Proteolytic Bacillus sp. Isolation and Identification from Tannery Alkaline Baths
Abstract
1. Introduction
2. Results
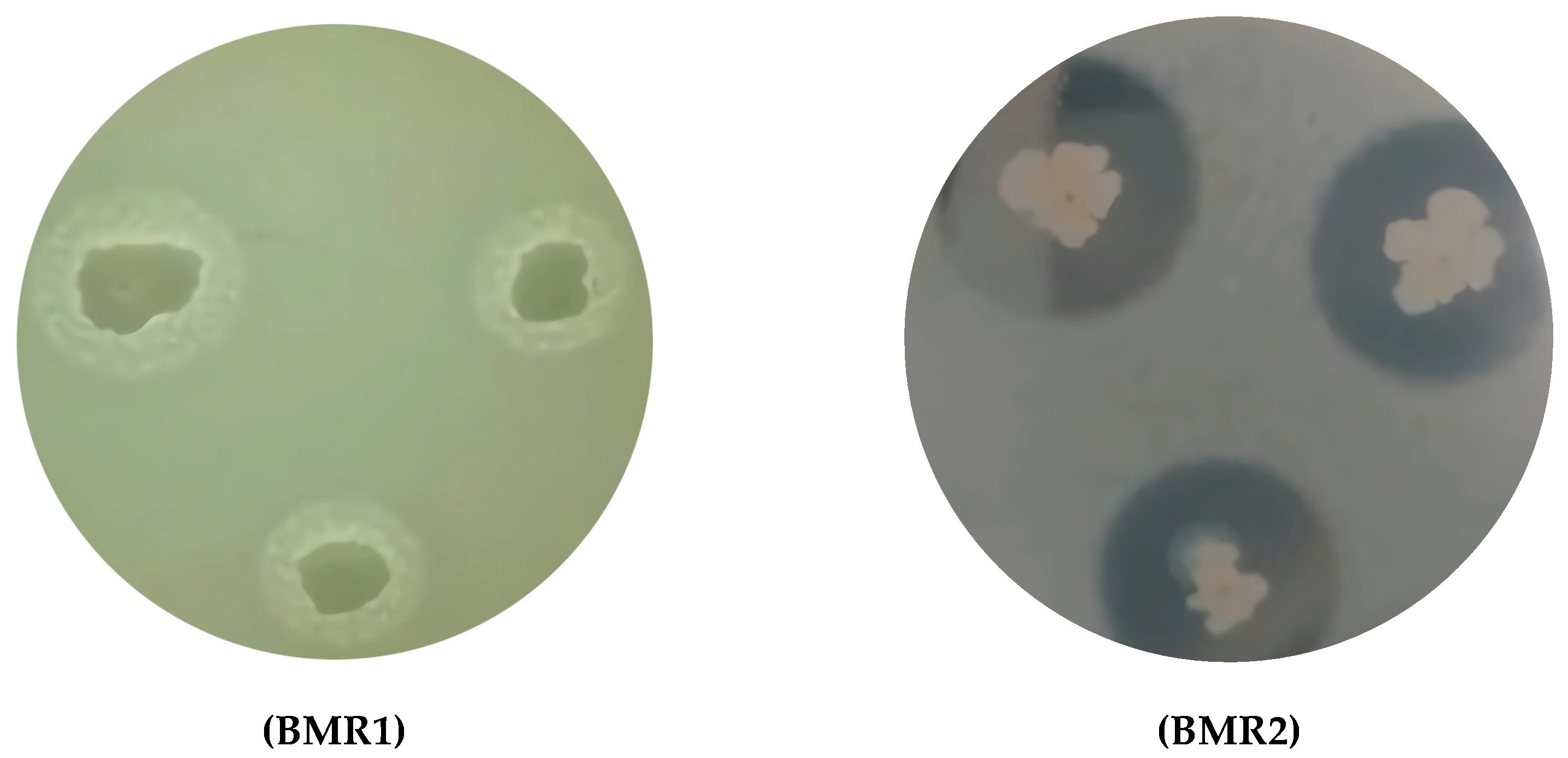
2.1. Screening of Proteolytic Microorganisms from Tanning Baths
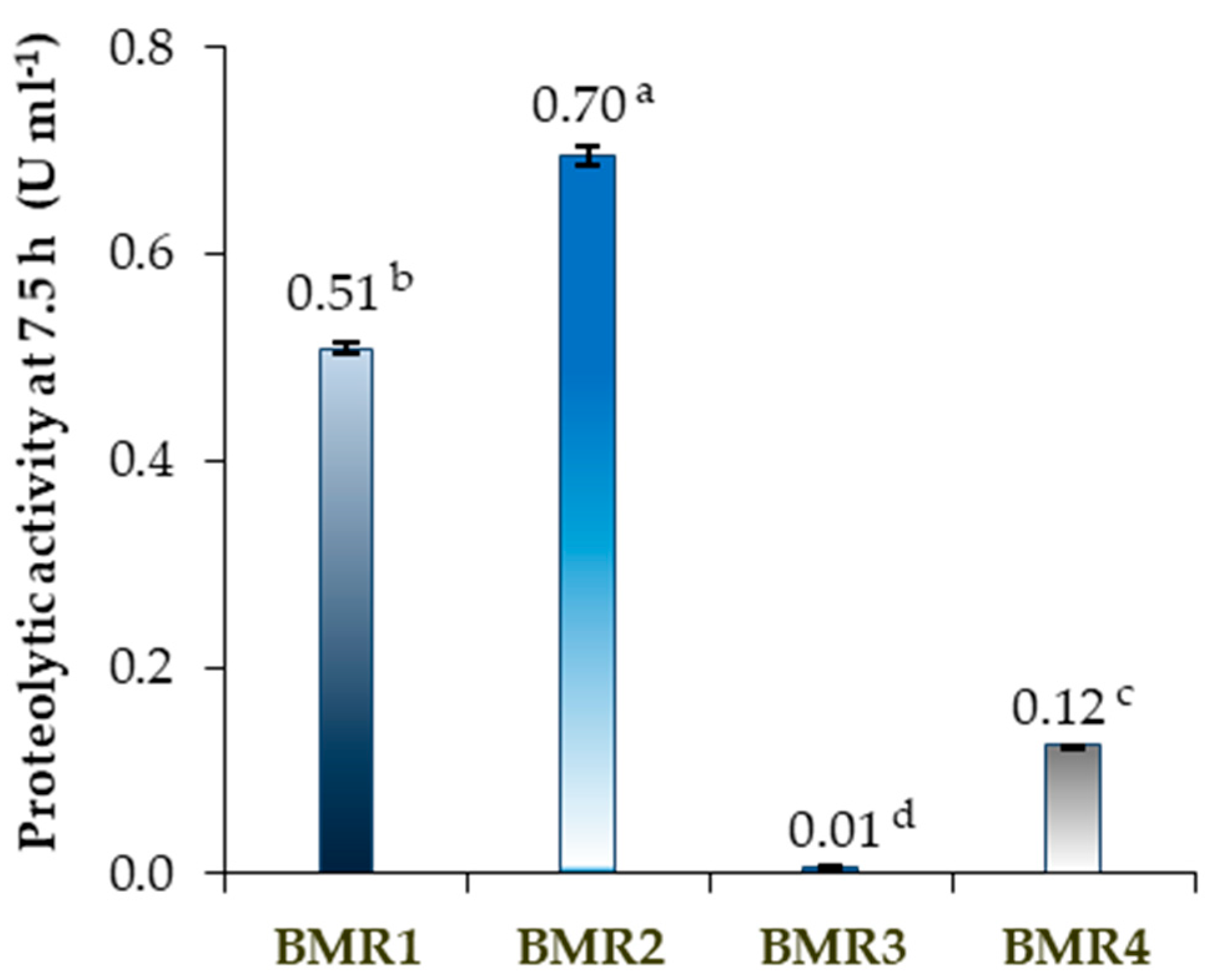
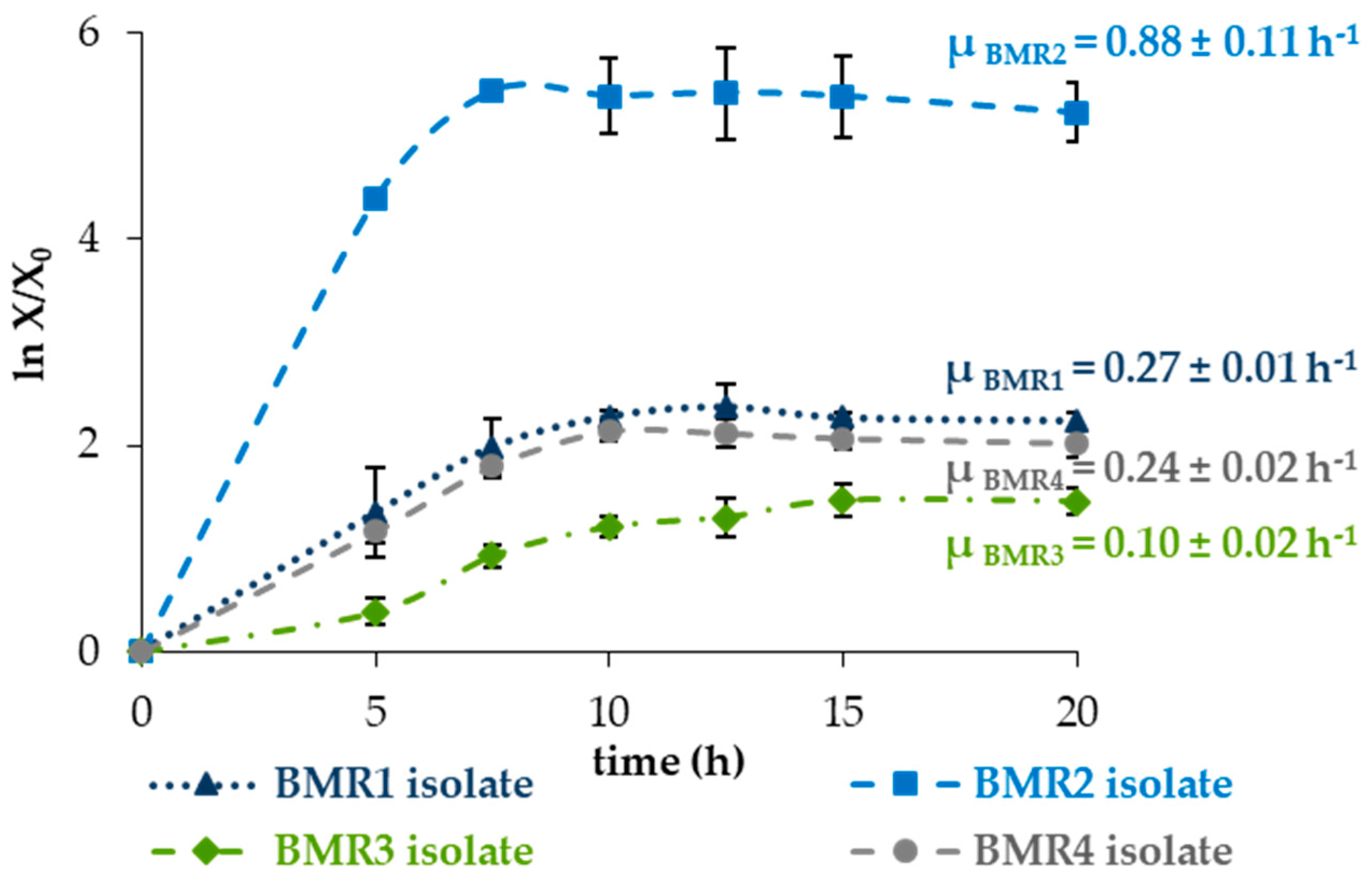
2.2. Proteolytic Activity and Growth of the Isolated Microorganisms in Solid and Liquid Medium
2.3. Microorganism Identification
2.3.1. Morphological and Physiological Characterisation
2.3.2. Biochemical Tests
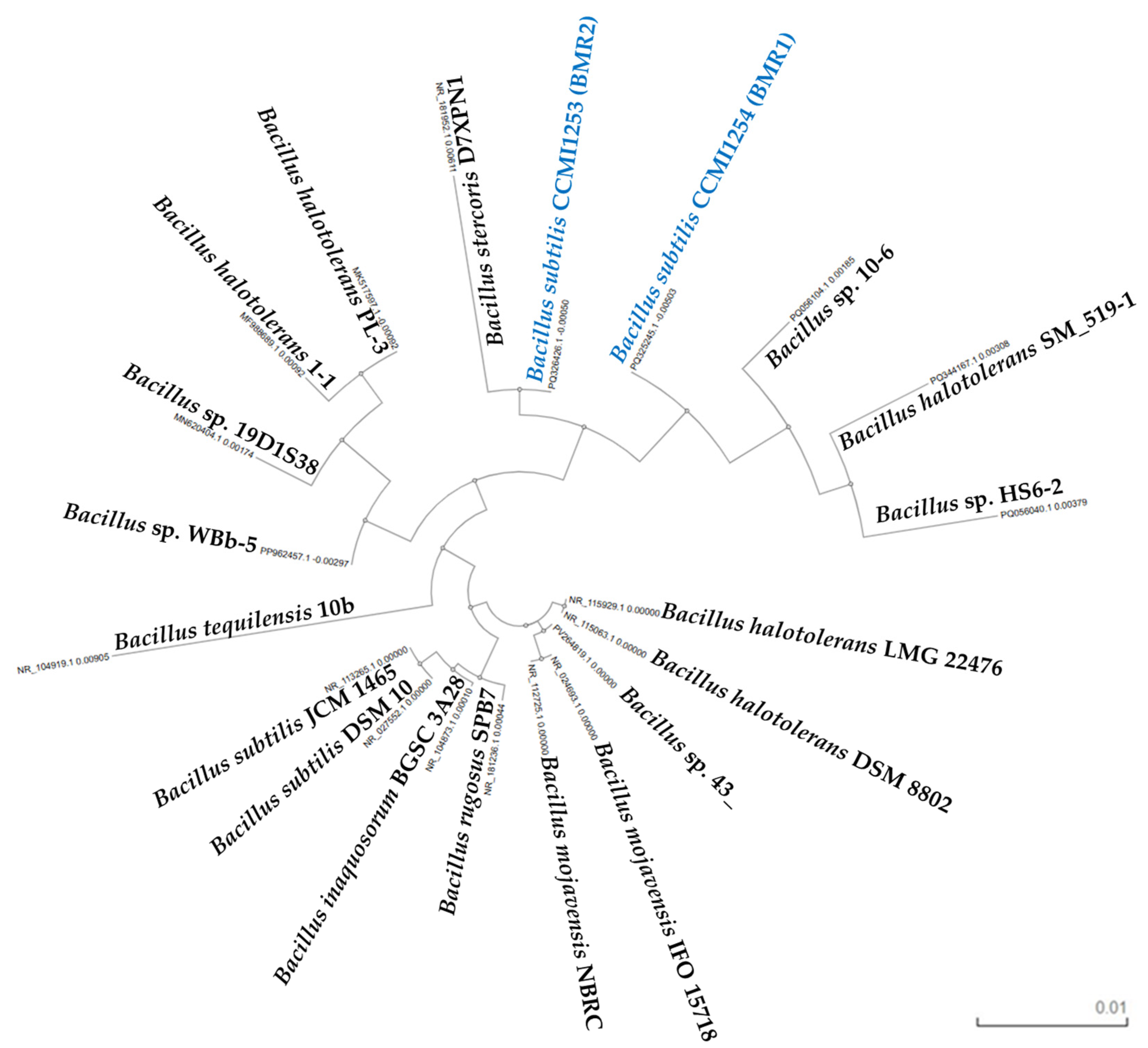
2.3.3. Molecular Identification by 16S rRNA Gene Sequencing
2.3.4. Fatty Acid Methyl Esters Analysis
3. Discussion
3.1. Screening of Proteolytic Microorganisms
3.2. Proteolytic Activity and Growth
3.3. Microorganism Identification
4. Materials and Methods
4.1. Screening and Isolation of Proteolytic Microorganisms
4.2. Proteolytic Activity and Growth of the Isolated Microorganisms
4.2.1. Protease Production in Solid Medium
4.2.2. Protease Production in Liquid Medium
4.3. Microorganism Identification
4.3.1. Morphological, Physiological and Biochemical Identification
4.3.2. Molecular Identification by 16S rRNA Gene Sequencing
4.3.3. Fatty Acid Methyl Esters Analysis Identification
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Madhavi, J.; Srilakshmi, J.; Rao, M.V.R.; Rao, K.R.S.S. Efficient Leather Dehairing by Bacterial Thermostable Protease. Int. J. Biosci. Biotechnol. 2011, 3, 11–26. [Google Scholar]
- Song, P.; Zhang, X.; Wang, S.; Xu, W.; Fu, R.; Wei, F. Microbial Proteases and Their Applications. Front. Microbiol. 2023, 14, 1236368. [Google Scholar] [CrossRef] [PubMed]
- Noreen, S.; Siddiqa, A.; Fatima, R.; Anwar, F.; Adnan, M.; Raza, A. Protease Production and Purification from Agro Industrial Waste by Utilizing Penicillium digitatum. Int. J. Appl. Biol. Forensics 2017, 1, 119–129. [Google Scholar]
- Mrudula, S. A Review on Microbial Alkaline Proteases: Optimization of Submerged Fermentative Production, Properties, and Industrial Applications. Appl. Biochem. Microbiol. 2024, 60, 383–401. [Google Scholar] [CrossRef]
- Niehaus, F.; Gabor, E.; Wieland, S.; Siegert, P.; Maurer, K.H.; Eck, J. Enzymes for the Laundry Industries: Tapping the Vast Metagenomic Pool of Alkaline Proteases. Microb. Biotechnol. 2011, 4, 767–776. [Google Scholar] [CrossRef]
- Krishnaveni, K.; Kalaichelvan, P.T. Production and Optimization of Extracellular Alkaline Protease from Bacillus subtilis Isolated from Dairy Effluent. Der Pharm. Lett. 2012, 4, 98–109. [Google Scholar]
- Porwal, S.; Lal, S.; Cheema, S.; Kalia, V.C. Phylogeny in Aid of the Present and Novel Microbial Lineages: Diversity in Bacillus. PLoS ONE 2009, 4, e4438. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Z.; Gao, C.; Jiang, Z.; Huang, S.; Li, X.; Yang, H. Bacillus subtilis as an Excellent Microbial Treatment Agent for Environmental Pollution: A Review. Biotechnol. J. 2025, 20, e70026. [Google Scholar] [CrossRef]
- Errington, J.; van der Aart, L.T. Microbe Profile: Bacillus subtilis: Model Organism for Cellular Development, and Industrial Workhorse. Microbiology 2020, 166, 425–427. [Google Scholar] [CrossRef]
- Hellany, H.; Assaf, J.C.; Barada, S.; el-Badan, D.; Hajj, R.E.; Abou Najem, S.; Abou Fayad, A.G.; Khalil, M.I. Isolation and Characterization of Bacillus subtilis BSP1 from Soil: Antimicrobial Activity and Optimization of Fermentation Conditions. Processes 2024, 12, 1621. [Google Scholar] [CrossRef]
- Kalwasińska, A.; Jankiewicz, U.; Felföldi, T.; Burkowska-But, A.; Brzezinska, M.S. Alkaline and Halophilic Protease Production by Bacillus luteus H11 and Its Potential Industrial Applications. Food Technol. Biotechnol. 2018, 56, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Bourdichon, F.; Casaregola, S.; Farrokh, C.; Frisvad, J.C.; Gerds, M.L.; Hammes, W.P.; Harnett, J.; Huys, G.; Laulund, S.; Ouwehand, A.; et al. Food Fermentations: Microorganisms with Technological Beneficial Use. Int. J. Food Microbiol. 2012, 154, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.; Sharma, V.; Kuila, A. Fermentation Technology: Current Status and Future Prospects. In Principles and Applications of Fermentation Technology; Kuila, A., Sharma, V., Eds.; Scrivener Publishing LLC: Beverly, MA, USA; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2018; Volume 1, pp. 1–13. [Google Scholar] [CrossRef]
