Recent Advance in Electrochemical Chiral Recognition Based on Biomaterials (2019–2024)
Abstract
1. Introduction
2. Mechanisms and Methods of Electrochemical Chiral Recognition
2.1. Mechanisms of Electrochemical Chiral Recognition
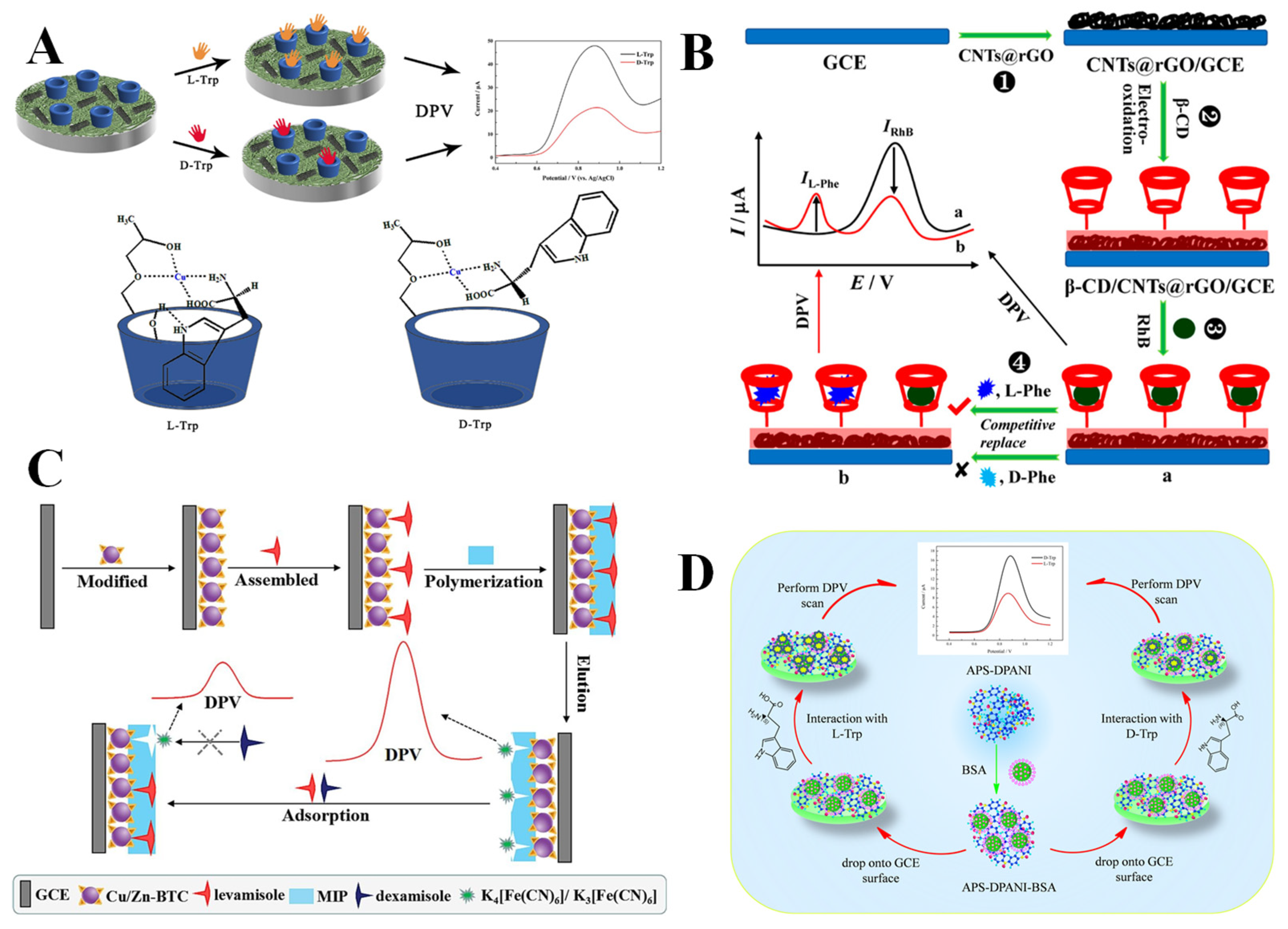
2.2. Methods of Electrochemical Chiral Recognition
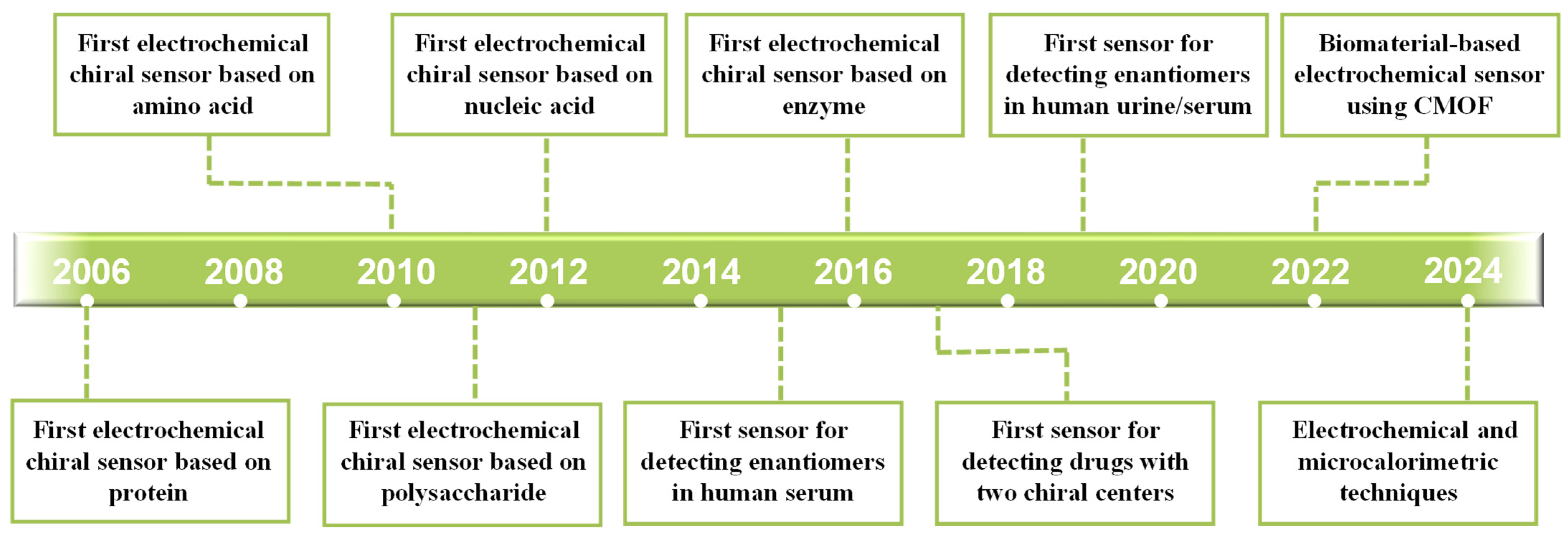
3. Biomaterials as Chiral Selectors for Electrochemical Chiral Recognition
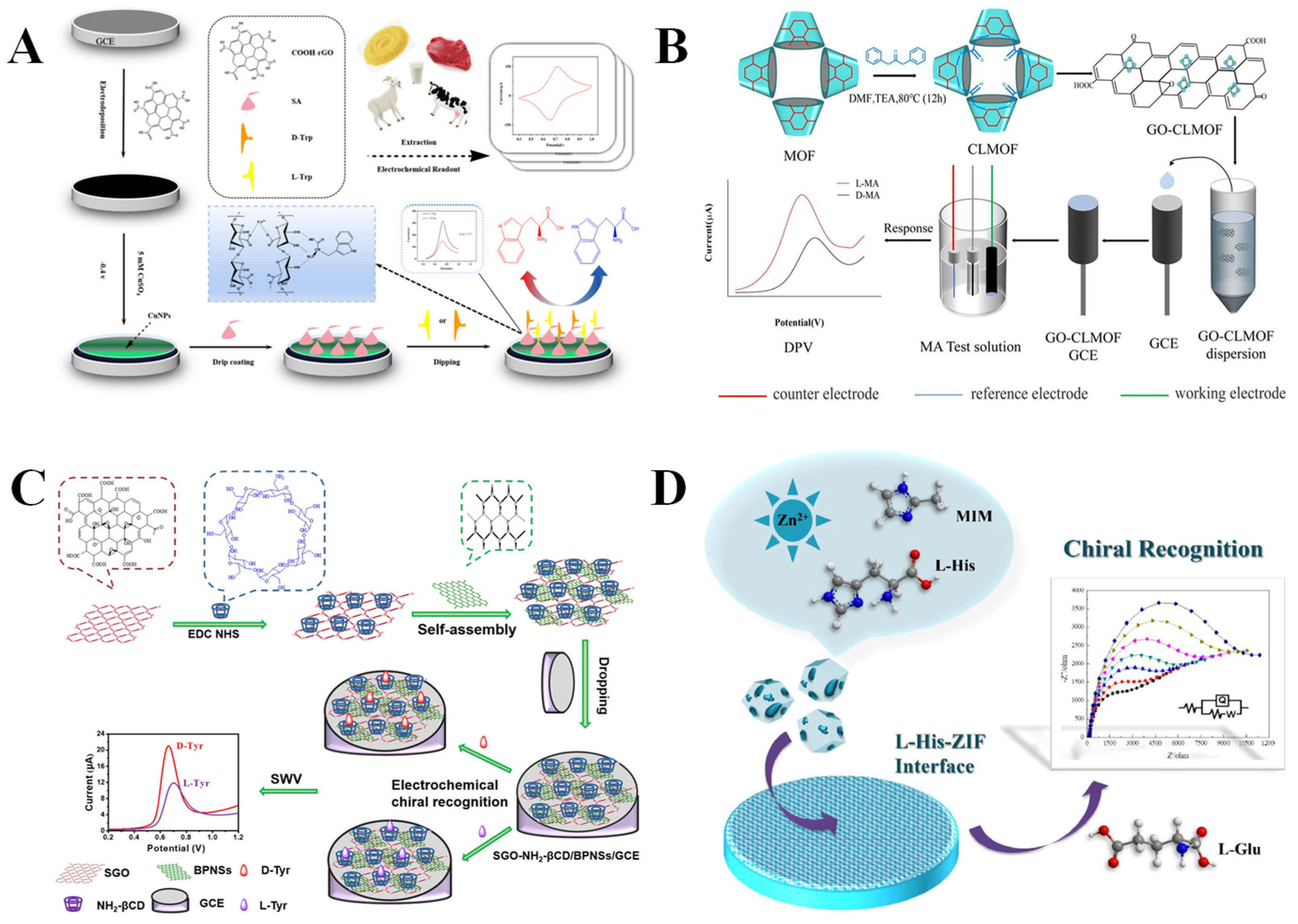
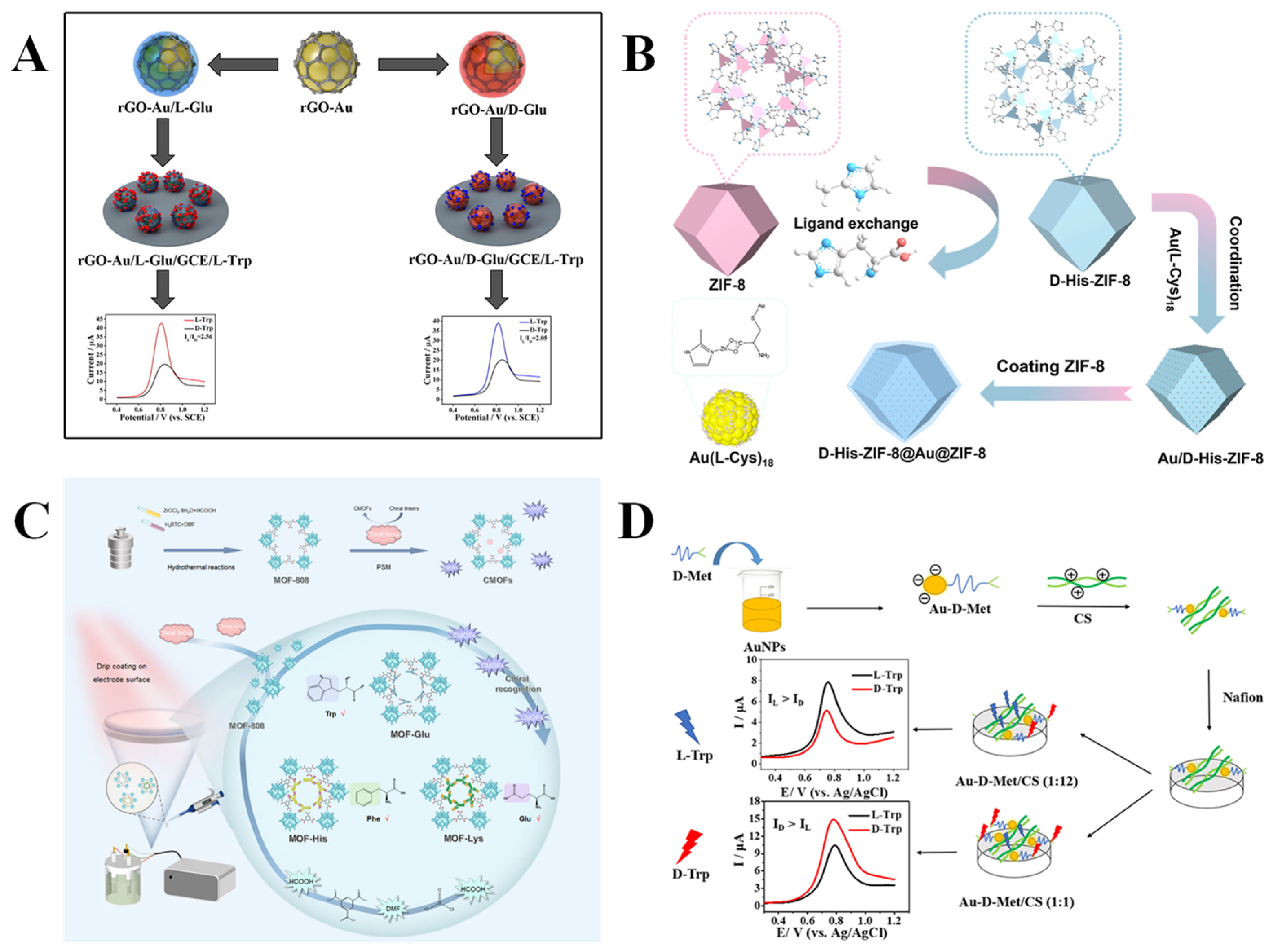
3.1. Amino Acids and Their Derivatives
3.2. Polysaccharides and Their Derivatives
3.3. Proteins
3.4. Enzymes
3.5. Nucleic Acids
4. Discussion and Prospects
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sekhon, B.S. Chiral pesticides. J. Pestic. Sci. 2009, 34, 1–12. [Google Scholar] [CrossRef]
- Shen, Q.; Wang, L.; Zhou, H.; Jiang, H.D.; Yu, L.S.; Zeng, S. Stereoselective binding of chiral drugs to plasma proteins. Acta Pharmacol. Sin. 2013, 34, 998–1006. [Google Scholar] [CrossRef]
- Hao, C.L. Recent progress in detecting enantiomers in food. Molecules 2024, 29, 1106. [Google Scholar] [CrossRef]
- Zhang, S.N.; Zheng, Y.L.; An, H.D.; Aguila, B.; Yang, C.X.; Dong, Y.Y.; Xie, W.; Cheng, P.; Zhang, Z.J.; Chen, Y.; et al. Covalent organic frameworks with chirality enriched by biomolecules for efficient chiral separation. Angew. Chem. Int. Ed. 2018, 57, 16754–16759. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Askarpour, A.N.; Sun, L.Y.; Shi, J.W.; Li, X.Q.; Alù, A. Chirality detection of enantiomers using twisted optical metamaterials. Nat. Commun. 