
3.3. Preparation of R. crenulata Extract
3.4. Expression and Purification of Recombinant LDHA and LDHB
3.5. Establishment and Optimization of the LDHA and LDHB Inhibitor Screening Platform
3.6. Evaluation of the LDHA and LDHB Inhibitor Screening Platform
3.7. Screening of Inhibitory Components from R. crenulata Against LDHA and LDHB
3.8. Molecular Docking Analysis of LDHA and LDHB Inhibitors
3.9. Cell Viability Assays of LDHA and LDHB Inhibitors
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HPLC-QTOF MS | High-Performance Liquid Chromatography System Coupled with Quadrupole Time-of-Flight Mass Spectrometer |
| LDH | Lactate dehydrogenase |
| LDHA | Lactate dehydrogenase A |
| LDHB | Lactate dehydrogenase B |
| NAD+ | Oxidized nicotinamide adenine dinucleotide |
| NADH | Reduced nicotinamide adenine dinucleotide |
| BPC | Base Peak Chromatogram |
| RT | Retention Time |
| IC50 | The half maximal inhibitory concentration |
| IPTG | Isopropyl β-D-thiogalactoside |
| DMSO | Dimethyl sulfoxide |
References
- Faubert, B.; Solmonson, A.; DeBerardinis, R.J. Metabolic reprogramming and cancer progression. Science 2020, 368, eaaw5473. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Oyang, L.; Lin, J.; Tan, S.; Han, Y.; Wu, N.; Yi, P.; Tang, L.; Pan, Q.; Rao, S.; et al. The cancer metabolic reprogramming and immune response. Mol. Cancer 2021, 20, 28. [Google Scholar] [CrossRef]
- Heiden, M.G.V.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Khan, A.A.; Allemailem, K.S.; Alhumaydhi, F.A.; Gowder, S.J.T.; Rahmani, A.H. The Biochemical and Clinical Perspectives of Lactate Dehydrogenase: An Enzyme of Active Metabolism. Endocr. Metab. Immune Disord.-Drug Targets 2020, 20, 855–868. [Google Scholar] [CrossRef]
- Mishra, D.; Banerjee, D. Lactate Dehydrogenases as Metabolic Links between Tumor and Stroma in the Tumor Microenvironment. Cancers 2019, 11, 750. [Google Scholar] [CrossRef]
- Forkasiewicz, A.; Dorociak, M.; Stach, K.; Szelachowski, P.; Tabola, R.; Augoff, K. The usefulness of lactate dehydrogenase measurements in current oncological practice. Cell. Mol. Biol. Lett. 2020, 25, 35. [Google Scholar] [CrossRef]
- Kocianova, E.; Piatrikova, V.; Golias, T. Revisiting the Warburg effect with focus on lactate. Cancers 2022, 14, 6028. [Google Scholar] [CrossRef]
- Comandatore, A.; Franczak, M.; Smolenski, R.T.; Morelli, L.; Peters, G.J.; Giovannetti, E. Lactate Dehydrogenase and its clinical significance in pancreatic and thoracic cancers. Semin Cancer Biol. 2022, 86, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Xiong, Y.; Qiao, T.; Li, X.; Jia, L.; Han, Y. Lactate dehydrogenase A: A key player in carcinogenesis and potential target in cancer therapy. Cancer Med. 2018, 7, 6124–6136. [Google Scholar] [CrossRef] [PubMed]
- Claps, G.; Faouzi, S.; Quidville, V.; Chehade, F.; Shen, S.; Vagner, S.; Robert, C. The multiple roles of LDH in cancer. Nat. Rev. Clin. Oncol. 2022, 19, 749–762. [Google Scholar] [CrossRef]
- Urbańska, K.; Orzechowski, A. Unappreciated role of LDHA and LDHB to control apoptosis and autophagy in tumor cells. Int. J. Mol. Sci. 2019, 20, 2085. [Google Scholar] [CrossRef]