- Joshi, P.P.; Ghike, P.B. Study of Protease Producing Bacteria and Their Enzymatic Activity at Different Parameters. Int. Res. J. Eng. Technol. 2019, 06, 2239. [Google Scholar]
- Christensen, L.F.; García-Béjar, B.; Bang-Berthelsen, C.H.; Hansen, E.B. Extracellular Microbial Proteases with Specificity for Plant Proteins in Food Fermentation. Int. J. Food Microbiol. 2022, 381, 109889. [Google Scholar] [CrossRef]
- Abe, S.; Yasumura, A.; Tanaka, T. Regulation of Bacillus subtilis aprE Expression by glnA through Inhibition of scoC and σD-Dependent degR Expression. J. Bacteriol. 2009, 191, 3050–3058. [Google Scholar] [CrossRef]
- Alshehri, W.A.; Alhothifi, S.A.; Khalel, A.F.; Alqahtani, F.S.; Hadrich, B.; Sayari, A. Production Optimization of a Thermostable Alkaline and Detergent Biocompatible Protease by Bacillus paramycoides WSA for the Green Detergent Industry. Sci. Rep. 2025, 15, 13205. [Google Scholar] [CrossRef]
- Yang, H.; Ren, X.; Zhao, Y.; Xu, T.; Xiao, J.; Chen, H. Enhancing Alkaline Protease Stability through Enzyme-Catalyzed Crosslinking and Its Application in Detergents. Processes 2024, 12, 624. [Google Scholar] [CrossRef]
- Alam, S.; Hasan, J.; Haque, P.; Rahman, M.M. Sustainable Leather Tanning: Enhanced Properties and Pollution Reduction through Crude Protease Enzyme Treatment. Int. J. Biol. Macromol. 2024, 268, 131858. [Google Scholar] [CrossRef]
- Hassan, M.M.; Harris, J.; Busfield, J.J.C.; Bilotti, E. A Review of the Green Chemistry Approaches to Leather Tanning in Imparting Sustainable Leather Manufacturing. Green. Chem. 2023, 25, 7441–7469. [Google Scholar] [CrossRef]
- Arunachalam, C.; Saritha, K. Protease Enzyme: An Eco-Friendly Alternative for Leather Industry. Indian. J. Sci. Technol. 2009, 2, 29–32. [Google Scholar] [CrossRef]
- Foroughi, F.; Keshavarz, T.; Evans, C.S. Specificities of Proteases for Use in Leather Manufacture. J. Chem. Technol. Biotechnol. 2006, 81, 257–261. [Google Scholar] [CrossRef]
- Ugbede, A.S.; Abioye, O.P.; Aransiola, S.A.; Oyewole, O.A.; Maddela, N.R.; Prasad, R. Production, Optimization and Partial Purification of Bacterial and Fungal Proteases for Animal Skin Dehairing: A Sustainable Development in Leather-Making Process. Bioresour. Technol. Rep. 2023, 24, 101632. [Google Scholar] [CrossRef]
- Naveed, M. Protease—A Versatile and Ecofriendly Biocatalyst with Multi-Industrial Applications: An Updated Review. Catal. Lett. 2021, 151, 307–323. [Google Scholar] [CrossRef]
- Pawar, K.S.; Singh, P.N.; Singh, S.K. Fungal Alkaline Proteases and Their Potential Applications in Different Industries. Front. Microbiol. 2023, 14, 11. [Google Scholar] [CrossRef] [PubMed]
- Adetunji, A.I.; Olaitan, M.O.; Erasmus, M.; Olaniran, A.O. Microbial Proteases: A next Generation Green Catalyst for Industrial, Environmental and Biomedical Sustainability. Food Mater. Res. 2023, 3, 15. [Google Scholar] [CrossRef]
- Demirkan, E.; Kut, D.; Sevgi, T.; Dogan, M.; Baygin, E. Investigation of Effects of Protease Enzyme Produced by Bacillus subtilis 168 E6-5 and Commercial Enzyme on Physical Properties of Woolen Fabric. J. Text. Inst. 2020, 111, 26–35. [Google Scholar] [CrossRef]
- Gautam, S. A Review of Bacillus Species Alkaline Protease Production and Industrial Applications. Int. J. Ther. Innov. 2024, 2, 266–273. [Google Scholar] [CrossRef]
- Mousavi Ghahfarrokhi, S.S.; Mahdigholi, F.S.; Amin, M. Collateral Beauty in the Damages: An Overview of Cosmetics and Therapeutic Applications of Microbial Proteases. Arch. Microbiol. 2023, 205, 375. [Google Scholar] [CrossRef]
