Screening of a Combinatorial Library of Triazine-Scaffolded Dipeptide-Mimic Affinity Ligands to Bind Plasmid DNA
Abstract
1. Introduction
2. Results
2.1. Preliminary Binding Assessment
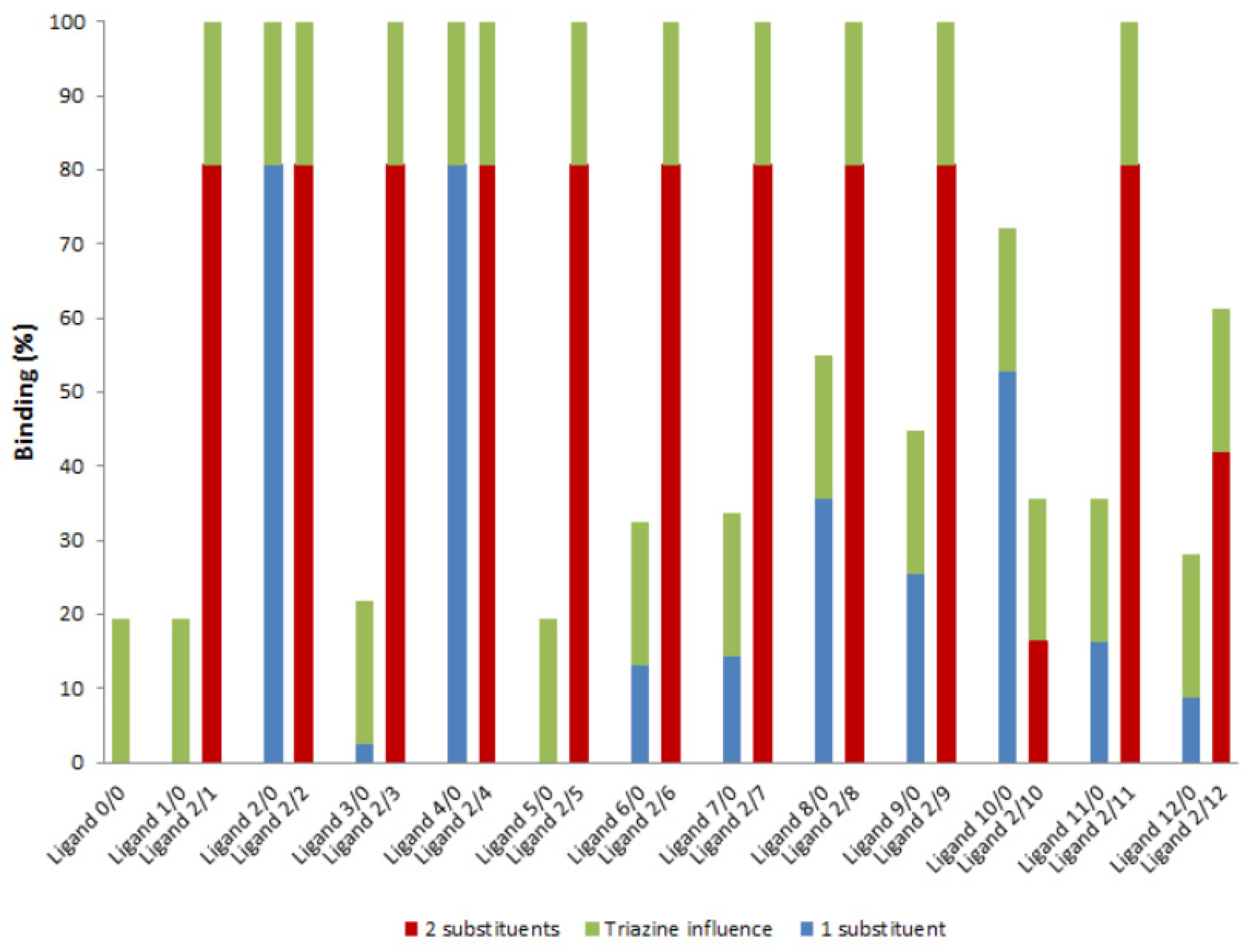
2.2. Screening with FITC-Labeled Poly T and Poly G Oligonucleotides
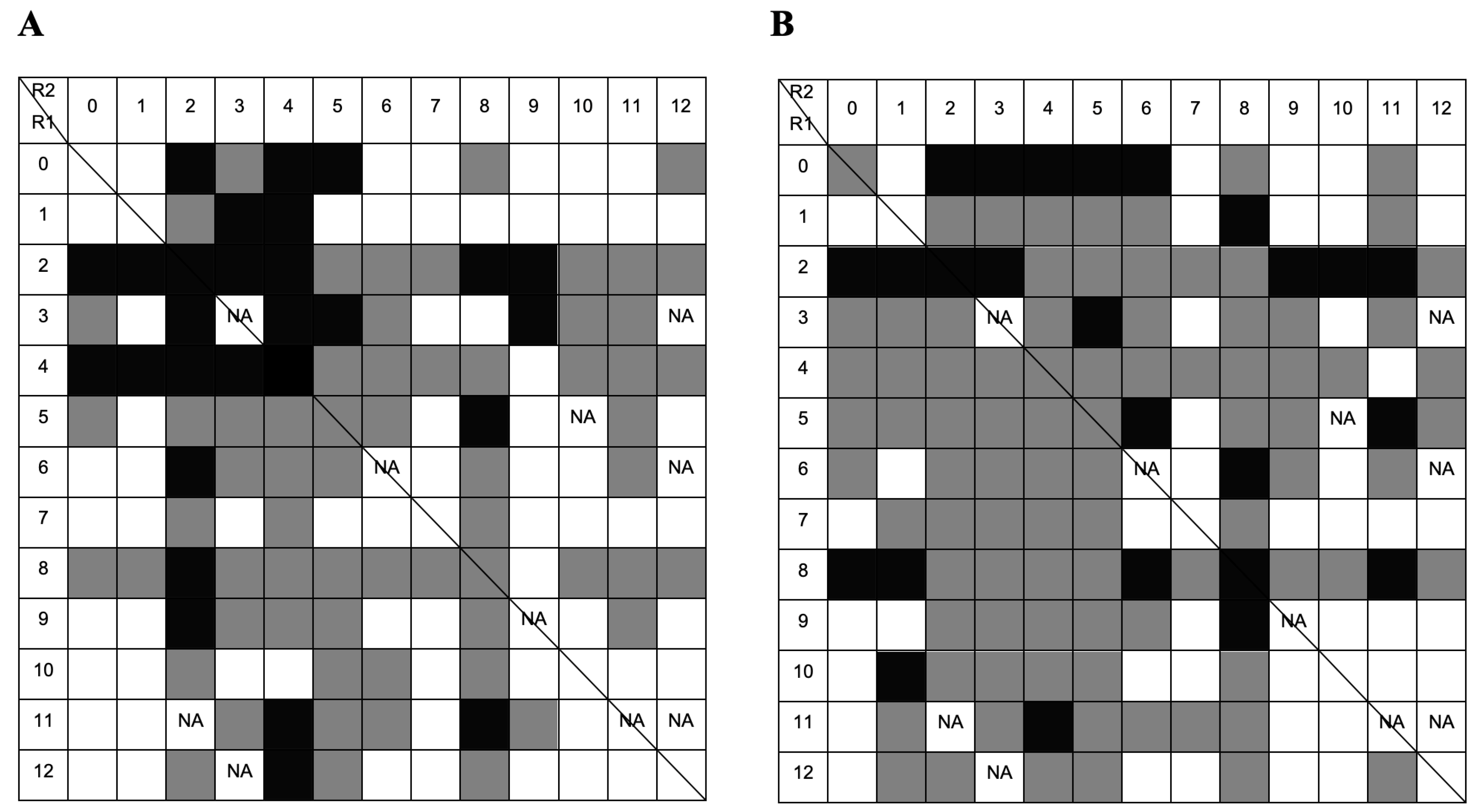
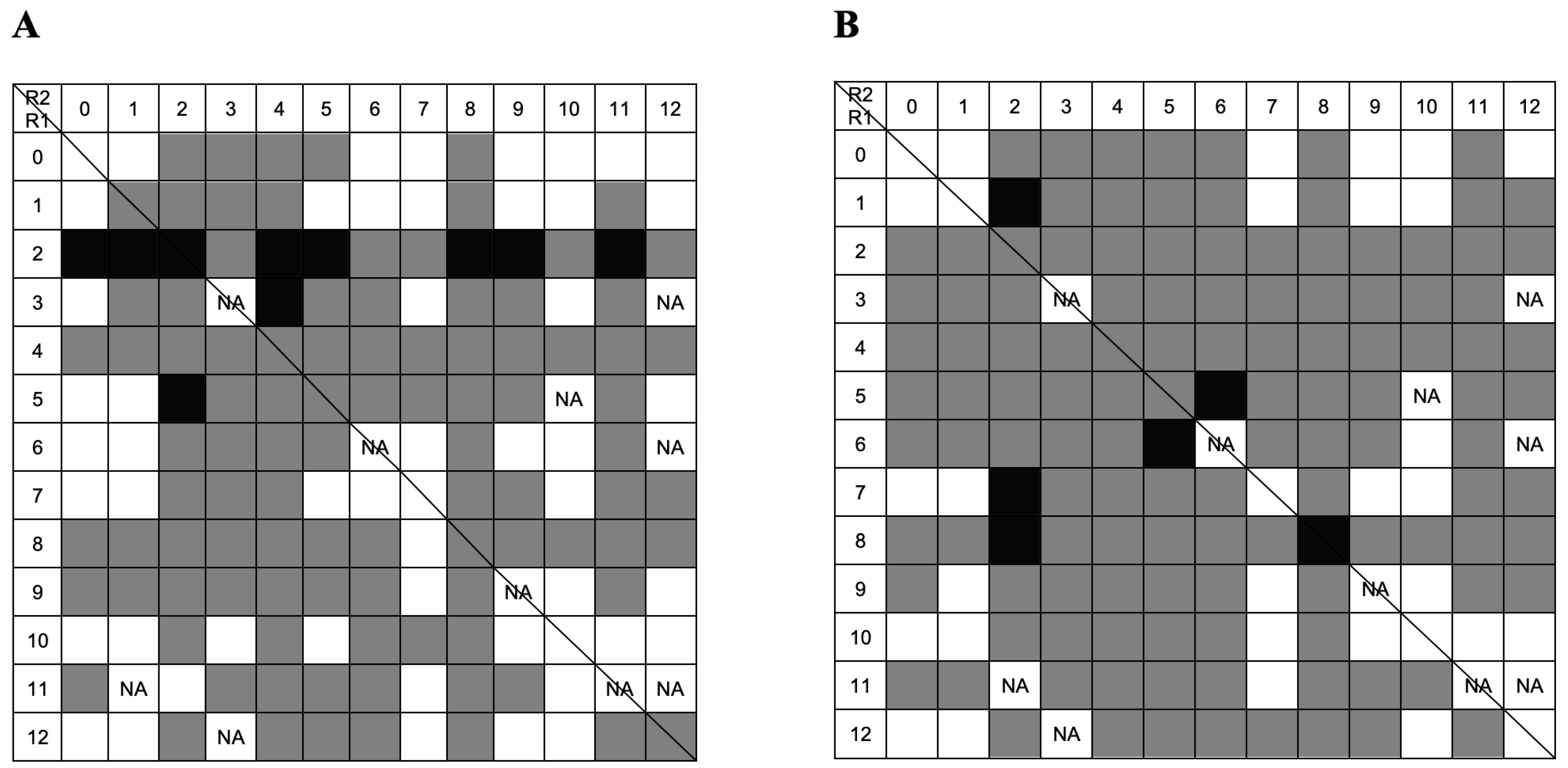
2.3. Screening by Microscale Affinity Chromatography with Single-Stranded Oligonucleotides
2.3.1. Screening with Thymine Homo-Oligonucleotide
2.3.2. Screening with Guanine Homo-Oligonucleotide
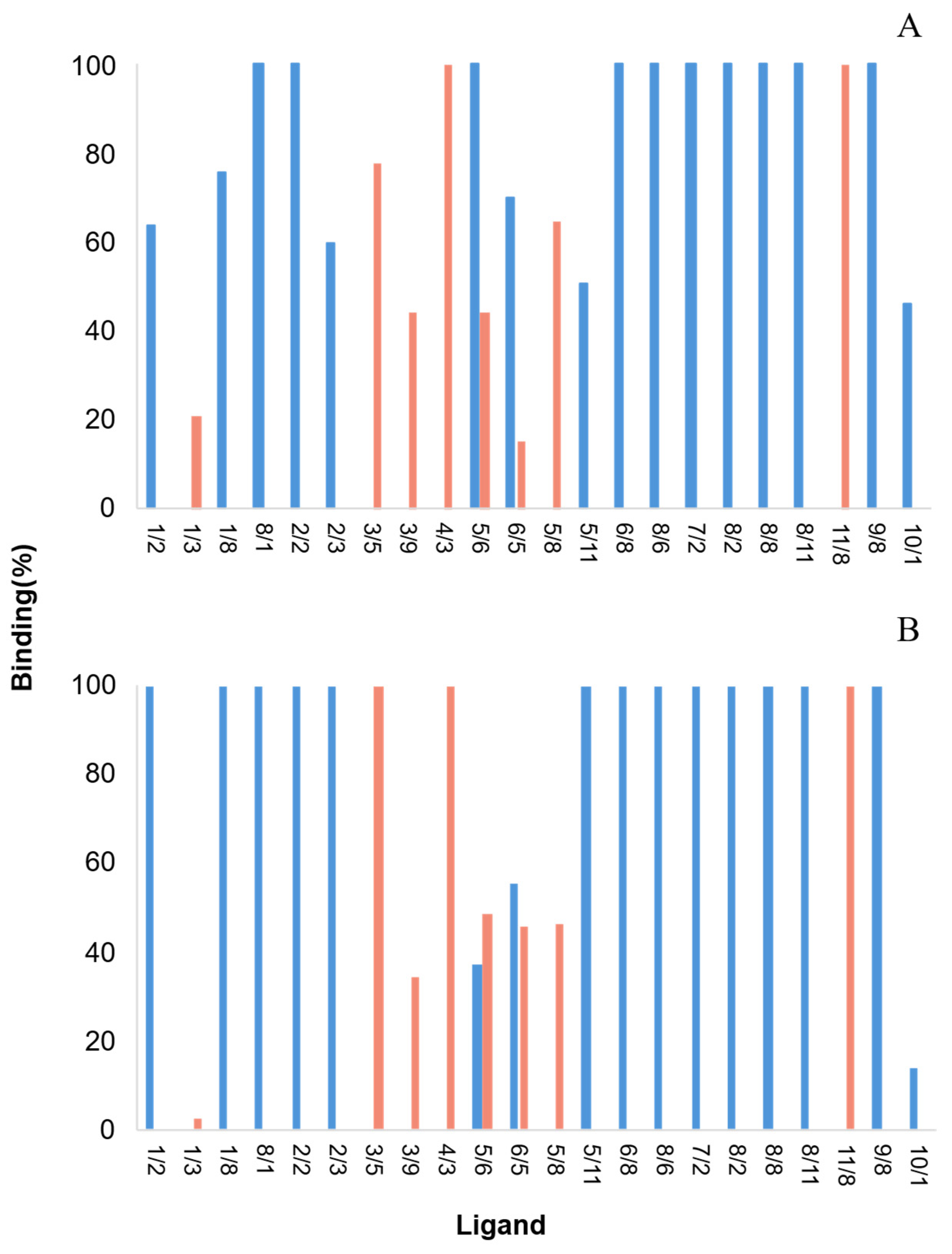
2.4. Screening by Microscale Affinity Chromatography with Double-Stranded Oligonucleotides (AT and GC)
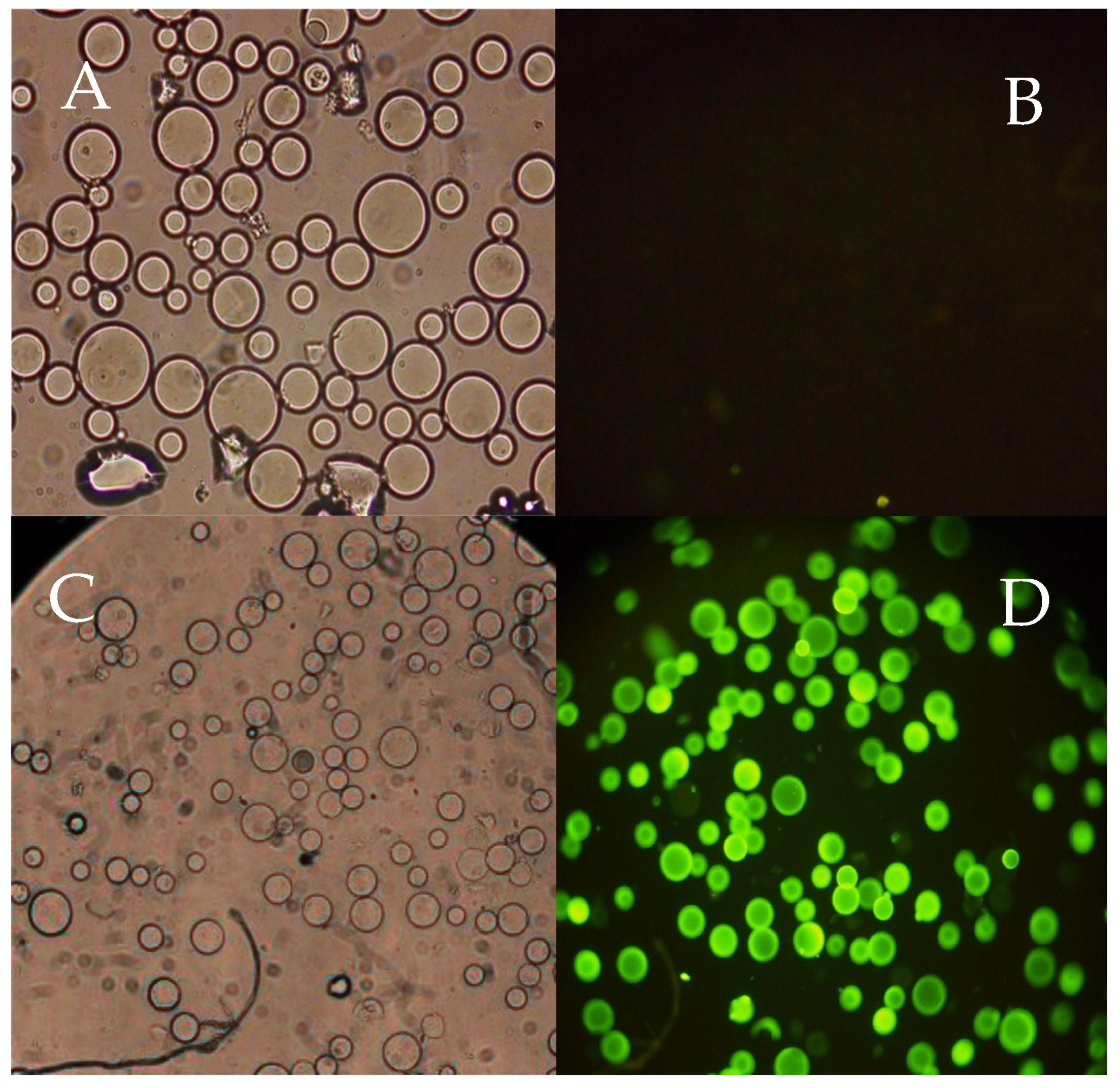
2.5. Screening Assays Using Plasmid DNA
2.6. Purification of Plasmid DNA from a Clarified E. coli Crude Extract
3. Materials and Methods
3.1. Materials
3.2. Fermentation
3.3. Cell Lysis
3.4. Concentration and Clarification of the Lysate
3.5. Synthetic Affinity Ligand Library
3.6. Screening of Ligands Using DNA Single-Stranded Oligonucleotides Conjugated to FITC
3.7. Chromatographic Binding Assays
3.8. Hybridization Assay with Homo-Oligonucleotides
3.9. Agarose Gel Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Leong, K.Y.; Tham, S.K.; Poh, C.L. Revolutionizing immunization: A comprehensive review of mRNA vaccine technology and applications. Virol. J. 2025, 22, 71. [Google Scholar] [CrossRef]
