Computational Discovery of Potent Nucleoprotein Inhibitors for Influenza A Virus: Validation Through QM/MM Analysis and Experimental Binding Assays
Abstract
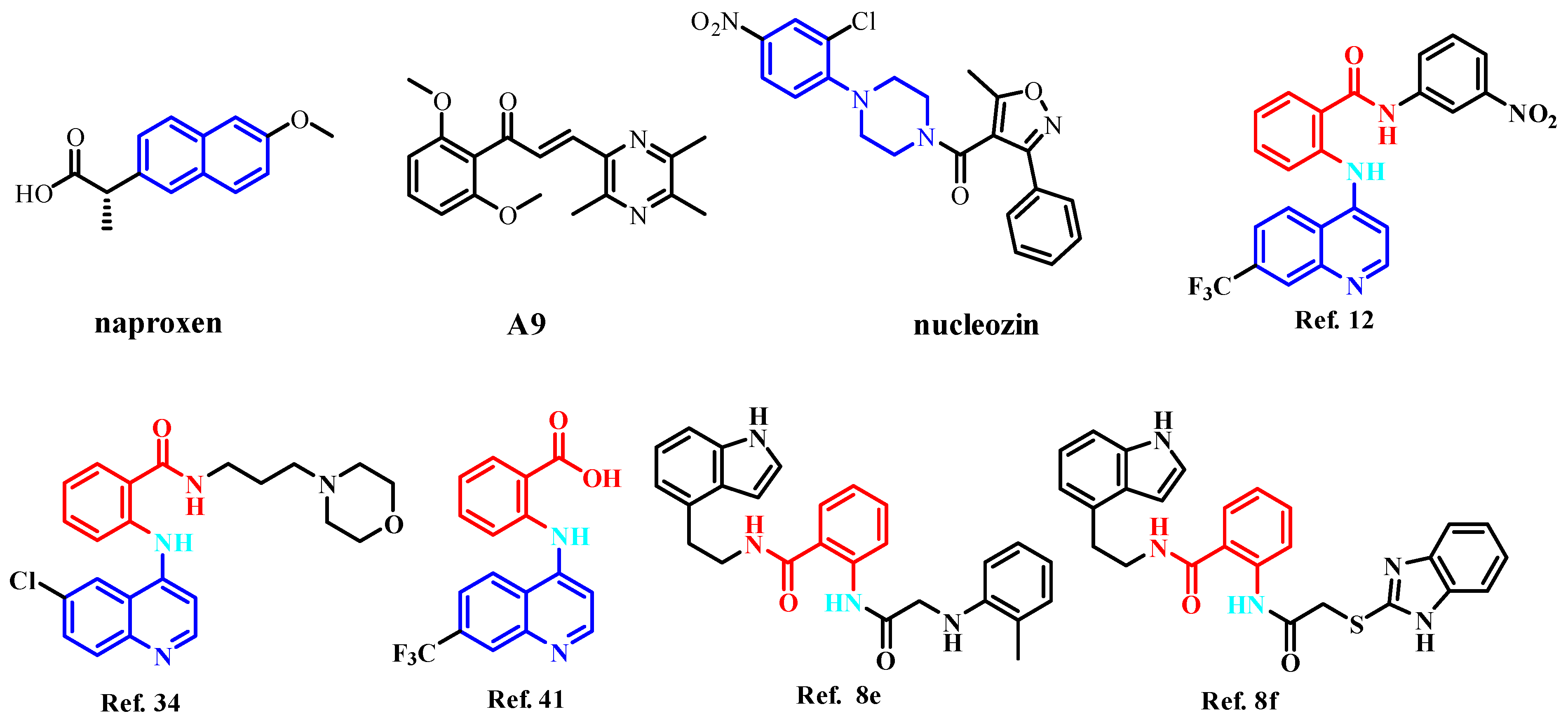
1. Introduction
2. Results and Discussion
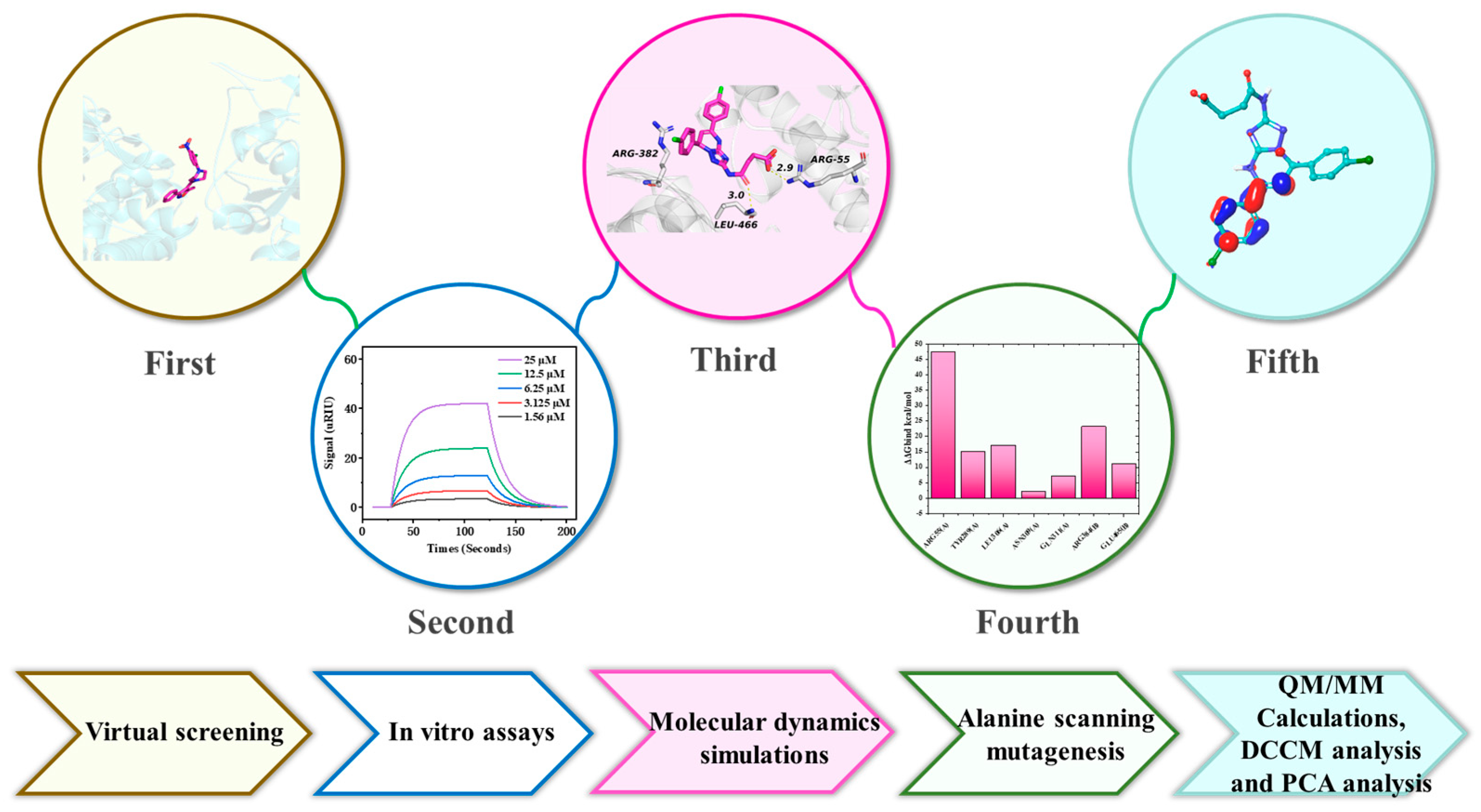
2.1. Virtual Screening
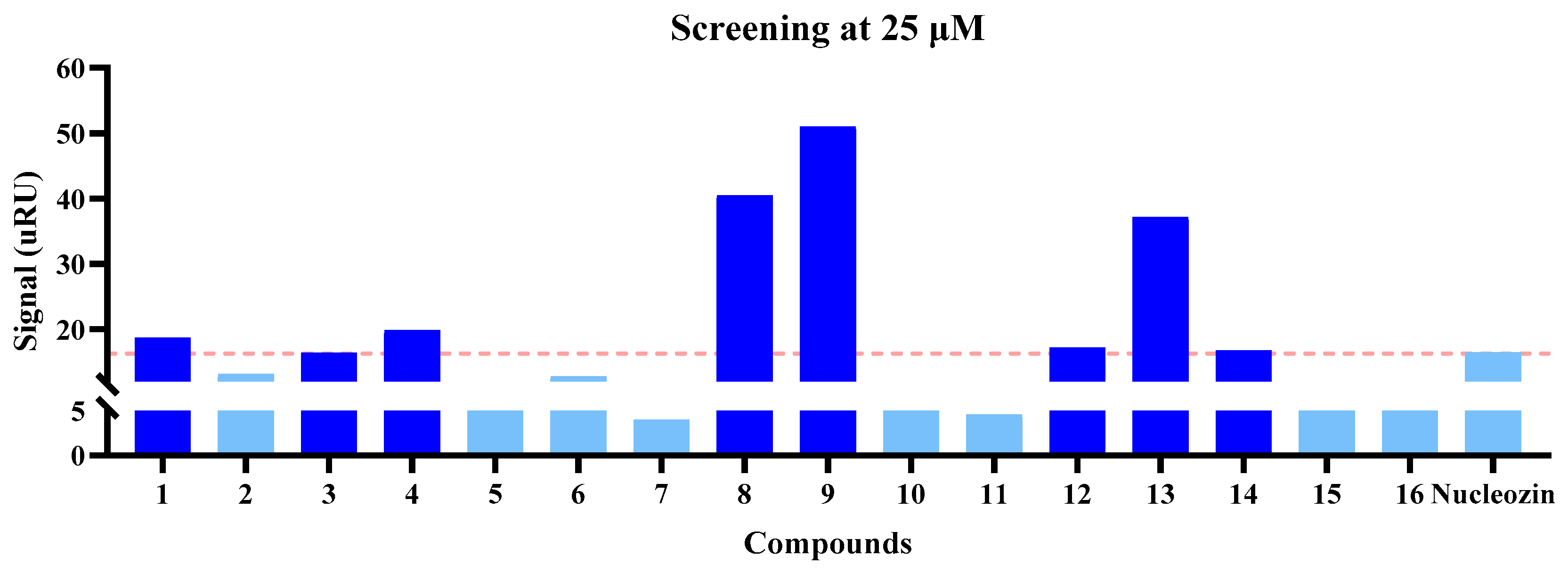
2.2. Analysis of In Vitro Assays
2.3. Molecular Dynamics Simulations
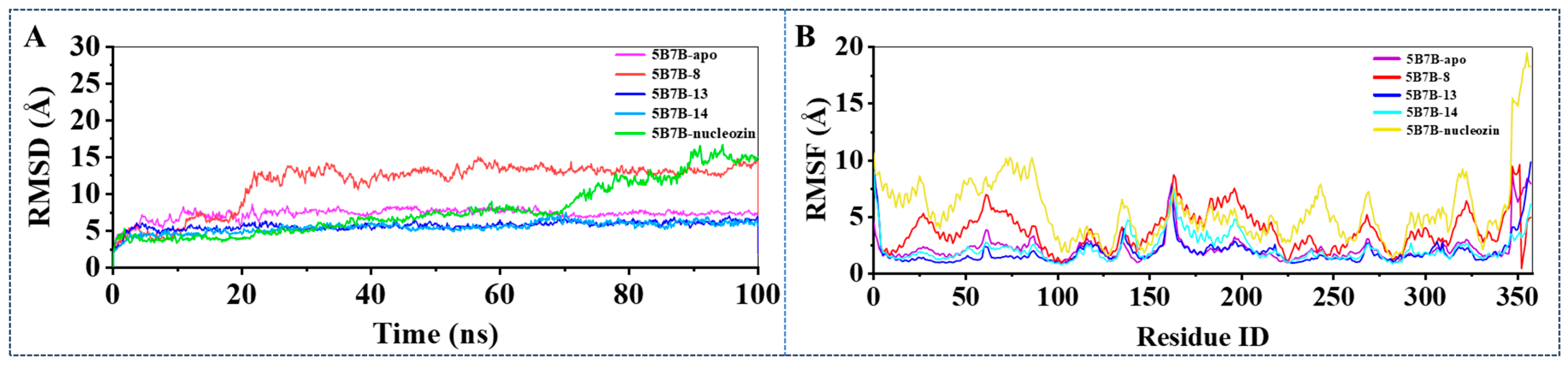
2.3.1. Stability of Dynamics Trajectory from Root Mean Square Deviation (RMSD) Analysis
2.3.2. Structural Flexibility Evaluation from Root Mean Square Fluctuation (RMSF) Analysis
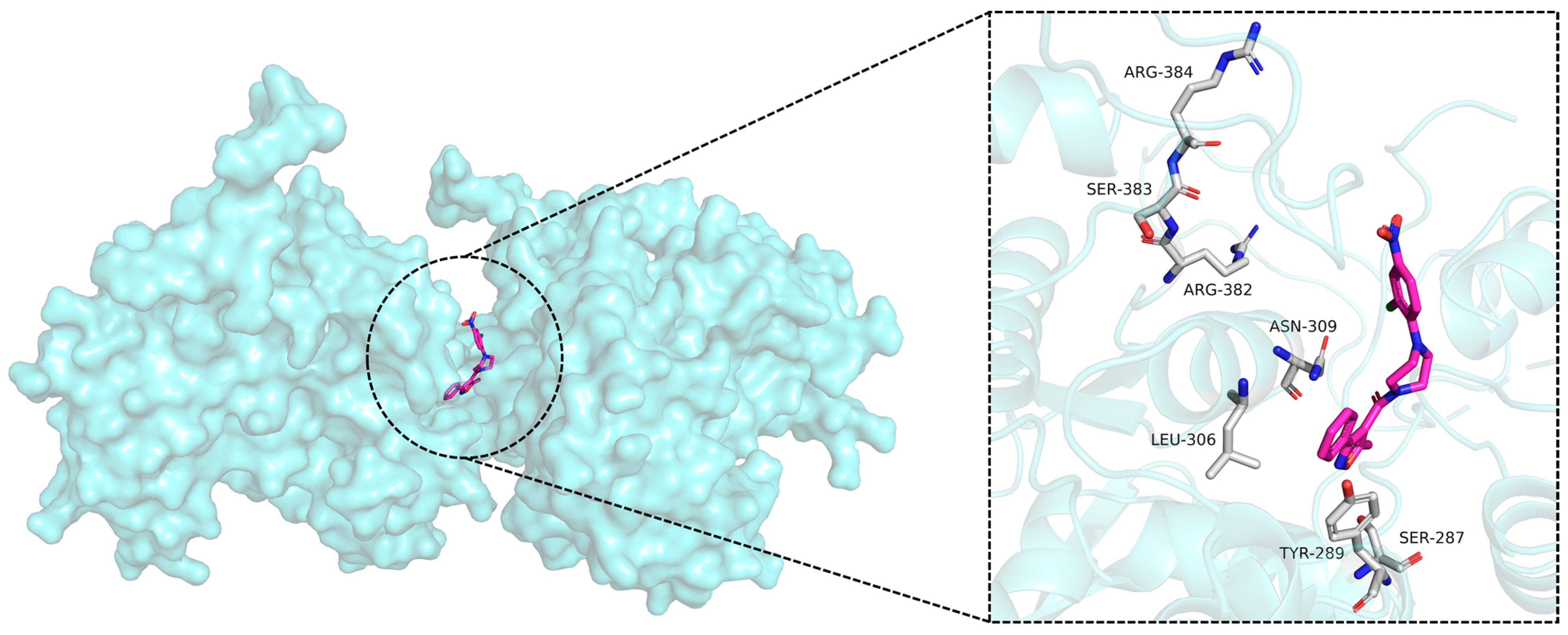
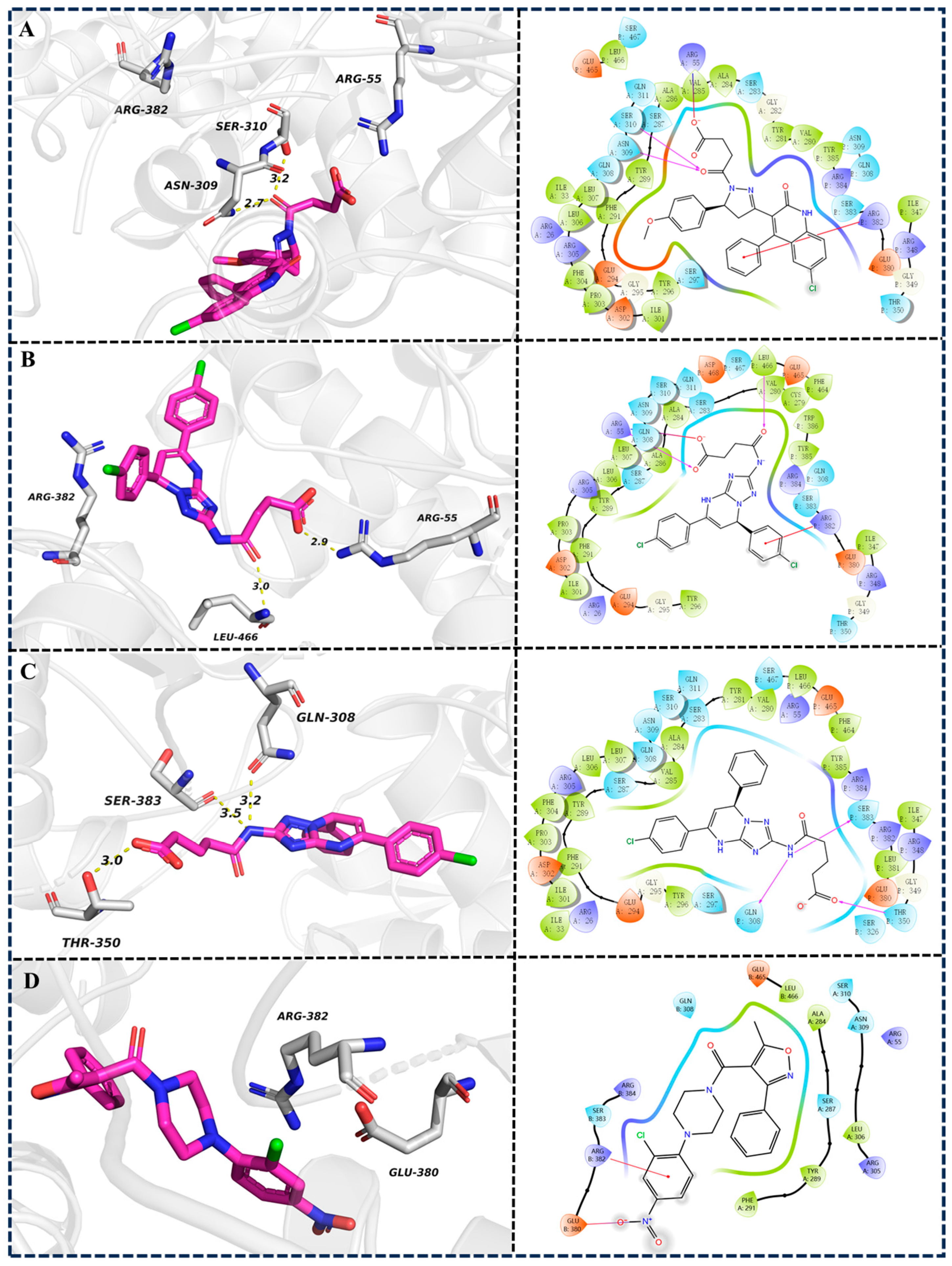
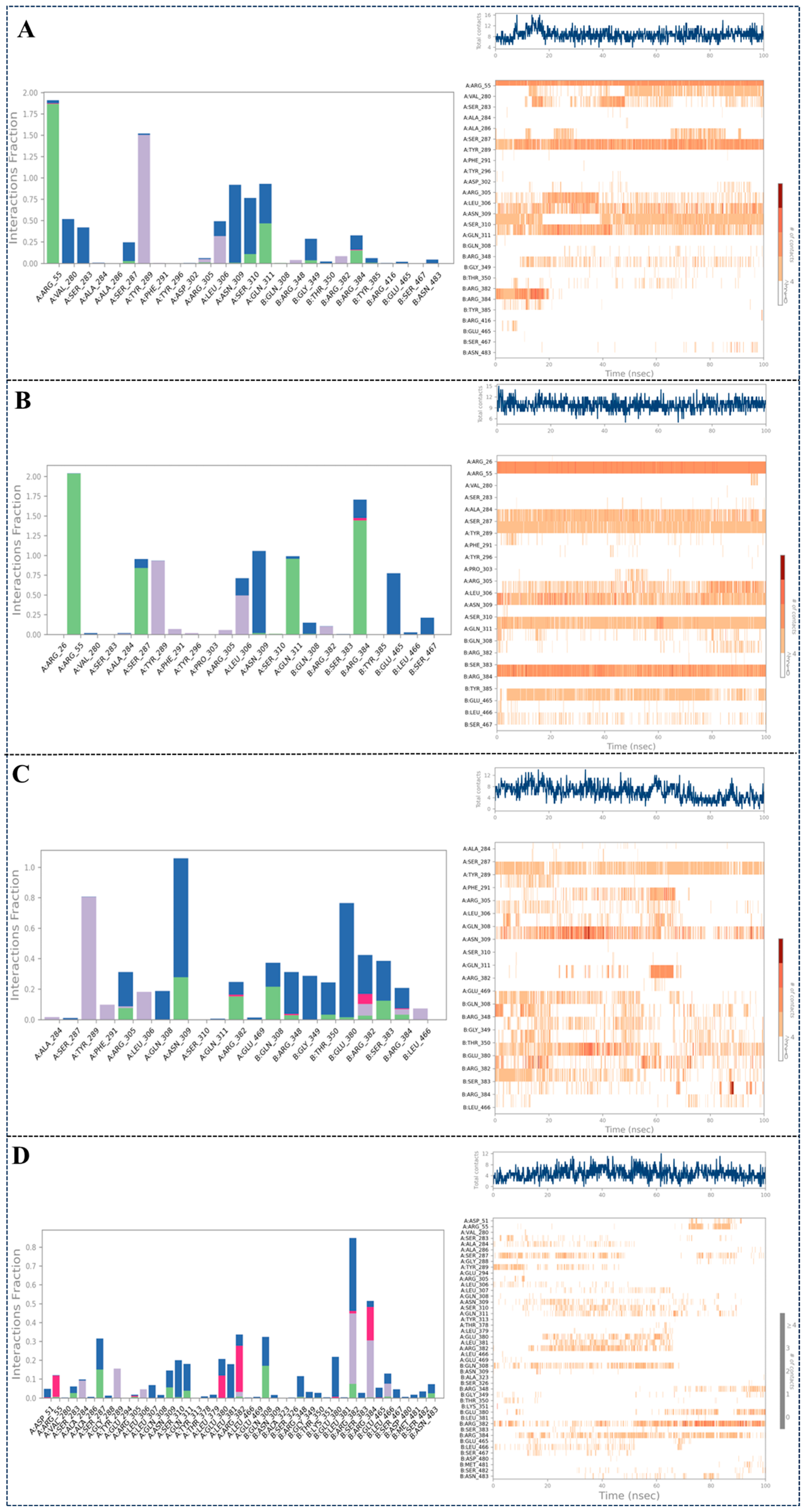
2.3.3. Intermolecular Interaction Analysis During the 100 ns MD Simulation
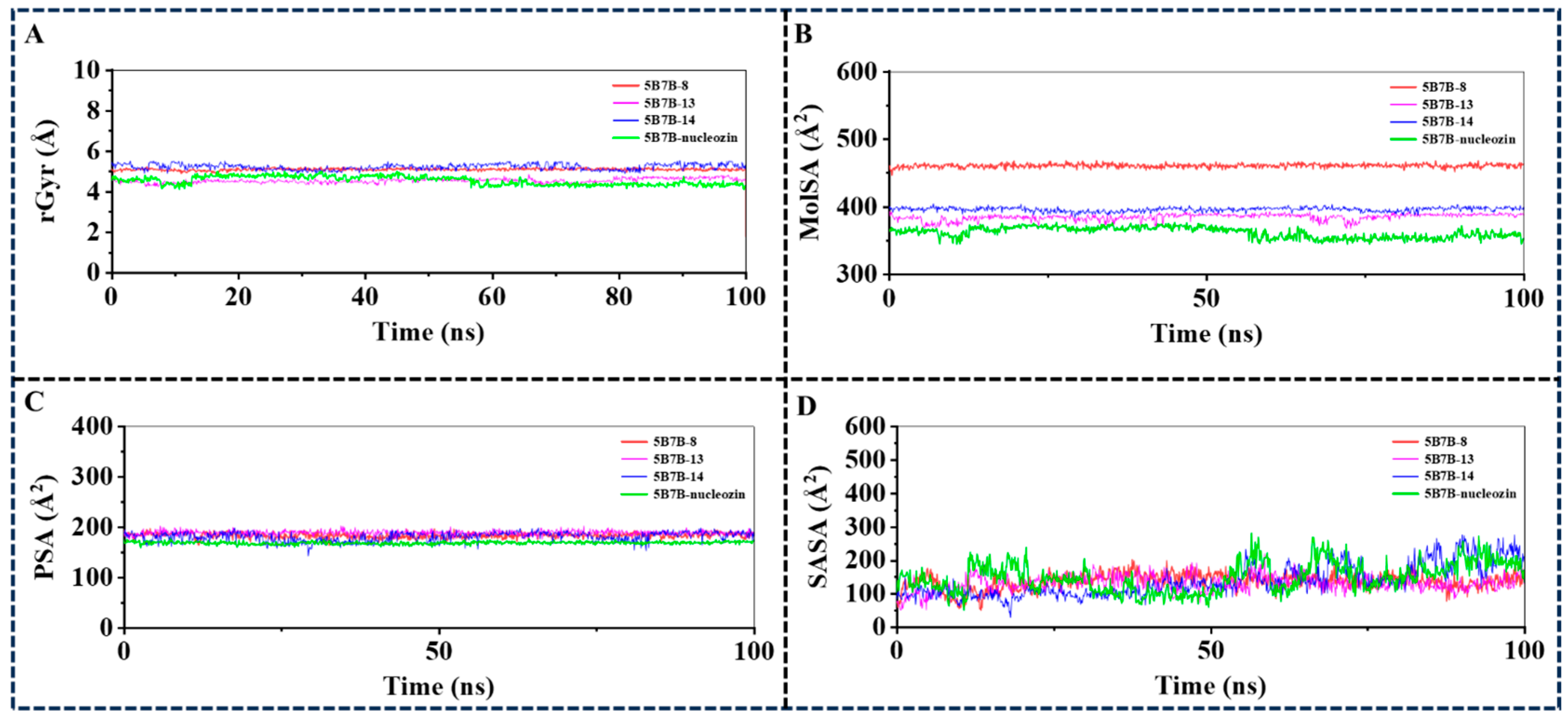
2.3.4. Analysis of Ligand Properties During the 100 ns MD Simulation
2.4. Alanine Scanning Mutagenesis
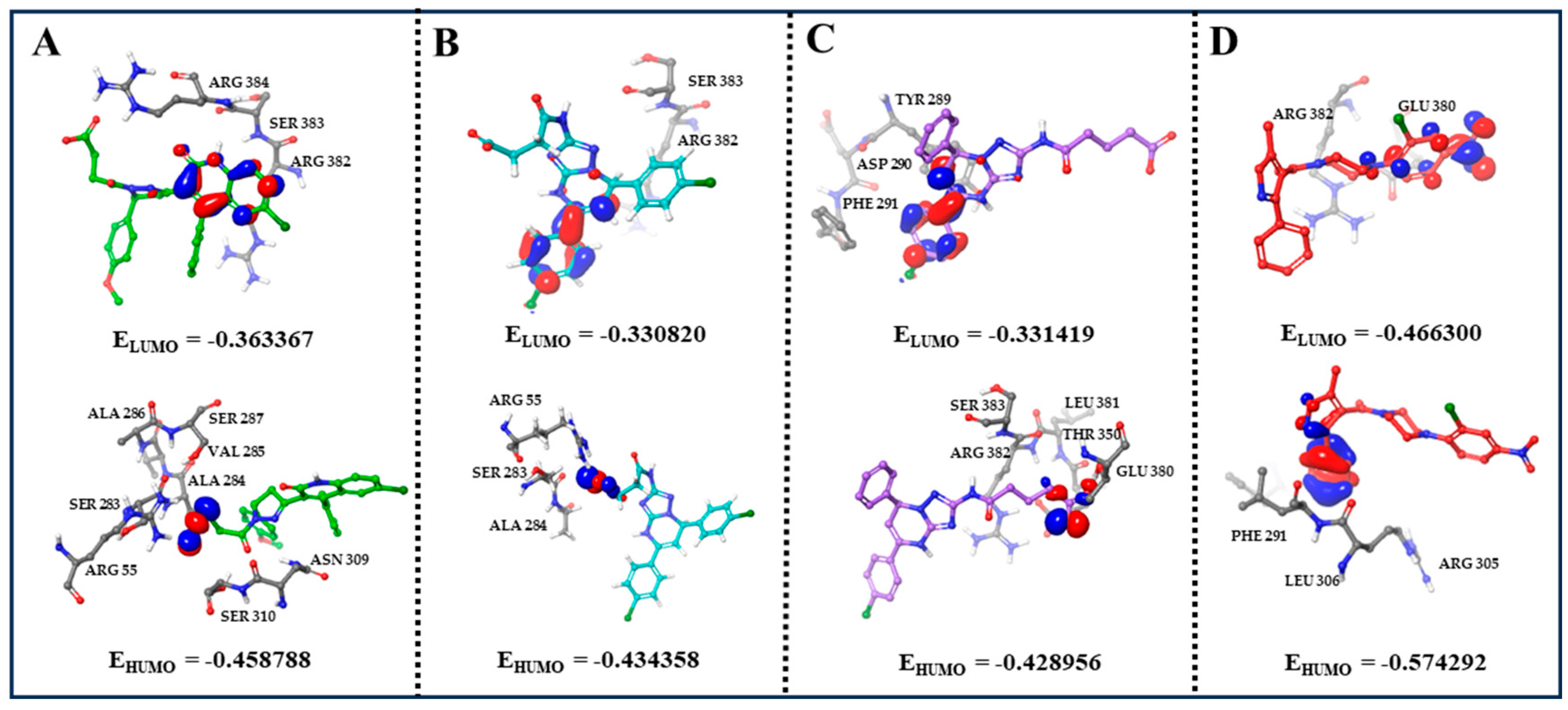
2.5. Quantum Mechanics/Molecular Mechanics (QM/MM) Analysis
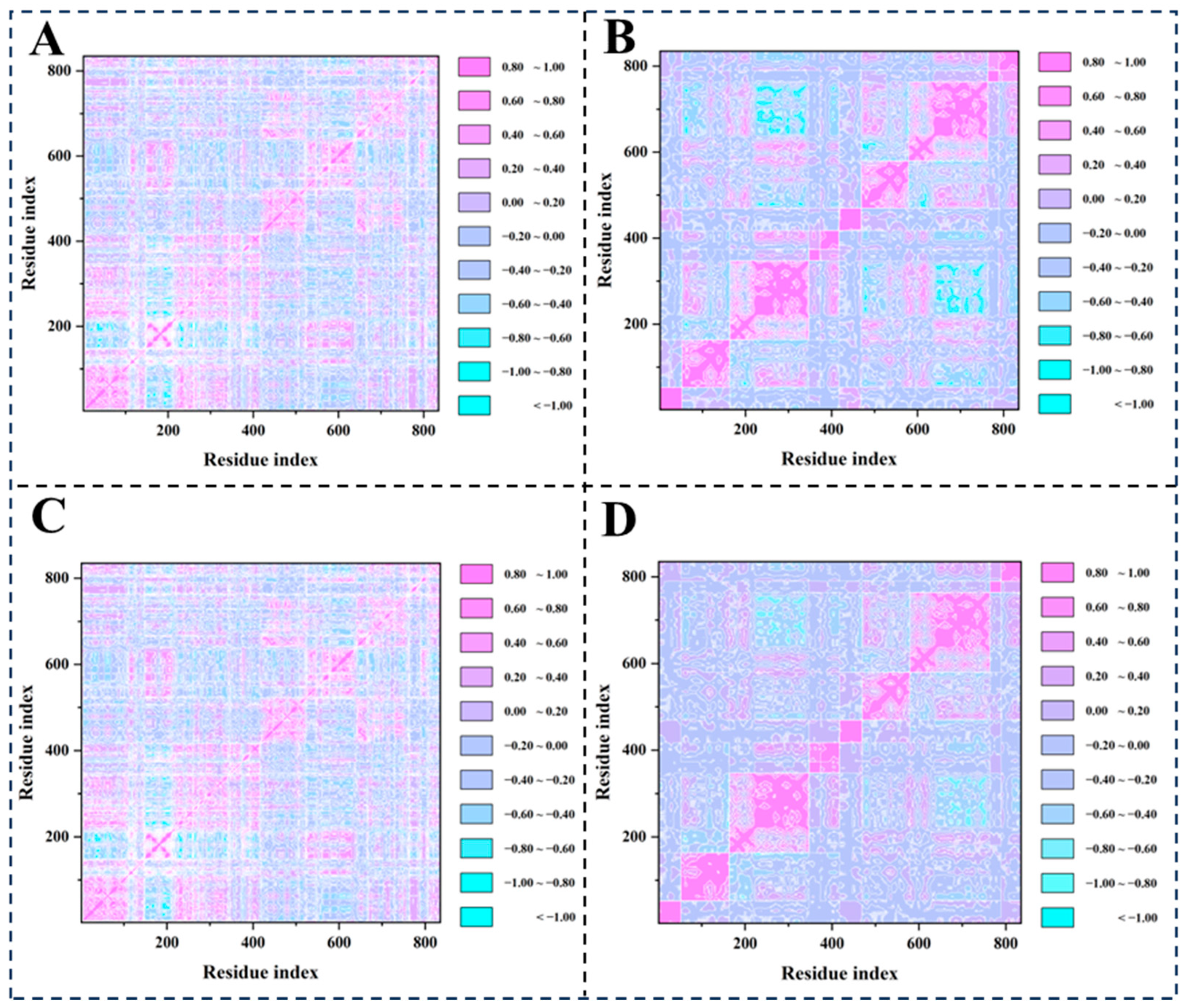
2.6. Dynamic Cross-Correlation Matrix (DCCM) Analysis and Principal Component Analysis (PCA)
3. Materials and Methods
3.1. Virtual Screening
3.2. Prime/MM–GBSA Simulation
3.3. Lipinski’s Rule and ADMET Prediction
3.4. Materials
3.5. Expression and Purification of NP
3.6. SPR Experiment
3.7. Molecular Dynamics Simulation
3.8. Alanine Scanning Mutagenesis
3.9. QM/MM Calculations
3.10. DCCM Analysis and PCA
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Taubenberger, J.K.; Kash, J.C. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 2010, 7, 440–451. [Google Scholar] [CrossRef]