- Slimane, M.; El-Hafid, N. Recent Status in Production, Biotechnological Applications, Commercial Aspects, and Future Prospects of Microbial Enzymes: A Comprehensive Review. Int. J. Agric. Sc. Food Technol. 2024, 10, 006–020. [Google Scholar] [CrossRef]
- Sun, F.; Hu, Y.; Chen, Q.; Kong, B.; Liu, Q. Purification and Biochemical Characteristics of the Extracellular Protease from Pediococcus pentosaceus Isolated from Harbin Dry Sausages. Meat Sci. 2019, 156, 156–165. [Google Scholar] [CrossRef]
- Singh, P.K.; Shrivastava, N.; Ojha, B.K. Enzymes in the Meat Industry. In Enzymes in Food Biotechnology; Kuddus, M., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 111–128. [Google Scholar] [CrossRef]
- Kumar, A.; Dhiman, S.; Krishan, B.; Samtiya, M.; Kumari, A.; Pathak, N.; Kumari, A.; Aluko, R.E.; Dhewa, T. Microbial Enzymes and Major Applications in the Food Industry: A Concise Review. Food Prod. Process. Nutr. 2024, 6, 85. [Google Scholar] [CrossRef]
- Mamo, J.; Assefa, F. The Role of Microbial Aspartic Protease Enzyme in Food and Beverage Industries. J. Food Qual. 2018, 2018, 15. [Google Scholar] [CrossRef]
- Heredia-Sandoval, N.G.; Valencia-Tapia, M.Y. Microbial Proteases in Baked Goods: Modification of Gluten and Effects on Immunogenicity and Product Quality. Foods 2016, 5, 59. [Google Scholar] [CrossRef]
- Ojo-Omoniyi, O.A.; Moro, D.D.; Afolabi, O.B. Microbial Proteases: Sources, Significance and Industrial Applications. Int. J. Curr. Microbiol. Appl. Sci. 2024, 13, 1–23. [Google Scholar] [CrossRef]
- Masi, C.; Gemechu, G.; Tafesse, M. Isolation, Screening, Characterization, and Identification of Alkaline Protease-Producing Bacteria from Leather Industry Effluent. Ann. Microbiol. 2021, 71, 24. [Google Scholar] [CrossRef]
- Rahman, S.; Islam, R.; Mondol, O.K.; Rahman, M.S.; Sabrin, F.; Zohora, U.S. Screening of Protease Producing Bacteria from Tannery Wastes of Leather Processing Industries at Hazaribag, Bangladesh. J. Biol. Sci. 2018, 7, 23–34. [Google Scholar] [CrossRef]
- Dettmer, A.; Cavalli, É.; Ayub, M.A.Z.; Gutterres, M. Optimization of the Unhairing Leather Processing with Enzymes and the Evaluation of Inter-Fibrillary Proteins Removal: An Environment-Friendly Alternative. Bioprocess. Biosyst. Eng. 2012, 35, 1317–1324. [Google Scholar] [CrossRef]
- Butt, M.Q.; Zeeshan, N.; Ashraf, N.M.; Akhtar, M.A.; Ashraf, H.; Afroz, A.; Shaheen, A.; Naz, S. Environmental Impact and Diversity of Protease-Producing Bacteria in Areas of Leather Tannery Effluents of Sialkot, Pakistan. Environ. Sci. Pollut. Res. 2021, 28, 54842–54851. [Google Scholar] [CrossRef] [PubMed]
- Feyissa, B.H. Screening, Identification, and Optimization of Protease Producing Bacillus pumilus Strain DT-15 From Tannery Waste Disposal Site in Addis Ababa, Ethiopia. Int. J. Microbiol. 2025, 2025, 7176092. [Google Scholar] [CrossRef] [PubMed]
- Marimuthu, A.; Sivasankari, B.; Nagarajan, S.; Parveen Rani, R.; Sivakumar, S. Production, Purification, and Biochemical Characterization of Thermostable Metallo-Protease from Novel Bacillus alkalitelluris TWI3 Isolated from Tannery Waste. Appl. Biochem. Biotechnol. 2016, 178, 1666–1686. [Google Scholar] [CrossRef] [PubMed]
- Zeb, J.; Zeeshan, N.; Yasmin, G.; Khan, U.J. Isolation and Characterization of Bacteria Associated with Tannery Effluent and Their Protease Producing Ability. Pak. J. Sci. Ind. Res. Ser. B Biol. Sci. 2024, 67B, 154–160. [Google Scholar]
- Desalegn, T.; Bacha, K.; Masi, C. The Effectiveness of Proteolytic Bacteria in the Leather and Detergent Industry Isolated Waste from the Modjo Tannery. Kuwait J. Sci. 2021, 50, 1–15. [Google Scholar] [CrossRef]
- Tari, C.; Genckal, H.; Tokatlı, F. Optimization of a Growth Medium Using a Statistical Approach for the Production of an Alkaline Protease from a Newly Isolated Bacillus sp. L21. Process Biochem. 2006, 41, 659–665. [Google Scholar] [CrossRef]
- Lageiro, M.; Alvarenga, N.; Lourenço, V.; Reis, A. Applicability Assessment of a Proteolytic Fermentation Broth to Leather Tanning and Protein Stain Removal. In Proceedings of the 5th International Electronic Conference on Applied Sciences, Online, Switzerland, 26–28 March 2024. [Google Scholar]