2017, 8, 14180. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, H.M.; Yang, C.X.; Fang, L.X.; Zheng, L.; Lv, H.H.; Stavropoulos, P.; Ai, L.; Zhang, J.X. Chiral recognition of chiral (hetero)cyclic derivatives probed by tetraaza macrocyclic chiral solvating agents via 1H NMR spectroscopy. Anal. Chem. 2024, 96, 5188–5194. [Google Scholar] [CrossRef]
- Ceramella, J.; Iacopetta, D.; Franchini, A.; De Luca, M.; Saturnino, C.; Andreu, I.; Sinicropi, M.S.; Catalano, A. A look at the importance of chirality in drug activity: Some significative examples. Appl. Sci. 2022, 12, 10909. [Google Scholar] [CrossRef]
- Furusho, A.; Koga, R.; Akita, T.; Mita, M.; Kimura, T.; Hamase, K. Three-dimensional high-performance liquid chromatographic determination of Asn, Ser, Ala, and Pro enantiomers in the plasma of patients with chronic kidney disease. Anal. Chem. 2019, 91, 11569–11575. [Google Scholar] [CrossRef]
- Tang, B.; Wang, W.; Hou, H.P.; Liu, Y.Q.; Liu, Z.K.; Geng, L.N.; Sun, L.Q.; Luo, A.Q. A β-cyclodextrin covalent organic framework used as a chiral stationary phase for chiral separation in gas chromatography. Chin. Chem. Lett. 2022, 33, 898–902. [Google Scholar] [CrossRef]
- Deng, S.Y.; Pan, J.M.; Wang, M.; Huang, Y.K.; Xia, Z.N. Study on improvement of chiral separation of capillary electrophoresis based on cyclodextrin by deep eutectic solvents. Talanta 2020, 220, 121419. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.X.; Zhang, S.Y.; Sun, D.A.; Li, X.X.; Meng, G.P.; Zhang, X.; Wang, B.D. Chiral gold nanorod vertical arrays for enantioselective chemiluminescence recognition of naproxen and mechanism revealing. Chem. Eng. J. 2024, 482, 148900. [Google Scholar] [CrossRef]