- Wang, R.; Li, J.; Zhang, C.; Guan, X.; Qin, B.; Jin, R.; Qin, L.; Xu, S.; Zhang, X.; Liu, R. Lactate dehydrogenase B is required for pancreatic cancer cell immortalization through activation of telomerase activity. Front. Oncol. 2022, 12, 821620. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.; Xiong, G.; Cao, Z.; Yang, G.; Zheng, S.; Qiu, J.; You, L.; Zheng, L.; Zhang, T.; Zhao, Y. LAT2 regulates glutamine-dependent mTOR activation to promote glycolysis and chemoresistance in pancreatic cancer. J. Exp. Clin. Cancer Res. 2018, 37, 274. [Google Scholar] [CrossRef]
- Arundhathi, J.D.; Mathur, S.R.; Gogia, A.; Deo, S.; Mohapatra, P.; Prasad, C.P. Metabolic changes in triple negative breast cancer-focus on aerobic glycolysis. Mol. Biol. Rep. 2021, 48, 4733–4745. [Google Scholar] [CrossRef] [PubMed]
- Khajah, M.A.; Khushaish, S.; Luqmani, Y.A. Lactate dehydrogenase A or B knockdown reduces lactate production and inhibits breast cancer cell motility in vitro. Front. Pharmacol. 2021, 12, 747001. [Google Scholar] [CrossRef]
- Marti, T.M.; Deng, H.; Gao, Y.; Trappetti, V.; Hertig, D.; Karatkevich, D.; Losmanova, T.; Urzi, C.; Ge, H.; Geest, G.A. Targeting Lactate Dehydrogenase B-dependent Mitochondrial Metabolism Affects Tumor Initiating Cells and Inhibits Tumorigenesis of Non-small Cell Lung Cancer by Inducing MtDNA Damage. Cell. Mol. Life Sci. 2022, 79, 445. [Google Scholar]
- Cai, S.; Xia, Q.; Duan, D.; Fu, J.; Wu, Z.; Yang, Z.; Yu, C. Creatine kinase mitochondrial 2 promotes the growth and progression of colorectal cancer via enhancing Warburg effect through lactate dehydrogenase B. PeerJ 2024, 12, e17672. [Google Scholar] [CrossRef]
- Liu, S.; Zhao, H.; Hu, Y.; Yan, C.; Mi, Y.; Li, X.; Tao, D.; Qin, J. Lactate promotes metastasis of normoxic colorectal cancer stem cells through PGC-1α-mediated oxidative phosphorylation. Cell Death Dis. 2022, 13, 651. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, G.; Liu, Z.; Liu, S.; Cai, Z.; You, P.; Ke, Y.; Lai, L.; Huang, Y.; Gao, H. Aberrant FGFR tyrosine kinase signaling enhances the warburg effect by reprogramming LDH isoform expression and activity in prostate cancer. Cancer Res. 2018, 78, 4459–4470. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Yang, F.; Gu, Z.; Li, W.; Yuan, Y.; Liu, S.; Zhou, L.; Han, B.; Zheng, R.; Cao, Z. Fibroblast growth factor pathway promotes glycolysis by activating LDHA and suppressing LDHB in a STAT1-dependent manner in prostate cancer. J. Transl. Med. 2024, 22, 474. [Google Scholar] [CrossRef]
- Zhang, P.; Wan, Y.; Ma, J.; Gong, J.; Zhong, Z.; Cui, Y.; Zhang, H.; Da, Y.; Ma, J.; Li, C. Epigenetic silencing of LDHB promotes hepatocellular carcinoma by remodeling the tumor microenvironment. Cancer Immunol. Immunother. 2024, 73, 127. [Google Scholar] [CrossRef]
- Jin, H.; Liu, Q.; Li, J.; Zhao, S.; Tuo, B. Multifaceted roles of lactate dehydrogenase in liver cancer. Int. J. Oncol. 2025, 66, 50. [Google Scholar] [CrossRef] [PubMed]