- Lageiro, M.; Alvarenga, N.; Lourenço, V.; Reis, A. Protease Application in Agroindustry: A Step towards the Bioeconomy. In Proceedings of the Encontro Ciência 2025, Oeiras, Portugal, 9–11 July 2025. [Google Scholar]
- Su, Y.; Liu, C.; Fang, H.; Zhang, D. Bacillus subtilis: A Universal Cell Factory for Industry, Agriculture, Biomaterials and Medicine. Microb. Cell Fact. 2020, 19, 173. [Google Scholar] [CrossRef] [PubMed]
- Anson, M.L. The Estimation of Pepsin, Trypsin, Papain, and Cathepsin with Hemoglobin. J. Gen. Physiol. 1938, 22, 79–89. [Google Scholar] [CrossRef]
- Folin, O.; Ciocalteu, V. On Tyrosine and Tryptophane Determinations in Proteins. J. Biol. Chem. 1927, 73, 627–650. [Google Scholar] [CrossRef]
- Logan, N.A.; Berkeley, R.C.W. Identification of Bacillus Strains Using the API System. Microbiology 1984, 130, 1871–1882. [Google Scholar] [CrossRef]
- Madeira, F.; Madhusoodanan, N.; Lee, J.; Eusebi, A.; Niewielska, A.; Tivey, A.R.N.; Lopez, R.; Butcher, S. The EMBL-EBI Job Dispatcher Sequence Analysis Tools Framework in 2024. Nucleic Acids Res. 2024, 52, W521–W525. [Google Scholar] [CrossRef]
- Ashraf, M.; Hussain, N.; Baqar, Z.; Kumar, A.; Ferreira, L.F.R.; Iqbal, H.M.N. Bioprospecting Microbial Proteases in Various Industries/Sectors. In Microbial Biomolecules; Kumar, A., Bilal, M., Ferreira, L.F.R., Kumari, M., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 301–324. [Google Scholar] [CrossRef]
- Pertiwiningrum, A.; Anggraini, F.D.; Fitrianto, N.A.; Rochijan, R. Isolation and Identification of Bacterial Protease Enzyme of Leather Waste. J. Indones. Trop. Anim. Agric. 2017, 42, 33. [Google Scholar] [CrossRef]
- Martinez, R.M. Bacillus subtilis. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Maloy, S., Hughes, K., Eds.; Academic Press: Cambridge, MA, USA, 2013; Volume 1, pp. 246–248. [Google Scholar]
- Schallmey, M.; Singh, A.; Ward, O.P. Developments in the Use of Bacillus Species for Industrial Production. Can. J. Microbiol. 2004, 50, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Contesini, F.J.; Melo, R.R.d.; Sato, H.H. An Overview of Bacillus Proteases: From Production to Application. Crit. Rev. Biotechnol. 2018, 38, 321–334. [Google Scholar] [CrossRef]
- Cupi, D.; Thorsen, M.; Elvig-Jørgensen, S.G.; Wulf-Andersen, L.; Berti-Sorbara, J.-O.; Cowieson, A.J.; Faruk, M.U. Efficacy and Safety Profile of a Subtilisin Protease Produced by Fermentation in Bacillus licheniformis to Be Used as a Feed Additive. Heliyon 2022, 8, e10030. [Google Scholar] [CrossRef]
- Khambhaty, Y. Applications of Enzymes in Leather Processing. Environ. Chem. Lett. 2020, 18, 747–769. [Google Scholar] [CrossRef]
- Solanki, P. Microbial Proteases: Ubiquitous Enzymes with Innumerable Uses. 3 Biotech 2021, 11, 428. [Google Scholar] [CrossRef]
- Fang, Z.; Yong, Y.-C.; Zhang, J.; Du, G.; Chen, J. Keratinolytic Protease: A Green Biocatalyst for Leather Industry. Appl. Microbiol. Biotechnol. 2017, 101, 7771–7779. [Google Scholar] [CrossRef]
- Al-Dhabi, N.A.; Esmail, G.A.; Ghilan, A.-K.M.; Arasu, M.V.; Duraipandiyan, V.; Ponmurugan, K. Characterization and Fermentation Optimization of Novel Thermo Stable Alkaline Protease from Streptomyces sp. Al-Dhabi-82 from the Saudi Arabian Environment for Eco-Friendly and Industrial Applications. J. King Saud Univ.—Sci. 2020, 32, 1258–1264. [Google Scholar] [CrossRef]
- Outtrup, H.; Jrgensen, S.T. The Importance of Bacillus Species in the Production of Industrial Enzymes. In Applications and Systematics of Bacillus and Relatives; Berkeley, R., Heyndrickx, M., Logan, N., De Vos, P., Eds.; Blackwell Science Ltd.: Oxford, UK, 2002; pp. 206–218. [Google Scholar] [CrossRef]
- Fu, L.L.; Xu, Z.R.; Li, W.F.; Shuai, J.B.; Lu, P.; Hu, C.X. Protein Secretion Pathways in Bacillus Subtilis: Implication for Optimization of Heterologous Protein Secretion. Biotechnol. Adv. 2007, 25, 1–12. [Google Scholar] [CrossRef]