- Simonsen, L. The global impact of influenza on morbidity and mortality. Vaccine 1999, 17 (Suppl. 1), S3–S10. [Google Scholar] [CrossRef]
- Zoungrana, T.D.; Yerbanga, A.; Ouoba, Y. Socio-economic and environmental factors in the global spread of COVID-19 outbreak. Res. Econ. 2022, 76, 325–344. [Google Scholar] [CrossRef]
- Wu, Y.; Wu, Y.; Tefsen, B.; Shi, Y.; Gao, G.F. Bat-derived influenza-like viruses H17N10 and H18N11. Trends Microbiol. 2014, 22, 183–191. [Google Scholar] [CrossRef]
- Hu, Y.; Sneyd, H.; Dekant, R.; Wang, J. Influenza A virus nucleoprotein: A highly conserved multi-functional viral protein as a hot antiviral drug target. Curr. Top. Med. Chem. 2017, 17, 2271–2285. [Google Scholar] [CrossRef]
- Krammer, F.; Smith, G.J.D.; Fouchier, R.A.M.; Peiris, M.; Kedzierska, K.; Doherty, P.C.; Palese, P.; Shaw, M.L.; Treanor, J.; Webster, R.G.; et al. Influenza. Nat. Rev. Dis. Primers. 2018, 4, 3. [Google Scholar] [CrossRef]
- Loregian, A.; Mercorelli, B.; Nannetti, G.; Compagnin, C.; Palù, G. Antiviral strategies against influenza virus: Towards new therapeutic approaches. Cell. Mol. Life Sci. 2014, 71, 3659–3683. [Google Scholar] [CrossRef] [PubMed]
- Leonov, H.; Astrahan, P.; Krugliak, M.; Arkin, I.T. How do aminoadamantanes block the influenza M2 channel, and how does resistance develop? J. Am. Chem. Soc. 2011, 133, 9903–9911. [Google Scholar] [CrossRef] [PubMed]
- Moss, R.B.; Davey, R.T.; Steigbigel, R.T.; Fang, F. Targeting pandemic influenza: A primer on influenza antivirals and drug resistance. J. Antimicrob. Chemother. 2010, 65, 1086–1093. [Google Scholar] [CrossRef] [PubMed]