- Jiang, W.J.; He, R.; Lv, H.; He, X.H.; Wang, L.; Wei, Y.L. Chiral sensing of tryptophan enantiomers based on the enzyme mimics of β-cyclodextrin-modified sulfur quantum dots. ACS Sens. 2023, 8, 4264–4271. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.X.; Chen, L.X.; Tian, T.T.; Zhang, Z.G.; Li, X.J. Identification and quantitation of enantiomers by capillary electrophoresis and circular dichroism independent of single enantiomer standard. Anal. Chem. 2019, 91, 13803–13809. [Google Scholar] [CrossRef]
- Salinas, G.; Niamlaem, M.; Kuhn, A.; Arnaboldi, S. Recent advances in electrochemical transduction of chiral information. Curr. Opin. Colloid Interface Sci. 2022, 61, 101626. [Google Scholar] [CrossRef]
- Zou, J.; Zhao, G.Q.; Zhao, G.L.; Yu, J.G. Fast and sensitive recognition of enantiomers by electrochemical chiral analysis: Recent advances and future perspectives. Coord. Chem. Rev. 2022, 471, 214732. [Google Scholar] [CrossRef]
- Wu, X.F.; Ge, Q.M.; Jiang, N.; Zhao, W.F.; Liu, M.; Cong, H.; Zhao, J.L. Research progress on chiral supramolecular sensors for enantiomer detection. Chemosensors 2023, 11, 269. [Google Scholar] [CrossRef]
- Chen, H.T.; Xia, L.; Li, G.K. Recent progress of chiral metal-organic frameworks in enantioselective separation and detection. Microchim. Acta 2024, 191, 640. [Google Scholar] [CrossRef]