- Schmeer, M.; Buchholz, T.; Schleef, M. Plasmid DNA Manufacturing for Indirect and Direct Clinical Applications. Hum. Gene Ther. 2017, 28, 856–861. [Google Scholar] [CrossRef]
- Ohlson, J. Plasmid manufacture is the bottleneck of the genetic medicine revolution. Drug Discov. Today 2020, 25, 1891–1893. [Google Scholar] [CrossRef] [PubMed]
- Prazeres, D. The supporting role of plasmids in gene & cell therapy. Cell Gene Ther. Insights 2023, 9, 755–762. [Google Scholar] [CrossRef]
- Nguyen, D.N.; Ko, P.; Roper, B.; Console, G.; Gao, Y.; Shaw, D.; Yu, C.; Yehl, P.; Zhang, K.; Goyon, A. Analytical and Functional Characterization of Plasmid DNA Topological Forms and Multimers. Anal. Chem. 2024, 96, 12801–12808. [Google Scholar] [CrossRef] [PubMed]
- Evans, R.K.; Xu, Z.; Bohannon, K.E.; Wang, B.; Bruner, M.W.; Volkin, D.B. Evaluation of Degradation Pathways for Plasmid DNA in Pharmaceutical Formulations via Accelerated Stability Studies. J. Pharm. Sci. 2000, 89, 76–87. [Google Scholar] [CrossRef]
- Diogo, M.M.; Queiroz, J.A.; Prazeres, D.M.F. Chromatography of plasmid DNA. J. Chromatogr. A 2005, 1069, 3–22. [Google Scholar] [CrossRef]
- Wils, P.; Escriou, V.; Warnery, A.; Lacroix, F.; Lagneaux, D.; Ollivier, M.; Crouzet, J.; Mayaux, J.-F.; Scherman, D. Efficient purification of plasmid DNA for gene transfer using triple-helix affinity chromatography. Gene Ther. 1997, 4, 323–330. [Google Scholar] [CrossRef]