- Bononi, G.; Di Bussolo, V.; Tuccinardi, T.; Minutolo, F.; Granchi, C. A patent review of lactate dehydrogenase inhibitors (2014–present). Expert Opin. Ther. Pat. 2024, 34, 1121–1135. [Google Scholar] [CrossRef]
- Balkrishna, A.; Sharma, N.; Srivastava, D.; Kukreti, A.; Srivastava, S.; Arya, V. Exploring the safety, efficacy, and bioactivity of herbal medicines: Bridging traditional wisdom and modern science in healthcare. Future Integr. Med. 2024, 3, 35–49. [Google Scholar] [CrossRef]
- Misra, R. Modern drug development from traditional medicinal plants using radioligand receptor-binding assays. Med. Res. Rev. 1998, 18, 383–402. [Google Scholar] [CrossRef]
- Zhang, B.; Zhao, S.; Yang, D.; Wu, Y.; Xin, Y.; Cao, H.; Huang, X.-P.; Cai, X.; Sun, W.; Ye, N. A Novel G Protein-Biased and Subtype-Selective Agonist for a G Protein-Coupled Receptor Discovered from Screening Herbal Extracts. ACS Cent. Sci. 2020, 6, 823–824. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Tassawer, A.; Khan, M.U.; Shehroz, M.; Nishan, U.; Sheheryar, S.; Dib, H.; Abdelfattah, M.A.; Shah, M. Identification of novel amides and alkaloids as putative inhibitors of dopamine transporter for schizophrenia using computer-aided virtual screening. Front. Pharmacol. 2025, 16, 1509263. [Google Scholar] [CrossRef]
- Nada, H.; Meanwell, N.A.; Gabr, M.T. Virtual screening: Hope, hype, and the fine line in between. Expert Opin. Drug Discov. 2025, 20, 145–162. [Google Scholar] [CrossRef]
- Tao, Y.; Yan, J.; Cai, B. Label-free bio-affinity mass spectrometry for screening and locating bioactive molecules. Mass Spectrom. Rev. 2021, 40, 53–71. [Google Scholar] [CrossRef]
- Hsu, S.-W.; Chang, T.-C.; Wu, Y.-K.; Lin, K.-T.; Shi, L.-S.; Lee, S.-Y. Rhodiola crenulata extract counteracts the effect of hypobaric hypoxia in rat heart via redirection of the nitric oxide and arginase 1 pathway. BMC Complement. Altern. Med. 2017, 17, 29. [Google Scholar] [CrossRef]
- Chang, P.-K.; Yen, I.-C.; Tsai, W.-C.; Chang, T.-C.; Lee, S.-Y. Protective Effects of Rhodiola crenulata Extract on Hypoxia-Induced Endothelial Damage via Regulation of AMPK and ERK Pathways. Int. J. Mol. Sci. 2018, 19, 2286. [Google Scholar] [CrossRef]
- Li, J.; Cui, Y.; Yao, J.; Guo, C.; Jiao, M. Mechanistic insights of Rhodiola crenulata in treating diabetic kidney disease via network pharmacology. J. Mol. Endocrinol. 2025, 75, e250006. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, Y.; Su, M.; Chu, X.; Li, S.; Yue, Y.; Zhang, X.; Wang, J.; Han, F. Brain metabolomics study for the protective effects of Rhodiola crenulata extract on Alzheimer’s disease by HPLC coupled with Fourier transform-ion cyclotron resonance mass spectrometry. J. Sep. Sci. 2020, 43, 3216–3223. [Google Scholar] [CrossRef]
- Zhu, R.; Fang, C.-F.; Zhang, S.-J.; Han, Z.; Zhu, G.-H.; Cai, S.-Z.; Zheng, C.; Tang, Y.; Wang, Y. Comprehensive Review on Rhodiola crenulata: Ethnopharmacology, Phytochemistry, Pharmacological Properties and Clinical Applications. Chin. J. Integr. Med. 2025, 31, 752–759. [Google Scholar] [CrossRef]