- Hayden, F.G.; Hoffman, H.E.; Spyker, D.A. Differences in side effects of amantadine hydrochloride and rimantadine hydrochloride relate to differences in pharmacokinetics. Antimicrob. Agent Chemother. 1983, 23, 458–464. [Google Scholar] [CrossRef]
- Ohigashi, Y.; Ma, C.; Jing, X.; Balannick, V.; Pinto, L.H.; Lamb, R.A. An amantadine-sensitive chimeric BM2 ion channel of influenza B virus has implications for the mechanism of drug inhibition. Proc. Natl. Acad. Sci. USA 2009, 106, 18775–18779. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.U.; Lew, W.; Williams, M.A.; Liu, H.; Zhang, L.; Swaminathan, S.; Bischofberger, N.; Chen, M.S.; Mendel, D.B.; Tai, C.Y.; et al. Influenza Neuraminidase Inhibitors Possessing a Novel Hydrophobic Interaction in the Enzyme Active Site: Design, Synthesis, and Structural Analysis of Carbocyclic Sialic Acid Analogues with Potent Anti-Influenza Activity. J. Am. Chem. Soc. 1997, 119, 681–690. [Google Scholar] [CrossRef]
- Hurt, A.C. The epidemiology and spread of drug resistant human influenza viruses. Curr. Opin. Virol. 2014, 8, 22–29. [Google Scholar] [CrossRef]
- Shi, F.; Xie, Y.; Shi, L.; Xu, W. Viral RNA Polymerase: A Promising Antiviral Target for Influenza A Virus. Curr. Med. Chem. 2013, 20, 3923–3934. [Google Scholar] [CrossRef]
- Lee, S.M.; Yen, H.L. Targeting the host or the virus: Current and novel concepts for antiviral approaches against influenza virus infection. Antivir. Res. 2012, 96, 391–404. [Google Scholar] [CrossRef]
- Kukol, A.; Hughes, D.J. Large-scale analysis of influenza A virus nucleoprotein sequence conservation reveals potential drug-target sites. Virology 2014, 454–455, 40–47. [Google Scholar] [CrossRef]