- Guo, J.L.; Hou, J.X.; Hu, J.T.; Geng, Y.J.; Li, M.X.; Wang, H.; Wang, J.L.; Luo, Q. Recent advances in β-cyclodextrin-based materials for chiral recognition. Chem. Commun. 2023, 59, 9157–9166. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Q.; Zhao, L.; Li, J.Y.; Xie, S.S.; Liang, N. Synthesis of cyclodextrin-based MOFs incorporating amino acid chiral ligands for chiral separation of naproxen enantiomers. Microchem. J. 2023, 190, 108684. [Google Scholar] [CrossRef]
- Scriba, G.K.E. Chiral recognition in separation sciences. Part I: Polysaccharide and cyclodextrin selectors. TrAC-Trends Anal. Chem. 2019, 120, 115639. [Google Scholar] [CrossRef]
- Haginaka, J. Recent progresses in protein-based chiral stationary phases for enantioseparations in liquid chromatography. J. Chromatogr. B-Anal. Technol. Biomed. Life Sci. 2008, 875, 12–19. [Google Scholar] [CrossRef]
- Zhang, J.L.; Liu, X.; Mu, Q.X.; Li, R.J.; Ji, Y.B. Construction and application of a pepsin-functionalized covalent organic framework with prominent chiral recognition ability. Chem.-A Eur. J. 2024, 30, e202303827. [Google Scholar] [CrossRef] [PubMed]
- Oukacine, F.; Ravelet, C.; Peyrin, E. Enantiomeric sensing and separation by nucleic acids. TrAC-Trends Anal. Chem. 2020, 122, 115733. [Google Scholar] [CrossRef]
- Wang, Y.X.; Yin, X.L.; Shi, M.H.; Li, W.; Zhang, L.; Kong, J.L. Probing chiral amino acids at sub-picomolar level based on bovine serum albumin enantio selective films coupled with silver-enhanced gold nanoparticles. Talanta 2006, 69, 1240–1245. [Google Scholar] [CrossRef]