- Yuan, C.; Jin, Y.; Yao, L.; Liu, L.; Li, J.; Li, H.; Lai, Y.; Chen, Z.; Pan, Z.; Han, T. Rhodiola crenulata root extract ameliorates fructose-induced hepatic steatosis in rats: Association with activating autophagy. Biomed. Pharmacother. 2020, 125, 109836. [Google Scholar] [CrossRef]
- Recio, M.-C.; Giner, R.-M.; Máñez, S. Immunmodulatory and antiproliferative properties of Rhodiola species. Planta Medica 2016, 82, 952–960. [Google Scholar] [CrossRef]
- Ravi, P.; Jasuja, H.; Sarkar, D.; Vahidi Pashaki, B.; Gaikwad, H.K.; Vahidi Pashaki, P.; Katti, D.R.; Shetty, K.; Katti, K.S. Rhodiola crenulata induces apoptosis in bone metastatic breast cancer cells via activation of caspase-9 and downregulation of MtMP activity. Sci. Rep. 2025, 15, 9341. [Google Scholar] [CrossRef] [PubMed]
- Dudek, M.C.; Wong, K.E.; Bassa, L.M.; Mora, M.C.; Ser-Dolansky, J.; Henneberry, J.M.; Crisi, G.M.; Arenas, R.B.; Schneider, S.S. Antineoplastic effects of Rhodiola crenulata treatment on B16-F10 melanoma. Tumor Biol. 2015, 36, 9795–9805. [Google Scholar] [CrossRef]
- Mora, M.C.; Bassa, L.M.; Wong, K.E.; Tirabassi, M.V.; Arenas, R.B.; Schneider, S.S. Rhodiola crenulata inhibits Wnt/β-catenin signaling in glioblastoma. J. Surg. Res. 2015, 197, 247–255. [Google Scholar] [CrossRef]
- Sun, A.-Q.; Ju, X.-L. Advances in research on anticancer properties of salidroside. Chin. J. Integr. Med. 2021, 27, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cui, X.; Chen, Z. Screening of lactate dehydrogenase inhibitor from bioactive compounds in natural products by electrophoretically mediated microanalysis. J. Chromatogr. A 2021, 1656, 462554. [Google Scholar] [CrossRef]
- Rogatzki, M.J.; Ferguson, B.S.; Goodwin, M.L.; Gladden, L.B. Lactate is always the end product of glycolysis. Front. Neurosci. 2015, 9, 22. [Google Scholar] [CrossRef]
- Zhang, J.H.; Chung, T.D.Y.; Oldenburg, K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999, 4, 67–73. [Google Scholar] [CrossRef]
- Tao, H.; Wu, X.; Cao, J.; Peng, Y.; Wang, A.; Pei, J.; Xiao, J.; Wang, S.; Wang, Y. Rhodiola species: A comprehensive review of traditional use, phytochemistry, pharmacology, toxicity, and clinical study. Med. Res. Rev. 2019, 39, 1779–1850. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Liu, H.; Sha, Y.; Sun, L. A comparative study of phytochemical metabolites and antioxidant properties of Rhodiola. Arab. J. Chem. 2023, 16, 104420. [Google Scholar] [CrossRef]
- Dong, T.; Sha, Y.; Liu, H.; Sun, L. Altitudinal variation of metabolites, mineral elements and antioxidant activities of Rhodiola crenulata (Hook. f. & Thomson) H. Ohba. Molecules 2021, 26, 7383. [Google Scholar]
- Yang, Y.-N.; Liu, Z.-Z.; Feng, Z.-M.; Jiang, J.-S.; Zhang, P.-C. Lignans from the root of Rhodiola crenulata. J. Agric. Food Chem. 2012, 60, 964–972. [Google Scholar] [CrossRef]
- Ma, D.; Wang, L.; Jin, Y.; Gu, L.; Yin, G.; Wang, J.; Yu, X.-A.; Huang, H.; Zhang, Z.; Wang, B. Chemical characteristics of Rhodiola crenulata and its mechanism in acute mountain sickness using UHPLC-Q-TOF-MS/MS combined with network pharmacology analysis. J. Ethnopharmacol. 2022, 294, 115345. [Google Scholar] [CrossRef]