- Woodgate, J.; Palfrey, D.; Nagel, D.A.; Hine, A.V.; Slater, N.K.H. Protein-mediated isolation of plasmid DNA by a zinc finger-glutathione S-transferase affinity linker. Biotech Bioeng. 2002, 79, 450–456. [Google Scholar] [CrossRef]
- Caramelo-Nunes, C.; Gabriel, M.F.; Almeida, P.; Marcos, J.C.; Tomaz, C.T. Purification of plasmid DNA from clarified and non-clarified Escherichia coli lysates by berenil pseudo-affinity chromatography. J. Chromatogr. B 2012, 904, 81–87. [Google Scholar] [CrossRef]
- Çimen, D.; Yılmaz, F.; Perçin, I.; Türkmen, D.; Denizli, A. Dye affinity cryogels for plasmid DNA purification. Mater. Sci. Eng. C 2015, 56, 318–324. [Google Scholar] [CrossRef]
- Sousa, F.; Tomaz, C.T.; Prazeres, D.M.F.; Queiroz, J.A. Separation of supercoiled and open circular plasmid DNA isoforms by chromatography with a histidine–agarose support. Anal. Biochem. 2005, 343, 183–185. [Google Scholar] [CrossRef]
- Sousa, A.; Sousa, F.; Queiroz, J.A. Biorecognition of supercoiled plasmid DNA isoform in lysine-affinity chromatography. J. Chromatogr. B 2009, 877, 3257–3260. [Google Scholar] [CrossRef]
- Carapito, R.; Valente, J.F.A.; Queiroz, J.A.; Sousa, F. Arginine-Affinity Chromatography for Nucleic Acid (DNA and RNA) Isolation. In Affinity Chromatography; Ayyar, B.V., Arora, S., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2022; Volume 2466, pp. 135–144. ISBN 978-1-07-162175-2. [Google Scholar]