- Kang, S.Z.; Chen, H.; Li, X.Q.; Mu, J. Preparation of L-alanine ethyl ester modified multiwalled carbon nanotubes and their chiral discrimination between D- and L-tryptophan. Diam. Relat. Mater. 2010, 19, 1221–1224. [Google Scholar] [CrossRef]
- Bian, Y.H.; Zhang, G.F.; Zhong, X.; Tian, D.M.; Li, H.B. Enantioselective recognition of electrochemically inactive phenylalanine by thiolated-cyclodextrin/ferrocene-coated gold nanoparticles. Supramol. Chem. 2011, 23, 455–461. [Google Scholar] [CrossRef]
- Feng, L.Y.; Xu, B.L.; Ren, J.S.; Zhao, C.Q.; Qu, X.G. A human telomeric DNA-based chiral biosensor. Chem. Commun. 2012, 48, 9068–9070. [Google Scholar] [CrossRef]
- Lin, L.; Lian, H.T.; Sun, X.Y.; Yu, Y.M.; Liu, B. An L-dopa electrochemical sensor based on a graphene doped molecularly imprinted chitosan film. Anal. Methods 2015, 7, 1387–1394. [Google Scholar] [CrossRef]
- Wang, L.L.; Gong, W.C.; Wang, F.; Yu, Z.Y.; Chen, Z.L. Efficient bienzyme nanocomposite film for chiral recognition of L-tryptophan, L-phenylalanine and L-tyrosine. Anal. Methods 2016, 8, 3481–3487. [Google Scholar] [CrossRef]
- Upadhyay, S.S.; Kalambate, P.K.; Srivastava, A.K. Enantioselective analysis of moxifloxacin hydrochloride enantiomers with graphene-β-cyclodextrin-nanocomposite modified carbon paste electrode using adsorptive stripping differential pulse voltammetry. Electrochim. Acta 2017, 248, 258–269. [Google Scholar] [CrossRef]