- Ariaeenejad, S.; Kavousi, K.; Mamaghani, A.S.A.; Ghasemitabesh, R.; Hosseini Salekdeh, G. Simultaneous Hydrolysis of Various Protein-Rich Industrial Wastes by a Naturally Evolved Protease from Tannery Wastewater Microbiota. Sci. Total Environ. 2022, 815, 152796. [Google Scholar] [CrossRef] [PubMed]
- Zambare, V.; Nilegaonkar, S.; Kanekar, P. A Novel Extracellular Protease from Pseudomonas aeruginosa MCM B-327: Enzyme Production and Its Partial Characterization. New Biotechnol. 2011, 28, 173–181. [Google Scholar] [CrossRef]
- Eggert, T.; Brockmeier, U.; Dröge, M.J.; Quax, W.J.; Jaeger, K.-E. Extracellular Lipases from Bacillus subtilis: Regulation of Gene Expression and Enzyme Activity by Amino Acid Supply and External pH. FEMS Microbiol. Lett. 2003, 225, 319–324. [Google Scholar] [CrossRef]
- Liu, Y.; Su, A.; Tian, R.; Li, J.; Liu, L.; Du, G. Developing Rapid Growing Bacillus subtilis for Improved Biochemical and Recombinant Protein Production. Metab. Eng. Commun. 2020, 11, e00141. [Google Scholar] [CrossRef]
- Turcios, A.E.; Weichgrebe, D.; Papenbrock, J. Effect of Salt and Sodium Concentration on the Anaerobic Methanisation of the Halophyte Tripolium pannonicum. Biomass Bioenergy 2016, 87, 69–77. [Google Scholar] [CrossRef]
- Navarro-Pérez, M.L.; Fernández-Calderón, M.C.; Vadillo-Rodríguez, V. Decomposition of Growth Curves into Growth Rate and Acceleration: A Novel Procedure To Monitor Bacterial Growth and the Time-Dependent Effect of Antimicrobials. Appl. Environ. Microbiol. 2022, 88, e01849-21. [Google Scholar] [CrossRef]
- Bergey, D.H.; Krieg, N.R. Bergey’s Manual® of Systematic Bacteriology: Volume Four, 2nd ed.; Springer: New York, NY, USA, 2010; pp. 656–668. [Google Scholar]
- Awais, M.; Shah, A.A.; Hameed, A.; Hasan, F. Isolation, Identification and Optimization of Bacitracin Produced by Bacillus sp. Pak. J. Bot. 2007, 39, 1303–1312. [Google Scholar]
- Carballido-López, R.; Formstone, A. Shape Determination in Bacillus subtilis. Curr. Opin. Microbiol. 2007, 10, 611–616. [Google Scholar] [CrossRef] [PubMed]
- Ravishankar, K.; Kumar, P.S.; Jacob, B.; Saravanan, K.; Kumar, M.A.; Jacob, A. Optimal Conditions for Production of Extracellular Alkaline Protease from a Newly Isolated Bacillus subtilis Strain AKRS3. Res. Biotechnol. 2012, 3, 45–54. [Google Scholar]
- Losick, R.; Youngman, P.; Piggot, P.J. Genetics of Endospore Formation in Bacillus subtilis. Ann. Rev. Genet. 1986, 20, 625–669. [Google Scholar] [CrossRef]
- Gibson, T. 306. A Study of Bacillus subtilis and Related Organisms. J. Dairy Res. 1944, 13, 248–260. [Google Scholar] [CrossRef]
- Gordon, R.E.; Haynes, W.C.; Pang, C.H.-N. The Genus Bacillus; Agricultural Research Service: Washington, DC, USA, 1973; pp. 36–48. [Google Scholar]
- Al-Dhabaan, F.A. Morphological, Biochemical and Molecular Identification of Petroleum Hydrocarbons Biodegradation Bacteria Isolated from Oil Polluted Soil in Dhahran, Saud Arabia. Saudi J. Biol. Sci. 2019, 26, 1247–1252. [Google Scholar] [CrossRef]
- Fritze, D. Taxonomy of the Genus Bacillus and Related Genera: The Aerobic Endospore-Forming Bacteria. Phytopathology 2004, 94, 1245. [Google Scholar] [CrossRef]
- Kunst, F.; Ogasawara, N.; Moszer, I.; Albertini, A.M.; Alloni, G.; Azevedo, V.; Bertero, M.G.; Bessières, P.; Bolotin, A.; Borchert, S.; et al. The Complete Genome Sequence of the Gram-Positive Bacterium Bacillus subtilis. Nature 1997, 390, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Slabbinck, B.; Waegeman, W.; Dawyndt, P.; De Vos, P.; De Baets, B. From Learning Taxonomies to Phylogenetic Learning: Integration of 16S rRNA Gene Data into FAME-Based Bacterial Classification. BMC Bioinform. 2010, 11, 69. [Google Scholar] [CrossRef]
- Huynh, T.; Vörös, M.; Kedves, O.; Turbat, A.; Sipos, G.; Leitgeb, B.; Kredics, L.; Vágvölgyi, C.; Szekeres, A. Discrimination between the Two Closely Related Species of the Operational Group B. amyloliquefaciens Based on Whole-Cell Fatty Acid Profiling. Microorganisms 2022, 10, 418. [Google Scholar] [CrossRef]