- Chen, K.; Liu, J.; Ma, Z.; Duan, F.; Guo, Z.; Xiao, H. Rapid identification of chemical constituents of Rhodiola crenulata using liquid chromatography-mass spectrometry pseudotargeted analysis. J. Sep. Sci. 2021, 44, 3747–3776. [Google Scholar] [CrossRef]
- Han, F.; Li, Y.; Mao, X.; Xu, R.; Yin, R. Characterization of chemical constituents in Rhodiola Crenulate by high-performance liquid chromatography coupled with Fourier-transform ion cyclotron resonance mass spectrometer (HPLC-FT-ICR MS). J. Mass Spectrom. 2016, 51, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Li, Y.; Ma, L.; Liu, T.; Wu, Y.; Xu, R.; Song, A.; Yin, R. A rapid and sensitive UHPLC-FT-ICR MS/MS method for identification of chemical constituents in Rhodiola crenulata extract, rat plasma and rat brain after oral administration. Talanta 2016, 160, 183–193. [Google Scholar] [CrossRef]
- Svoboda, P.; Vlčková, H.; Nováková, L. Development and validation of UHPLC-MS/MS method for determination of eight naturally occurring catechin derivatives in various tea samples and the role of matrix effects. J. Pharm. Biomed. Anal. 2015, 114, 62–70. [Google Scholar] [CrossRef]
- Thabault, L.; Liberelle, M.; Koruza, K.; Yildiz, E.; Joudiou, N.; Messens, J.; Brisson, L.; Wouters, J.; Sonveaux, P.; Frédérick, R. Discovery of a novel lactate dehydrogenase tetramerization domain using epitope mapping and peptides. J. Biol. Chem. 2021, 296, 100422. [Google Scholar] [CrossRef]
- Xu, B.; Zhang, H.L.; Shen, B.; Wu, J.M.; Shi, M.T.; Li, X.D.; Guo, Q. Identification biomarkers and therapeutic targets of disulfidptosis-related in rheumatoid arthritis via bioinformatics, molecular dynamics simulation, and experimental validation. Sci. Rep. 2025, 15, 8779. [Google Scholar] [CrossRef]
- Bissantz, C.; Kuhn, B.; Stahl, M. A Medicinal Chemist’s Guide to Molecular Interactions. J. Med. Chem. 2010, 53, 5061–5084. [Google Scholar] [CrossRef]
- Yırtıcı, Ü. Natural flavonoids as promising lactate dehydrogenase A inhibitors: Comprehensive in vitro and in silico analysis. Arch. Der Pharm. 2024, 357, 2400455. [Google Scholar] [CrossRef]
- Bae, J.; Kim, N.; Shin, Y.; Kim, S.-Y.; Kim, Y.-J. Activity of catechins and their applications. Biomed. Dermatol. 2020, 4, 8. [Google Scholar] [CrossRef]
- Mishra, S.; Vinayak, M. Role of ellagic acid in regulation of apoptosis by modulating novel and atypical PKC in lymphoma bearing mice. BMC Complement. Altern. Med. 2015, 15, 281. [Google Scholar] [CrossRef]
- McCleland, M.L.; Adler, A.S.; Shang, Y.; Hunsaker, T.; Truong, T.; Peterson, D.; Torres, E.; Li, L.; Haley, B.; Stephan, J.-P. An integrated genomic screen identifies LDHB as an essential gene for triple-negative breast cancer. Cancer Res. 2012, 72, 5812–5823. [Google Scholar] [CrossRef]
- Huang, X.; Xie, X.; Wang, H.; Xiao, X.; Yang, L.; Tian, Z.; Guo, X.; Zhang, L.; Tang, H.; Xie, X. PDL1 And LDHA act as ceRNAs in triple negative breast cancer by regulating miR-34a. J. Exp. Clin. Cancer Res. 2017, 36, 129.61. [Google Scholar] [CrossRef]
- Wu, S.; Liu, J.; Jin, K. Expression of human LDHB in prokaryote and establishment of its inhibitor screening model. J. Shanxi Med. Univ. 2018, 49, 591–595. [Google Scholar] [CrossRef]