- Cardoso, S.; De Alcântara Pessoa Filho, P.; Sousa, F.; Rodrigues Azzoni, A. Arginine and di-arginine ligands for plasmid DNA purification using negative chromatography. Sep. Purif. Technol. 2018, 202, 281–289. [Google Scholar] [CrossRef]
- Cardoso, S.; Sousa, F.; Pessoa Filho, P.A.; Azzoni, A.R. Understanding the adsorption of plasmid DNA and RNA molecules onto arginine-agarose chromatographic resin. Mol. Biol. Rep. 2022, 49, 3893–3901. [Google Scholar] [CrossRef] [PubMed]
- Luscombe, N.M. Amino acid-base interactions: A three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001, 29, 2860–2874. [Google Scholar] [CrossRef] [PubMed]
- Lowe, C.R.; Lowe, A.R.; Gupta, G. New developments in affinity chromatography with potential application in the production of biopharmaceuticals. J. Biochem. Biophys. Methods 2001, 49, 561–574. [Google Scholar] [CrossRef]
- Taipa, M.Â. AFFINITY SEPARATION | Rational Design, Synthesis, and Evaluation: Affinity Ligands. In Reference Module in Chemistry, Molecular Sciences and Chemical Engineering; Elsevier: Amsterdam, The Netherlands, 2014; p. 9780124095472107383. ISBN 978-0-12-409547-2. [Google Scholar]
- Kaleta, C.; Schäuble, S.; Rinas, U.; Schuster, S. Metabolic costs of amino acid and protein production in Escherichia coli. Biotechnol. J. 2013, 8, 1105–1114. [Google Scholar] [CrossRef]