- Roberts, M.S.; Nakamura, L.K.; Cohan, F.M. Bacillus mojavensis sp. Nov., Distinguishable from Bacillus subtilis by Sexual Isolation, Divergence in DNA Sequence, and Differences in Fatty Acid Composition. Int. J. Syst. Bacteriol. 1994, 44, 256–264. [Google Scholar] [CrossRef]
- Blanco Crivelli, X.; Cundon, C.; Bonino, M.P.; Sanin, M.S.; Bentancor, A. The Complex and Changing Genus Bacillus: A Diverse Bacterial Powerhouse for Many Applications. Bacteria 2024, 3, 256–270. [Google Scholar] [CrossRef]
- Lee, G.; Heo, S.; Kim, T.; Na, H.-E.; Park, J.; Lee, E.; Lee, J.-H.; Jeong, D.-W. Discrimination of Bacillus subtilis from Other Bacillus Species Using Specific Oligonucleotide Primers for the Pyruvate Carboxylase and Shikimate Dehydrogenase Genes. J. Microbiol. Biotechnol. 2022, 32, 1011–1016. [Google Scholar] [CrossRef]
- Kaneda, T. Fatty Acids of the Genus Bacillus: An Example of Branched-Chain Preference. Bacteriol. Rev. 1977, 41, 391–418. [Google Scholar] [CrossRef] [PubMed]
- Kämpfer, P. Limits and Possibilities of Total Fatty Acid Analysis for Classification and Identification of Bacillus Species. Syst. Appl. Microbiol. 1994, 17, 86–98. [Google Scholar] [CrossRef]
- Kaneda, T. Iso- and Anteiso-Fatty Acids in Bacteria: Biosynthesis, Function, and Taxonomic Significance. Microbiol. Rev. 1991, 55, 288–302. [Google Scholar] [CrossRef] [PubMed]
- Gudzenko, T.V.; Voliuvach, O.V.; Gorshkova, O.G.; Ostapchuk, А.М.; Ivanytsia, V.O. Fatty Acids Composition of Bacillus subtilis ONU551 Lipids. Ukr. Biochem. J. 2019, 91, 96–102. [Google Scholar] [CrossRef]
- Väisänen, O.; Salkinoja-Salonen, M. Use of Phage Typing and Fatty Acid Analysis for the Identification of Bacilli Isolated from Food Packaging Paper and Board Machines. Syst. Appl. Microbiol. 1989, 12, 103–111. [Google Scholar] [CrossRef]
- Beranová, J.; Jemioła-Rzemińska, M.; Elhottová, D.; Strzałka, K.; Konopásek, I. Metabolic Control of the Membrane Fluidity in Bacillus subtilis during Cold Adaptation. Biochim. Biophys. Acta (BBA)—Biomembr. 2008, 1778, 445–453. [Google Scholar] [CrossRef]
- Kunitsky, C.; Osterhout, G.; Sasser, M. Identification of Microorganisms Using Fatty Acid Methyl Ester (FAME) Analysis and the MIDI Sherlock® Microbial Identification System. Encycl. Rapid Microbiol. Methods 2006, 3, 17. [Google Scholar]
- Priest, F.G. Extracellular Enzyme Synthesis in the Genus Bacillus. Bacteriol. Rev. 1977, 41, 711–753. [Google Scholar] [CrossRef]
- Westers, L.; Westers, H.; Quax, W.J. Bacillus subtilis as Cell Factory for Pharmaceutical Proteins: A Biotechnological Approach to Optimize the Host Organism. Biochim. Biophys. Acta (BBA)—Mol. Cell Res. 2004, 1694, 299–310. [Google Scholar] [CrossRef]
- Gupta, R.; Beg, Q.; Khan, S.; Chauhan, B. An Overview on Fermentation, Downstream Processing and Properties of Microbial Alkaline Proteases. Appl. Microbiol. Biotechnol. 2002, 60, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Alam, G.; Uddin, E.; Rahman, S.; Ahmad, T.; Karim, R.; Mahmood, D.M.S.; Alam, M.S.; Hossain, J.; Maitra, P.; Islam, S. Protease Activity of Extracellular Enzyme Produced by B. Subtilis Isolated from Soil. Int. J. Environ. 2017, 2, 382–388. [Google Scholar] [CrossRef]
- Gurumallesh, P.; Alagu, K.; Ramakrishnan, B.; Muthusamy, S. A Systematic Reconsideration on Proteases. Int. J. Biol. Macromol. 2019, 128, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Salgado, V.; Fonseca, C.; Lopes Da Silva, T.; Roseiro, J.C.; Eusébio, A. Isolation and Identification of Magnusiomyces capitatus as a Lipase-Producing Yeast from Olive Mill Wastewater. Waste Biomass Valor. 2020, 11, 3207–3221. [Google Scholar] [CrossRef]
- Neves, K.C.S.; Porto, A.L.F.; Teixeira, M.F.S. Seleção de leveduras da Região Amazônica para produção de protease extracelular. Acta Amaz. 2006, 36, 299–306. [Google Scholar] [CrossRef][Green Version]
- Maczulak, A.E. Encyclopedia of Microbiology; Facts on File: New York, NY, USA, 2011; pp. 75–87, 349–364, 813–825. [Google Scholar]