- Li, Z.; Watanabe, T.; Hatta, M.; Watanabe, S.; Nanbo, A.; Ozawa, M.; Kakugawa, S.; Shimojima, M.; Yamada, S.; Neumann, G.; et al. Mutational analysis of conserved amino acids in the influenza A virus nucleoprotein. J. Virol. 2009, 83, 4153–4162. [Google Scholar] [CrossRef]
- Compans, R.W.; Content, J.; Duesberg, P.H. Structure of the Ribonucleoprotein of Influenza Virus. J. Virol. 1972, 10, 795–800. [Google Scholar] [CrossRef] [PubMed]
- Ortega, J.; Martín-Benito, J.; Zürcher, T.; Valpuesta, J.M.; Carrascosa, J.L.; Ortín, J. Ultrastructural and Functional Analyses of Recombinant Influenza Virus Ribonucleoproteins Suggest Dimerization of Nucleoprotein during Virus Amplification. J. Virol. 2000, 74, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Cianci, C.; Gerritz, S.W.; Deminie, C.; Krystal, M. Influenza nucleoprotein: Promising target for antiviral chemotherapy. Antivir. Chem. Chemother. 2013, 23, 77–91. [Google Scholar] [CrossRef]
- Lejal, N.; Tarus, B.; Bouguyon, E.; Chenavas, S.; Bertho, N.; Delmas, B.; Ruigrok, R.W.; Di Primo, C.; Slama-Schwok, A. Structure-based discovery of the novel antiviral properties of naproxen against the nucleoprotein of influenza A virus. Antimicrob. Agents Chemother. 2013, 57, 2231–2242. [Google Scholar] [CrossRef]
- Panthi, S.; Hong, J.Y.; Satange, R.; Yu, C.C.; Li, L.Y.; Hou, M.H. Antiviral drug development by targeting RNA binding site, oligomerization and nuclear export of influenza nucleoprotein. Int. J. Biol. Macromol. 2024, 282 Pt 4, 136996. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Ju, H.; Zhang, Y.; Achi, J.G.; Kang, D.; Zou, J.; Du, R.; Cui, Q.; Liu, X.; Rong, L.; et al. Discovery of ligustrazine and chalcone derivatives as novel viral nucleoprotein nuclear export inhibitors against influenza viruses. J. Med. Virol. 2023, 95, e28968. [Google Scholar] [CrossRef]