- Sousa, I.T.; Taipa, M.Â. Biomimetic Affinity Ligands for Protein Purification. In Protein Downstream Processing; Labrou, N.E., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2021; Volume 2178, pp. 167–199. ISBN 978-1-07-160774-9. [Google Scholar]
- Roque, A.C.A.; Taipa, M.Â.; Lowe, C.R. Synthesis and screening of a rationally designed combinatorial library of affinity ligands mimicking protein L from Peptostreptococcus magnus. J. Mol. Recognit. 2005, 18, 213–224. [Google Scholar] [CrossRef]
- Sousa, F.; Cruz, C.; Queiroz, J.A. Amino acids–nucleotides biomolecular recognition: From biological occurrence to affinity chromatography. J. Mol. Recognit. 2010, 23, 505–518. [Google Scholar] [CrossRef]
- Mandel-Gutfreund, Y.; Schueler, O.; Margalit, H. Comprehensive Analysis of Hydrogen Bonds in Regulatory Protein DNA-Complexes: In Search of Common Principles. J. Mol. Biol. 1995, 253, 370–382. [Google Scholar] [CrossRef]
- Marabotti, A.; Spyrakis, F.; Facchiano, A.; Cozzini, P.; Alberti, S.; Kellogg, G.E.; Mozzarelli, A. Energy-based prediction of amino acid-nucleotide base recognition. J. Comput. Chem. 2008, 29, 1955–1969. [Google Scholar] [CrossRef] [PubMed]
- Rooman, M.; Liévin, J.; Buisine, E.; Wintjens, R. Cation–π/H-bond Stair Motifs at Protein–DNA Interfaces. J. Mol. Biol. 2002, 319, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Mandel-Gutfreund, Y.; Margalit, H.; Jernigan, R.L.; Zhurkin, V.B. A role for CH…O interactions in protein-DNA recognition. J. Mol. Biol. 1998, 277, 1129–1140. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, P.; Bhattacharyya, R. Geometry of nonbonded interactions involving planar groups in proteins. Prog. Biophys. Mol. Biol. 2007, 95, 83–137. [Google Scholar] [CrossRef]
- Roque, A.C.A.; Taipa, M.Â.; Lowe, C.R. A new method for the screening of solid-phase combinatorial libraries for affinity chromatography. J. Mol. Recognit. 2004, 17, 262–267. [Google Scholar] [CrossRef]
- Li, W.; Xi, B.; Yang, W.; Hawkins, M.; Schubart, U.K. Complex DNA melting profiles of small PCR products revealed using SYBR® Green I. BioTechniques 2003, 35, 702–706. [Google Scholar] [CrossRef]
- Diogo, M.M.; Queiroz, J.A.; Prazeres, D.M.F. Assessment of purity and quantification of plasmid DNA in process solutions using high-performance hydrophobic interaction chromatography. J. Chromatogr. A 2003, 998, 109–117. [Google Scholar] [CrossRef]
- Antoni, G.; Presentini, R.; Neri, P. A simple method for the estimation of amino groups on insoluble matrix beads. Anal. Biochem. 1983, 129, 60–63. [Google Scholar] [CrossRef]
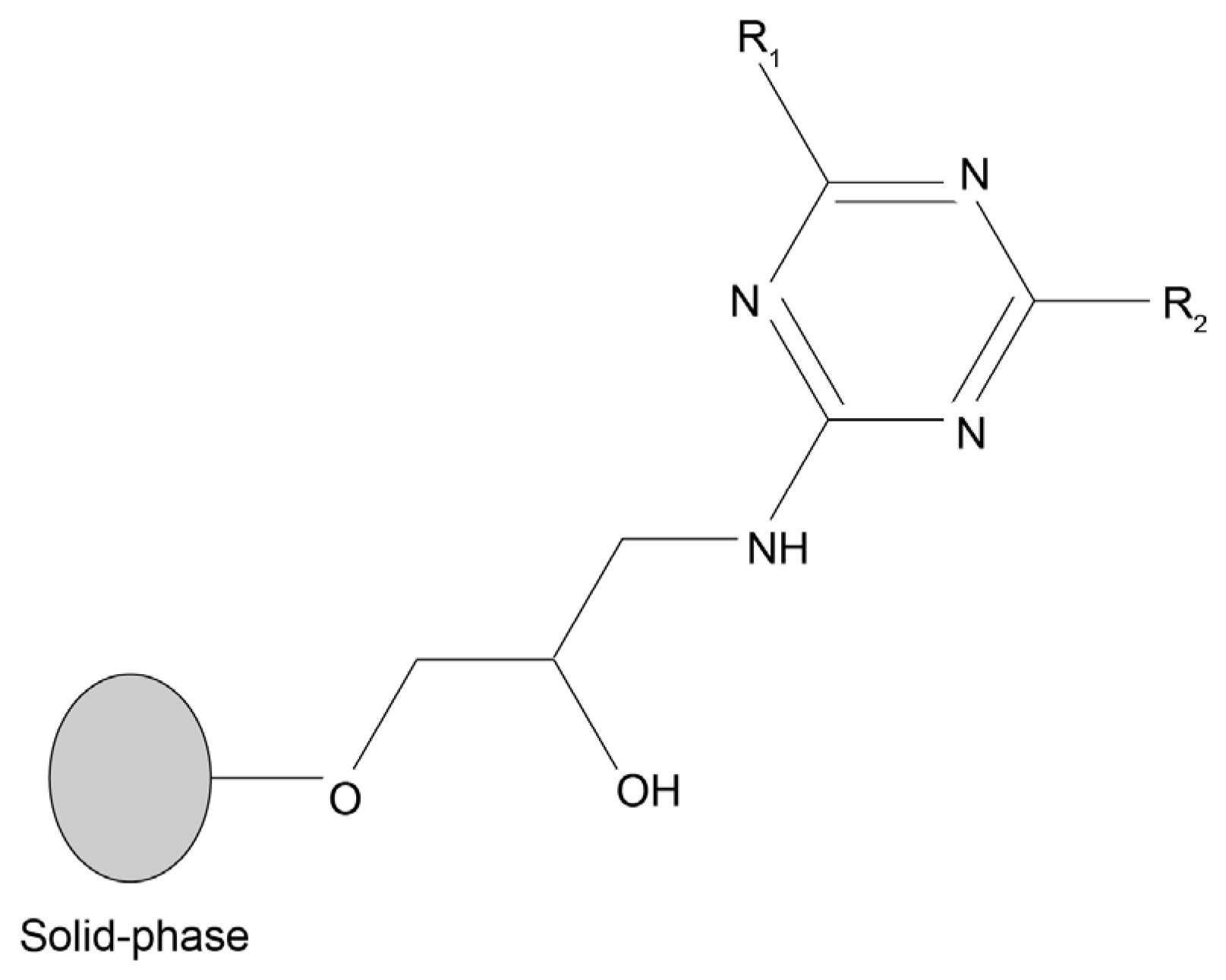

| Number | Structure and Chemical Name | Analog of | Number | Structure and Chemical Name | Analog of |
|---|---|---|---|---|---|