- Sudha, J.; Ramakrishnan, V.; Madhusudhanan, N.; Mandal, A.B.; Gurunathan, T. Studies on Industrially Significant Haloalkaline Protease from Bacillus sp. JSGT Isolated from Decaying Skin of Tannery. J. Adv. Lab. Res. Biol. 2010, 1, 6. [Google Scholar]
- Camacho-Fernández, C.; Hervás, D.; Rivas-Sendra, A.; Marín, M.P.; Seguí-Simarro, J.M. Comparison of Six Different Methods to Calculate Cell Densities. Plant Methods 2018, 14, 30. [Google Scholar] [CrossRef]
- Monod, J. The Growth of Bacterial Cultures. Annu. Rev. Microbiol. 1949, 3, 371–394. [Google Scholar] [CrossRef]
- Doran, P.M. Bioprocess Engineering Principles; Reprint; Elsevier/Acad. Press: Amsterdam, The Netherlands, 2004; pp. 277–278. [Google Scholar]
- Amirian, M.M.; Irwin, A.J.; Finkel, Z.V. Extending the Monod Model of Microbal Growth with Memory. Front. Mar. Sci. 2022, 9, 963734. [Google Scholar] [CrossRef]
- Coico, R. Gram Staining. Curr. Protoc. Microbiol. 2006, A, A.3C.1–A.3C.2. [Google Scholar] [CrossRef]
- Beveridge, T.J.; Davies, J.A. Cellular Responses of Bacillus subtilis and Escherichia coli to the Gram Stain. J. Bacteriol. 1983, 156, 846–858. [Google Scholar] [CrossRef]
- Bartholomew, J.W.; Mittwer, T. The Gram Stain. Bacteriol. Rev. 1952, 16, 1–29. [Google Scholar] [CrossRef]
- Schaeffer, A.B.; Fulton, M.D. A Simplified Method of Staining Endospores. Science 1933, 77, 194. [Google Scholar] [CrossRef]
- Frank, J.A.; Reich, C.I.; Sharma, S.; Weisbaum, J.S.; Wilson, B.A.; Olsen, G.J. Critical Evaluation of Two Primers Commonly Used for Amplification of Bacterial 16S rRNA Genes. Appl. Environ. Microbiol. 2008, 74, 2461–2470. [Google Scholar] [CrossRef]
- Regueira-Iglesias, A.; Vázquez-González, L.; Balsa-Castro, C.; Vila-Blanco, N.; Blanco-Pintos, T.; Tamames, J.; Carreira, M.J.; Tomás, I. In Silico Evaluation and Selection of the Best 16S rRNA Gene Primers for Use in Next-Generation Sequencing to Detect Oral Bacteria and Archaea. Microbiome 2023, 11, 58. [Google Scholar] [CrossRef] [PubMed]
- Heuer, H.; Krsek, M.; Baker, P.; Smalla, K.; Wellington, E.M. Analysis of Actinomycete Communities by Specific Amplification of Genes Encoding 16S rRNA and Gel-Electrophoretic Separation in Denaturing Gradients. Appl. Environ. Microbiol. 1997, 63, 3233–3241. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a Prokaryotic Universal Primer for Simultaneous Analysis of Bacteria and Archaea Using Next-Generation Sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef]
- Li, F.; Xie, Y.; Gao, X.; Shan, M.; Sun, C.; Niu, Y.D.; Shan, A. Screening of Cellulose Degradation Bacteria from Min Pigs and Optimization of Its Cellulase Production. Electron. J. Biotechnol. 2020, 48, 29–35. [Google Scholar] [CrossRef]
- Marynowska, M.; Goux, X.; Sillam-Dussès, D.; Rouland-Lefèvre, C.; Halder, R.; Wilmes, P.; Gawron, P.; Roisin, Y.; Delfosse, P.; Calusinska, M. Compositional and Functional Characterisation of Biomass-Degrading Microbial Communities in Guts of Plant Fibre- and Soil-Feeding Higher Termites. Microbiome 2020, 8, 96. [Google Scholar] [CrossRef]
- Zheng, X.; Zhao, L.; Wu, F.; Zhou, H.; Shi, F. Screening and Identification of Protease-Producing Microorganisms in the Gut of Gryllotalpa orientalis (Orthoptera: Gryllotalpidae). Insects 2024, 15, 629. [Google Scholar] [CrossRef]
- George, N.; Sondhi, S.; Soni, S.K.; Gupta, N. Lime and Sulphide-Free Dehairing of Animal Skin Using Collagenase-Free Alkaline Protease from Vibrio metschnikovii NG155. Indian. J. Microbiol. 2014, 54, 139–142. [Google Scholar] [CrossRef][Green Version]
- Afridi, S.; Ali, N.; Niaz, M.; Hussain, S.I.; Farman, F.U.; Kim, B.C. Molecular Characterization and Production of Bacterial Amylases from Shahdara Spring, Pakistan. Appl. Ecol. Environ. Res. 2020, 18, 2611–2620. [Google Scholar] [CrossRef]
- Buyer, J.S. Rapid Sample Processing and Fast Gas Chromatography for Identification of Bacteria by Fatty Acid Analysis. J. Microbiol. Methods 2002, 51, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Buyer, J.S. Improved Fast Gas Chromatography for FAME Analysis of Bacteria. J. Microbiol. Methods 2003, 54, 117–120. [Google Scholar] [CrossRef]