- Bisswanger, H. Practical Enzymology, 2nd ed.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2011. [Google Scholar]
- Zhou, Y.; Tao, P.; Wang, M.; Xu, P.; Lu, W.; Lei, P.; You, Q. Development of novel human lactate dehydrogenase A inhibitors: High-throughput screening, synthesis, and biological evaluations. Eur. J. Med. Chem. 2019, 177, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Shibata, S.; Sogabe, S.; Miwa, M.; Fujimoto, T.; Takakura, N.; Naotsuka, A.; Kitamura, S.; Kawamoto, T.; Soga, T. Identification of the first highly selective inhibitor of human lactate dehydrogenase B. Sci. Rep. 2021, 11, 21353. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Li, X.; Li, Y.; Zhang, X.; Li, X.; Li, X.; Shi, Z.; Bao, J. Screening of novel inhibitors targeting lactate dehydrogenase A via four molecular docking strategies and dynamics simulations. J. Mol. Model. 2015, 21, 133. [Google Scholar] [CrossRef] [PubMed]
- Fiume, L.; Manerba, M.; Vettraino, M.; Di Stefano, G. Inhibition of lactate dehydrogenase activity as an approach to cancer therapy. Future Med. Chem. 2014, 6, 429–445. [Google Scholar] [CrossRef]
- Vlasiou, M.; Nicolaidou, V.; Papaneophytou, C. Targeting lactate dehydrogenase-B as a strategy to fight cancer: Identification of potential inhibitors by in silico analysis and in vitro screening. Pharmaceutics 2023, 15, 2411. [Google Scholar] [CrossRef]

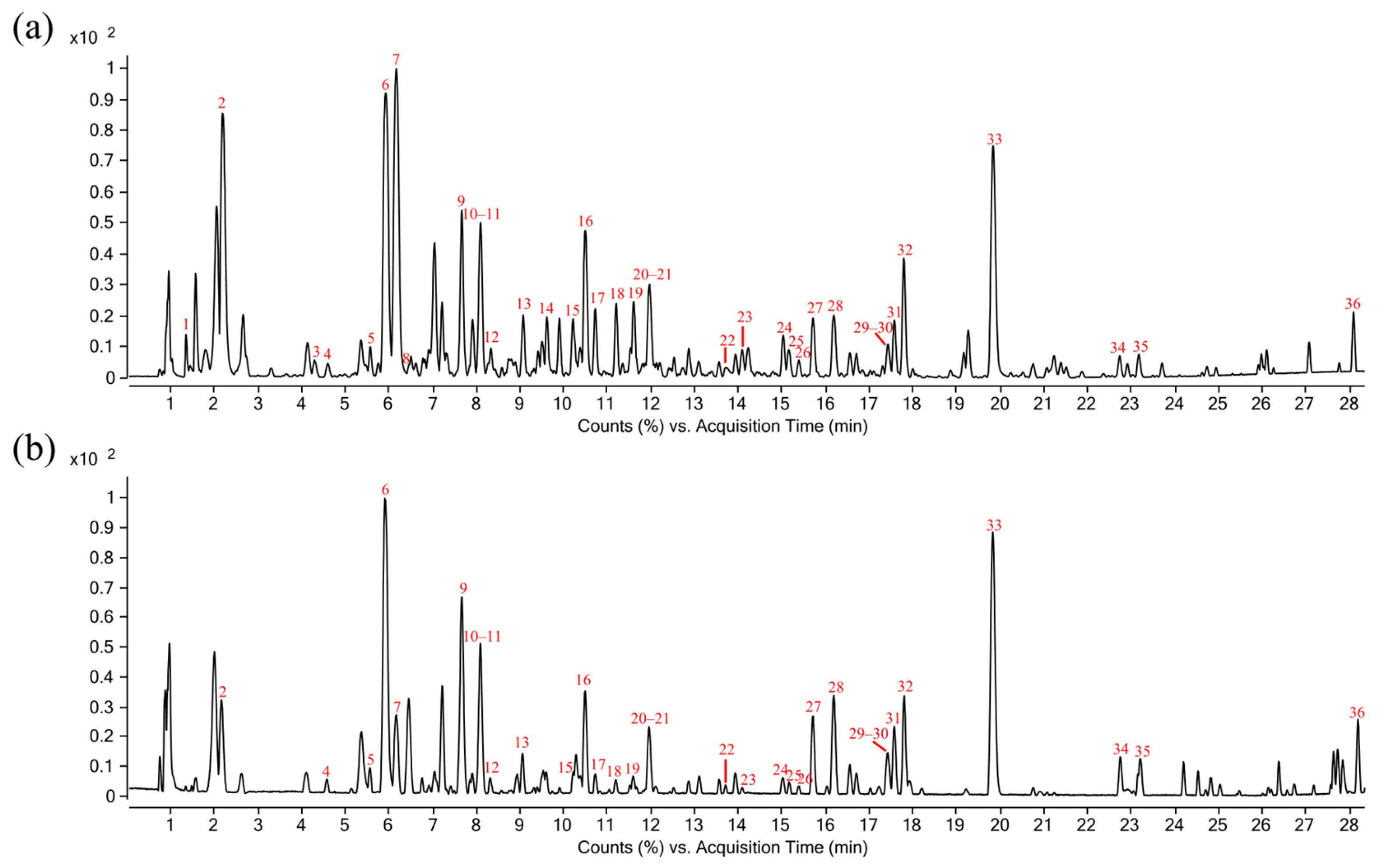
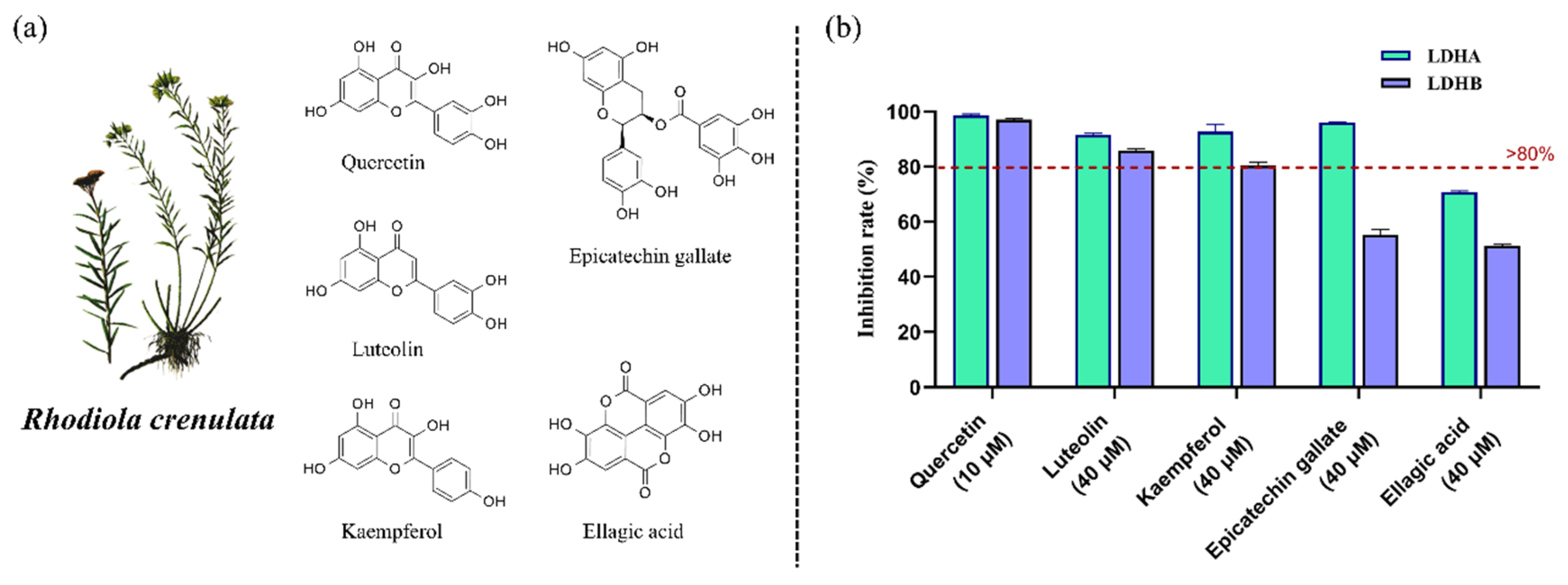
| No. | RT (min) | Precursor Ion | MS/MS (Da) | Formula | Identification | Reference | |
|---|---|---|---|---|---|---|---|
| Observed Mass (Da) | Error (ppm) | ||||||
| 1 | 1.349 | 191.0194 [M−H]− | 1.7 | (−): 129.0188, 111.0086, 87.0084, 85.0292, 67.0187, 57.0346 | C6H8O7 | Citric acid | standard |
| 2 | 2.19 | 169.0145 [M−H]−/171.0291 [M+H]+ | −1.49/−1.77 | (−): 125.0243, 79.0191, 51.0244; (+): 153.0181, 127.0389, 125.0229, 109.0285, 107.0126, 81.0335, 53.0388 | C7H6O5 | Gallic acid | standard |
| 3 | 4.296 | 153.0190 [M−H]−/155.0341 [M+H]+ | 2.16/−0.75 | (−): 108.0212, 109.0294, 110.0325, 91.0189, 81.0345, 53.0398; (+): 137.0227, 93.0335, 111.0440, 81.0333, 65.0387, 53.0383 | C7H6O4 | Protocatechuic acid | standard |
| 4 | 4.596 | 313.0924 [M−H]− | 1.56 | (−): 151.0394, 161.0448, 107.0501 | C14H18O8 | 2-(β-D-glucopyranosyloxy)-1-(4-hydroxyphenyl)ethanone | [46] |
| 5 | 5.570 | 461.1660 [M−H]− | 0.97 | (−): 299.1124, 179.0554, 119.0345, 101.0243, 89.0244 | C20H30O12 | 2-(4-hydroxyphenyl)ethyl-6-0-β-D-glucopyranosyl-β-D-glucopyranoside | [47] |
| 6 | 5.927 | 345.1199 [M+HCOO]−/318.1556 [M+NH4]+ | −2.65/−2.9 | (−): 299.1132, 89.0245, 119.0496, 179.0559, 59.0141, 71.0138, 101.0244, 113.0240; (+):69.0335, 85.0285, 121.0649, 145.0496, 187.0751, 205.0856, 229.0858, 247.0961, 265.1070 | C14H20O7 | Salidroside | standard |
| 7 | 6.169 | 293.1252 [M+HCOO]−/271.1156 [M+Na]+ | −4.07/−1.57 | (−): 247.1174, 161.0448, 101.0234, 71.0134, 59.0147; (+): 201.0367, 203.0514 | C11H20O6 | Crenulatin | standard |