| 1 | 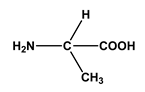 L-alanine | Alanine (Ala) Glycine (Gly) | 7 | 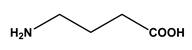 4-aminobutanoic acid | Glutamic acid (Glu) |
| 2 |  1,5-diaminopentane | Lysine (Lys) | 8 | 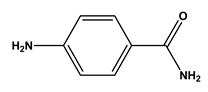 4-aminobenzamide | Glutamine (Gln) Asparagine (Asn) |
| 3 | 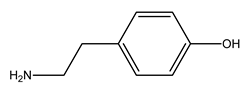 tyramine | Tyrosine (Tyr) | 9 | 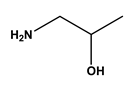 1-amino-2-propanol | Threonine (Thr) |
| 4 | 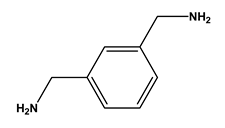 m-xylylenediamine | Lysine (Lys) | 10 | 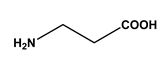 β-alanine | Aspartic acid (Asp) |
| 5 |  phenethylamine | Phenylalanine (Phe) | 11 | 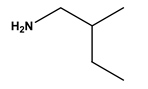 2-methylbutylamine | Isoleucine (Ile) |
| 6 | 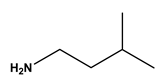 isoamylamine | Leucine (Leu) | 12 | 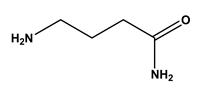 4-aminobutyramide | Glutamine (Gln) |
| Mass Load (µg) | Bound Mass (µg) | Binding (%) | |
|---|---|---|---|
| Single-strand homo-oligo (G) | 2.5 | 2.5 ± 0 | 100 |
| Double stranded oligo (GC) | 4.0 | 4 ± 0 | 100 |
| Double strand plasmid | 1.6 | 0.03 ± 0.07 | 1.9 |
| Single-stranded (denaturated) plasmid | 1.6 | 0 ± 0.12 | 0 |
| Name | DNA Sequence | Molecular Weight (g·mol−1) |
|---|---|---|
| FITC-Poly G | 5′-FITC-GGGGGGGGGGGGGGGGGGGG-3′ | 7060 |
| FITC-Poly T | 5′-FITC-TTTTTTTTTTTTTTTTTTTT-3′ | 6560 |
| Name | DNA Sequence | Molecular Weight (g·mol−1) |