- Tang, Y.S.; Zhang, C.; Lo, C.Y.; Jin, Z.; Kong, B.L.; Xiao, M.J.; Huang, E.F.; Hu, C.; Shaw, P.C. Anti-influenza virus activities and mechanism of antrafenine analogs. Eur. J. Med. Chem. 2023, 260, 115775. [Google Scholar] [CrossRef]
- Zhang, C.; Li, J.B.; Zhang, Y.W.; Tang, Y.S.; Yu, X.F.; Zhang, Q.G.; Jin, Z.; Hou, S.C.; Shaw, P.C.; Hu, C. Novel benzamide derivatives with indole moiety as dual-target antiviral agents: Rational design, efficient synthesis, and potent anti-influenza activity through concurrent binding to PAC terminal domain and viral nucleoprotein. Eur. J. Med. Chem. 2025, 293, 117681. [Google Scholar] [CrossRef]
- Su, C.Y.; Cheng, T.J.; Lin, M.I.; Wang, S.Y.; Huang, W.I.; Lin-Chu, S.Y.; Chen, Y.H.; Wu, C.Y.; Lai, M.M.; Cheng, W.C.; et al. High-throughput identification of compounds targeting influenza RNA-dependent RNA polymerase activity. Proc. Natl. Acad. Sci. USA 2010, 107, 19151–19156. [Google Scholar] [CrossRef] [PubMed]
- Kao, R.Y.; Yang, D.; Lau, L.S.; Tsui, W.H.; Hu, L.; Dai, J.; Chan, M.P.; Chan, C.M.; Wang, P.; Zheng, B.J.; et al. Identification of influenza A nucleoprotein as an antiviral target. Nat. Biotechnol. 2010, 28, 600–605. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Liu, J.; Zhang, R.; Lu, Y.; Cui, G.; Cui, Z.; Ding, Y. A review of deep learning methods for ligand based drug virtual screening. Fundam. Res. 2024, 4, 715–737. [Google Scholar] [CrossRef]