- Yang, X.; Niu, X.H.; Mo, Z.L.; Wang, J.; Shuai, C.; Pan, Z.; Liu, Z.Y.; Liu, N.J.; Guo, R.B. 3D nitrogen and sulfur Co -Doped graphene/integrated polysaccharides for electrochemical recognition tryptophan enantiomers. J. Electrochem. Soc. 2019, 166, B1053–B1062. [Google Scholar] [CrossRef]
- Niu, X.H.; Yan, S.M.; Chen, J.L.; Li, H.X.; Wang, K.J. Enantioselective recognition of L/D-amino acids in the chiral nanochannels of a metal-organic framework. Electrochim. Acta 2022, 405, 139809. [Google Scholar] [CrossRef]
- Stoian, I.A.; Iacob, B.C.; Oprean, R.; Bodoki, E. Chiral electrochemical sensing of propranolol enantiomers on D/L-Cys modified gold nanoparticles. Electrochim. Acta 2024, 498, 144608. [Google Scholar] [CrossRef]
- Huang, H.; Hu, L.L.; Sun, Y.; Liu, Y.; Kang, Z.H.; MacFarlane, D.R. Preparation of chiral graphene oxides by covalent attachment of chiral cysteines for voltammetric recognition of tartrates. Microchim. Acta 2019, 186, 298. [Google Scholar] [CrossRef]
- Pei, H.B.; Wang, J.; Jin, X.N.; Zhang, X.J.; Liu, W.J.; Guo, R.B.; Liu, N.J.; Mo, Z.L. An electrochemical chiral sensor based on glutamic acid functionalized graphene-gold nanocomposites for chiral recognition of tryptophan enantiomers. J. Electroanal. Chem. 2022, 913, 116283. [Google Scholar] [CrossRef]
- Pei, H.B.; Chen, F.; Guo, W.; Jia, Q.Q.; Guo, R.B.; Liu, N.J.; Mo, Z.L. Chiral nitrogen-doped graphene quantum dot electrochemical sensor for recognition of tartaric acid isomers. J. Electrochem. Soc. 2021, 168, 067515. [Google Scholar] [CrossRef]