| 8 | 6.468 | 137.0597 [M−H]− | 7.99 | (−): 119.0501, 106.0420 | C8H10O2 | Tyrosol | standard |
| 9 | 7.667 | 322.1866 [M+NH4]+ | −1.88 | (+): 125.0962, 107.0856 | C14H24O7 | Creoside I or Isomer | [48] |
| 10 | 8.050 | 179.0348 [M−H]− | 1.01 | (−): 134.0368, 135.0445, 107.0501 | C9H8O4 | caffeic acid | standard |
| 11 | 8.100 | 322.1866 [M+NH4]+ | −1.88 | (+): 125.0962, 107.0856 | C14H24O7 | Creoside I or Isomer | [48] |
| 12 | 8.333 | 577.1347 [M−H]− | 0.78 | (−): 407.0768, 289.0705, 125.0241 | C30H26O12 | Procyanidin B3 | [49] |
| 13 | 9.073 | 289.0715 [M−H]−/291.0868 [M+H]+ | 0.9/−1.67 | (−): 245.0811, 221.0818, 203.0702, 151.0391, 123.0446, 109.0290; (+): 207.0648, 165.0545, 147.0438, 139.0387, 123.0440, 111.0437 | C15H14O6 | (−)-epicatechin | standard |
| 14 | 9.615 | 635.0885 [M−H]− | 0.77 | (−): 483.0785, 423.0574, 313.0560, 169.0139, 125.0242 | C27H24O18 | trigalloyl-β-D-glucopyranose/isomer | [50] |
| 15 | 10.239 | 729.1455 [M−H]− | 0.83 | (−): 577.1325, 451.1040, 407.0774, 289.0718, 169.0143, 125.0248 | C37H30O16 | catechin-(4α-8)-catechin-3′0-gallate/epicatechin-(4β-8)epicatechingallate | [49] |
| 16 | 10.489 | 609.1461 [M−H]− | 0.01 | (−): 446.0854, 283.0242, 151.0031 | C27H30O16 | kaempferol 3,7-digalactoside/Isomer | [50] |
| 17 | 10.722 | 399.1296 [M−H]− | 0.18 | (−): 329.0515, 271.0453, 169.0141, 125.0241 | C18H24O10 | 3-O-galloyl quinic acid butyl ester/Isomer | [51] |
| 18 | 11.201 | 787.0993/[M−H]− | 0.82 | (−): 635.0906, 617.0788, 465.0676, 169.0139, 295.0453 | C34H28O22 | tetragalloyl-β-D-glucopyranose/isomer | [50] |
| 19 | 11.604 | 881.1566/[M−H]− | 0.53 | (−): 729.1447, 711.1360, 577.1328, 559.1244, 541.1143, 407.0768, 289.0715, 169.0136 | C44H34O20 | epicatechingallate-(4β-8)-epicatechingallate | [50] |
| 20 | 11.962 | 300.9989/[M−H]− | 0.3 | (−):283.9956, 257.0090, 245.0088, 228.0048, 201.0182, 185.0236, 161.0231, 145.0289, 129.0347, 117.0342 | C14H6O8 | Ellagic acid | standard |
| 21 | 11.970 | 441.0825 [M−H]−/443.0979 [M+H]+ | 0.5/−1.42 | (−): 331.0459, 289.0708, 271.0605, 245.0817, 169.0140, 125.0243; (+): 291.0857, 273.0755, 207.0646, 165.0548, 153.0179, 139.0390, 123.0442, 111.0437 | C22H18O10 | epicatechin gallate | standard |
| 22 | 13.702 | 447.0931 [M−H]−/449.1085 [M+H]+ | 0.41/−1.48 | (−): 284.0318, 255.0292, 227.0339; (+): 287.0553, 153.0174, 85.0282 | C21H20O11 | Astragalin | standard |
| 23 | 14.084 | 187.0976 [M−H]− | −0.09 | (−): 169.0872, 143.1072, 125.0969, 102.9481, 97.0658, 57.0341 | C9H16O4 | Azelaic acid | standard |
| 24 | 15.025 | 881.1566 [M−H]− | 0.53 | (−): 729.1447, 711.1348, 577.0993, 559.1236, 541.1141, 433.0921, 407.0768 | C44H34O20 | epicatechingallate-(4β-8)-epicatechingallate | [50] |
| 25 | 15.158 | 491.2131 [M+HCOO]− | 0.67 | (−): 313.1649, 161.0460, 149.0454, 131.0346, 101.0243 | C21H34O10 | sachaloside II | [44] |
| 26 | 15.383 | 431.0983 [M−H]−/433.1136 [M+H]+ | 0.16/−1.57 | (−): 284.0322, 285.0390, 255.0289, 227.0339; (+): 287.0550, 147.0649, 129.0546, 111.0434, 85.0282, 71.0492, 57.0333 | C21H20O10 | Afzelin | standard |
| 27 | 15.708 | 609.1459 [M−H]−/611.1613 [M+H]+ | 0.34/−1.05 | (−): 301.0351; (+): 449.1076, 303.0502, 145.0489, 127.0381, 97.0282, 85.0281 | C27H30O16 | rhodiosin | standard |