|---|---|---|
| MRS2rev | 5′-TGCAAGCAACGCGTTAGCACATATGTG-3′ | 8309 |
| MRS2fwd | 5′-CATGCATTCGCGATTGGTCAAATTGGG-3′ | 8331 |
| Poly A | 5′-AAAAAAAAAAAAAAAAAAAA-3′ | 6202 |
| Poly G | 5′-GGGGGGGGGGGGGGGGGGGG-3′ | 6522 |
| Poly T | 5′-TTTTTTTTTTTTTTTTTTTT-3′ | 6022 |
| Poly C | 5′-CCCCCCCCCCCCCCCCCCCC-3′ | 5722 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belchior, J.F.R.; Monteiro, G.A.; Prazeres, D.M.; Taipa, M.Â. Screening of a Combinatorial Library of Triazine-Scaffolded Dipeptide-Mimic Affinity Ligands to Bind Plasmid DNA. Molecules 2025, 30, 3423. https://doi.org/10.3390/molecules30163423
Belchior JFR, Monteiro GA, Prazeres DM, Taipa MÂ. Screening of a Combinatorial Library of Triazine-Scaffolded Dipeptide-Mimic Affinity Ligands to Bind Plasmid DNA. Molecules. 2025; 30(16):3423. https://doi.org/10.3390/molecules30163423
Chicago/Turabian StyleBelchior, João F. R., Gabriel A. Monteiro, D. Miguel Prazeres, and M. Ângela Taipa. 2025. "Screening of a Combinatorial Library of Triazine-Scaffolded Dipeptide-Mimic Affinity Ligands to Bind Plasmid DNA" Molecules 30, no. 16: 3423. https://doi.org/10.3390/molecules30163423
APA StyleBelchior, J. F. R., Monteiro, G. A., Prazeres, D. M., & Taipa, M. Â. (2025). Screening of a Combinatorial Library of Triazine-Scaffolded Dipeptide-Mimic Affinity Ligands to Bind Plasmid DNA. Molecules, 30(16), 3423. https://doi.org/10.3390/molecules30163423