- Descheemaeker, P.; Swings, J. The Application of Fatty Acid Methyl Ester Analysis (FAME) for the Identification of Heterotrophic Bacteria Present in Decaying Lede-Stone of the St. Bavo Cathedral in Ghent. Sci. Total Environ. 1995, 167, 241–247. [Google Scholar] [CrossRef]
- Dias, A.C.F.; Andreote, F.D.; Dini-Andreote, F.; Lacava, P.T.; Sá, A.L.B.; Melo, I.S.; Azevedo, J.L.; Araújo, W.L. Diversity and Biotechnological Potential of Culturable Bacteria from Brazilian Mangrove Sediment. World J. Microbiol. Biotechnol. 2009, 25, 1305–1311. [Google Scholar] [CrossRef]
- Slabbinck, B.; De Baets, B.; Dawyndt, P.; De Vos, P. Towards Large-Scale FAME-Based Bacterial Species Identification Using Machine Learning Techniques. Syst. Appl. Microbiol. 2009, 32, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Bacterial Identification by Gas Chromatographic Analysis of Fatty Acid Methyl Esters (GC-FAME). Available online: http://midi-inc.com/pdf/MIS_Technote_101.pdf (accessed on 1 February 2020).

| Medium Type (Conditions) (1) | Soaking Bath pH 9.03 | Purge Bath pH 9.45 | Liming Bath pH 12.62 |
|---|---|---|---|
| NA (pH 7.4–37 °C) (2) | + | + | − |
| ATCC 661 (pH 7.4–37 °C) (2) | − | − | + |
| SMA (pH 7.0–37 °C) (3) | >300 CFU (7 CFU with halos) | 11 CFU (4 CFU with big halos) | − |
| YM agar (pH 6.2–28 °C) (2) | − | − | − |
| Isolate | Halo Diameter (mm) |
|---|---|
| BMR1 | 16.9 ± 2.2 b |
| BMR2 | 30.1 ± 1.4 a |
| BMR3 | 9.2 ± 1.3 c |
| BMR4 | 12.3 ± 1.3 c |
| Test | BMR1 (1) | BMR2 (1) |
|---|---|---|
| Cell shape | rod | rod |
| Chains of cells | yes | yes |
| Gram staining | + | + |
| Colony in NA | yes | yes |
| Motility | yes | yes |
| Endospores | yes, apical | yes |
| Casein hydrolysis | yes | yes |
| API Test | Test * | BMR1 (1) | BMR2 (1) |
|---|---|---|---|
| 20E | ONPG (2) | + | + |
| L-arginine | + | − | |
| Citrate (Simmons’) | + | − | |
| Urease | + | + | |
| Sodium pyruvate | + | + | |
| Gelatinase | + | + | |
| Oxidase | + | + | |
| API Test | Test * | BMR1 (1) | BMR2 (1) |
| 50 CHB/E | Glycerol | + | + |
| L-Arabinose | + | + | |
| Ribose | + | + | |
| D-Xylose | + | + | |
| D-Glucose | + | + | |
| D-Fructose | + | + | |
| D-Mannose | + | + | |
| Inositol | + | + | |
| Mannitol | + | + | |
| Sorbitol | + | + | |
| α-Methyl-D-glucoside | + | + | |
| Amygdalin | + | + | |
| Arbutin | + | + | |
| Aesculin | + | + | |
| Salicin | + | + | |
| Cellobiose | + | + | |
| Maltose | + | + | |
| Melibiose | + | + | |
| Sucrose | + | + | |
| Trehalose | + | + | |
| D-Raffinose | + | + | |
| D-Turanose | + | + |
| Retention Time (min) | Reference Peak | BMR2 Isolate (%) |
|---|---|---|
| 5.647 | 13:0 anteiso | 0.13 |
| 6.812 | 14:0 iso | 0.80 |
| 7.316 | 14:0 | 0.28 |
| 8.261 | 15:0 iso | 14.06 |
| 8.398 | 15:0 anteiso | 45.10 |
| 9.858 | 16:0 iso | 3.24 |
| 10.470 | 16:0 | 2.47 |
| 11.548 | 17:0 iso | 7.88 |
| 11.707 | 17:0 anteiso | 18.75 |
| Σ iso, anteiso | 90% | |
| C15 iso/C15 anteiso | 0.31 | |
| BMR2 Similarity Index * with Bacillus subtilis | 0.893 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lageiro, M.; Simões, F.; Alvarenga, N.; Reis, A. Proteolytic Bacillus sp. Isolation and Identification from Tannery Alkaline Baths. Molecules 2025, 30, 3632. https://doi.org/10.3390/molecules30173632
Lageiro M, Simões F, Alvarenga N, Reis A. Proteolytic Bacillus sp. Isolation and Identification from Tannery Alkaline Baths. Molecules. 2025; 30(17):3632. https://doi.org/10.3390/molecules30173632
Chicago/Turabian StyleLageiro, Manuela, Fernanda Simões, Nuno Alvarenga, and Alberto Reis. 2025. "Proteolytic Bacillus sp. Isolation and Identification from Tannery Alkaline Baths" Molecules 30, no. 17: 3632. https://doi.org/10.3390/molecules30173632
APA StyleLageiro, M., Simões, F., Alvarenga, N., & Reis, A. (2025). Proteolytic Bacillus sp. Isolation and Identification from Tannery Alkaline Baths. Molecules, 30(17), 3632. https://doi.org/10.3390/molecules30173632