| 28 | 16.182 | 447.0933 [M−H]−/449.1086 [M+H]+ | −0.03/−1.7 | (−): 301.0346, 255.0289, 229.0505, 166.9975; (+): 303.0503, 169.0124, 121.0276 | C21H20O11 | Rhodionin | standard |
| 29 | 17.422 | 285.0403 [M−H]−/287.0555 [M+H]+ | 0.56/−1.7 | (−): 199.0391, 133.0294; (+): 269.0429, 241.0494, 213.0539, 153.0179, 135.0438, 117.0328, 89.0389, 67.0179 | C15H10O6 | luteolin | standard |
| 30 | 17.439 | 301.0350 [M−H]−/303.0502 [M+H]+ | 1.25/−0.9 | (−): 178.9978, 151.0034, 121.0288, 107.0136, 83.0136, 65.0034; (+): 257.0453, 229.0498, 201.0548, 153.0182, 137.0238 | C15H10O7 | Quercetin | standard |
| 31 | 17.572 | 431.0984 [M−H]− | −0.07 | (−): 285.0394, 257.0457, 151.0039 | C21H20O10 | kaempferol-7-0-α-L-rhamnoside | [52] |
| 32 | 17.788 | 469.2291 [M+HCOO]− | −0.12 | (−): 423.2232, 291.1811, 161.0430, 101.0243 | C19H36O10 | rhodiooctanoside | [44] |
| 33 | 19.828 | 285.0410 [M−H]−/287.0554 [M+H]+ | −1.88/−1.35 | (−): 257.0431, 239.0342, 211.0396, 185.0606, 171.0445, 159.0449, 107.0137, 93.0347, 63.0245; (+): 258.0527, 213.0547, 185.0595, 153.0182, 137.0231, 121.0281, 93.0335, 68.9973 | C15H10O6 | kaempferol | standard |
| 34 | 22.733 | 319.1188 [M−H]−/321.1338 [M+H]+ | −0.28/−1.67 | (−): 287.0915, 275.1281, 245.0814, 205.0503, 191.0348, 179.0340, 161.0594, 148.0522, 135.0427; (+): 303.1229, 275.1275, 223.0597, 207.0650, 177.0542, 159.0442 | C17H20O6 | Mycophenolic acid | standard |
| 35 | 23.165 | 479.0986 [M−H]− | −0.48 | (−): 299.0193, 271.0246, 165.9900 | C25H20O10 | crenulatin A/crenulatin B/rhodiolin | [51] |
| 36 | 28.085 | 295.2278 [M−H]− | 0.23 | (−): 277.2168, 195.1385, 59.0137 | C18H32O3 | 9-hydroxy-10,12-octadecadienoic acid | GNPS |
| Component | Binding Energy (kcal/mol) | |
|---|---|---|
| LDHA (1I10) | LDHB (7DBK) | |
| Quercetin | −8.7 | −6.8 |
| Luteolin | −8.8 | −7.1 |
| Kaempferol | −8.6 | −7.2 |
| Epicatechin gallate | −7.7 | −7.0 |
| Ellagic acid | −8.5 | −6.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Y.-X.; Asgari, M.; Zhao, Q.-F.; Zhang, S.-S.; Alifujiang, Z.; Zhang, F.; Chen, X.-P.; Feng, C.-G. Inhibition Profiling of Rhodiola crenulata on Lactate Dehydrogenase Isoenzymes. Molecules 2025, 30, 4199. https://doi.org/10.3390/molecules30214199
Wei Y-X, Asgari M, Zhao Q-F, Zhang S-S, Alifujiang Z, Zhang F, Chen X-P, Feng C-G. Inhibition Profiling of Rhodiola crenulata on Lactate Dehydrogenase Isoenzymes. Molecules. 2025; 30(21):4199. https://doi.org/10.3390/molecules30214199
Chicago/Turabian StyleWei, Yi-Xin, Motahareh Asgari, Qun-Fei Zhao, Shu-Sheng Zhang, Zuliayi Alifujiang, Fang Zhang, Xiu-Ping Chen, and Chen-Guo Feng. 2025. "Inhibition Profiling of Rhodiola crenulata on Lactate Dehydrogenase Isoenzymes" Molecules 30, no. 21: 4199. https://doi.org/10.3390/molecules30214199
APA StyleWei, Y.-X., Asgari, M., Zhao, Q.-F., Zhang, S.-S., Alifujiang, Z., Zhang, F., Chen, X.-P., & Feng, C.-G. (2025). Inhibition Profiling of Rhodiola crenulata on Lactate Dehydrogenase Isoenzymes. Molecules, 30(21), 4199. https://doi.org/10.3390/molecules30214199