- Saxena, S.; Abdullah, M.; Sriram, D.; Guruprasad, L. Discovery of novel inhibitors of Mycobacterium tuberculosis MurG: Homology modelling, structure based pharmacophore, molecular docking, and molecular dynamics simulations. J. Biomol. Struct. Dyn. 2018, 36, 3184–3198. [Google Scholar] [CrossRef]
- Zhang, C.; Tang, Y.S.; Meng, C.R.; Xu, J.; Zhang, D.L.; Wang, J.; Huang, E.F.; Shaw, P.C.; Hu, C. Design, synthesis, molecular docking analysis and biological evaluations of 4-[(Quinolin-4-yl)amino]benzamide derivatives as novel anti-Influenza virus agents. Int. J. Mol. Sci. 2022, 23, 6307. [Google Scholar] [CrossRef]
- Li, Z.Y.; Su, C.Y.; Ding, B. Molecular dynamics simulation of β-adrenoceptors and their coupled G proteins. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 6346–6351. [Google Scholar] [CrossRef] [PubMed]
- Kikiowo, B.; Ahmad, I.; Alade, A.A.; T Ijatuyi, T.; Iwaloye, O.; Patel, H.M. Molecular dynamics simulation and pharmacokinetics studies of ombuin and quercetin against human pancreatic α-amylase. J. Biomol. Struct. Dyn. 2023, 41, 10388–10395. [Google Scholar] [CrossRef] [PubMed]
- Patan, A.; Aanandhi, M.V.; Gopinath, P. Molecular dynamics simulation approach of hybrid chalcone-thiazole complex derivatives for DNA gyrase B inhibition: Lead generation. RSC Adv. 2023, 13, 24291–24308. [Google Scholar] [CrossRef]
- Chen, W.; Lee, J.Y.; Kim, J.S.; Shin, J.S.; Fung, T.S.; Yeh, J.-Y.; Chen, Z.; Zhou, B.; Song, J.J.; Go, Y.Y. Critical amino acid residues in human ACE2 for SARS-CoV-2 spike protein binding and virus entry. Microbiol. Spectr. 2025, 13, e0324424. [Google Scholar] [CrossRef]
- Stampelou, M.; Suchankova, A.; Tzortzini, E.; Dhingra, L.; Barkan, K.; Lougiakis, N.; Marakos, P.; Pouli, N.; Ladds, G.; Kolocouris, A. Dual A1/A3 adenosine receptor antagonists: Binding kinetics and structure-activity relationship studies using mutagenesis and alchemical binding free energy calculations. J. Med. Chem. 2022, 65, 13305–13327. [Google Scholar] [CrossRef] [PubMed]
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L.; et al. OPLS3: A force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput. 2016, 12, 281–296. [Google Scholar] [CrossRef]
- Warshel, A.; Levitt, M. Theoretical studies of enzymic reactions: Dielectric, electrostatic and steric stabilization of the carbonium ion in the reaction of lysozyme. J. Mol. Biol. 1976, 103, 227–249. [Google Scholar] [CrossRef]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010, 132, 154104. [Google Scholar] [CrossRef]
- Grant, B.J.; Rodrigues, A.P.; ElSawy, K.M.; McCammon, J.A.; Caves, L.S. Bio3d: An R package for the comparative analysis of protein structures. Bioinformatics 2006, 22, 2695–2696. [Google Scholar] [CrossRef]




| No. | Glide Score | MMGBSA ΔGBind (kcal/mol) | MMGBSA ΔGBind vdW (kcal/mol) | MMGBSA ΔGBind Solv GB (kcal/mol) |
|---|---|---|---|---|
| nucleozin | −4.835 | −41.83 | −47.54 | 9.19 |
| 1 | −7.832 | −38.95 | −50.49 | 109.15 |
| 2 | −6.515 | −37.77 | −40.26 | 105.12 |
| 3 | −6.341 | −36.90 | −55.11 | 409.67 |
| 4 | −6.572 | −52.60 | −61.46 | 99.66 |
| 5 | −6.912 | −35.32 | −58.04 | 102.68 |
| 6 | −6.246 | −57.63 | −62.54 | 39.88 |
| 7 | −6.436 | −44.76 | −53.07 | 239.10 |
| 8 | −10.940 | −46.79 | −59.25 | 108.27 |
| 9 | −6.653 | −39.94 | −57.16 | 211.07 |
| 10 | −6.648 | −55.10 | −62.14 | 95.06 |
| 11 | −6.473 | −51.54 | −50.27 | 100.60 |
| 12 | −6.060 | −44.21 | −52.59 | 210.74 |
| 13 | −7.161 | −52.84 | −54.04 | 105.39 |
| 14 | −5.560 | −50.16 | −53.87 | 119.75 |
| 15 | −6.654 | −68.02 | −67.37 | 36.61 |
| 16 | −5.492 | −59.16 | −45.97 | 25.81 |
| Compd. | KD (M) | Compd. | KD (M) | Compd. | KD (M) |
|---|---|---|---|---|---|
| nucleozin | 9.73 × 10−6 | 3 | 2.10 × 10−4 | 13 | 3.82 × 10−5 |
| 1 | 1.50 × 10−4 | 8 | 7.85 × 10−5 | 14 | 6.97 × 10−5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Guo, J.; Zhang, C.; Ding, Y.; Sun, S.; Yao, B.; Xing, C.; Liu, X.; Hu, C.; Xiao, J. Computational Discovery of Potent Nucleoprotein Inhibitors for Influenza A Virus: Validation Through QM/MM Analysis and Experimental Binding Assays. Molecules 2025, 30, 3960. https://doi.org/10.3390/molecules30193960
Liu Z, Guo J, Zhang C, Ding Y, Sun S, Yao B, Xing C, Liu X, Hu C, Xiao J. Computational Discovery of Potent Nucleoprotein Inhibitors for Influenza A Virus: Validation Through QM/MM Analysis and Experimental Binding Assays. Molecules. 2025; 30(19):3960. https://doi.org/10.3390/molecules30193960
Chicago/Turabian StyleLiu, Zixiao, Jialin Guo, Chao Zhang, Yongzhao Ding, Shiyang Sun, Binrong Yao, Cheng Xing, Xiaoping Liu, Chun Hu, and Junhai Xiao. 2025. "Computational Discovery of Potent Nucleoprotein Inhibitors for Influenza A Virus: Validation Through QM/MM Analysis and Experimental Binding Assays" Molecules 30, no. 19: 3960. https://doi.org/10.3390/molecules30193960
APA StyleLiu, Z., Guo, J., Zhang, C., Ding, Y., Sun, S., Yao, B., Xing, C., Liu, X., Hu, C., & Xiao, J. (2025). Computational Discovery of Potent Nucleoprotein Inhibitors for Influenza A Virus: Validation Through QM/MM Analysis and Experimental Binding Assays. Molecules, 30(19), 3960. https://doi.org/10.3390/molecules30193960