- Niu, X.H.; Yan, S.M.; Zhao, R.; Han, S.; Cao, K.J.; Li, H.X.; Wang, K.J. Chiral template-induced porphyrin-based self-assembled materials for electrochemical chiral sensing. Microchim. Acta 2023, 190, 61. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.H.; Yan, S.M.; Zhao, R.; Li, H.X.; Liu, X.Y.; Wang, K.J. Design and electrochemical chiral sensing of the robust sandwich chiral composite D-His-ZIF-8@Au@ZIF-8. ACS Appl. Mater. Interfaces 2023, 15, 22435–22444. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.H.; Zhao, R.; Yan, S.M.; Li, H.X.; Yang, J.; Cao, K.J.; Liu, X.Y.; Wang, K.J. Chiral MOFs encapsulated by polymers with poly-metallic coordination as chiral biosensors. Microchim. Acta 2023, 190, 230. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.H.; Liu, Y.Q.; Zhao, R.; Yuan, M.; Wang, Y.W.; Zhang, J.Y.; Li, H.X.; Yang, X.; Wang, K.J. Regulating catalytic oxidation enantiomers behavior by imparting chiral microenvironment in Zr-based metal-organic frameworks. Small 2024, 20, 2404554. [Google Scholar] [CrossRef]
- Niu, X.H.; Yuan, M.; Yang, X.; Li, H.X.; Zhao, R.; Liu, Y.Q.; Zhao, H.F.; Wang, K.J. Chiral metal-organic framework nanostructures for chiral channel-dependent enantioselective sensing. ACS Appl. Nano Mater. 2024, 7, 19175–19183. [Google Scholar] [CrossRef]
- Zhao, L.; Kuang, X.; Kuang, R.; Tong, L.; Liu, Z.X.; Hou, Y.; Sun, X.; Wang, Z.L.; Wei, Q. MOF-based supramolecule helical nanomaterials: Toward highly enantioselective electrochemical recognition of penicillamine. ACS Appl. Mater. Interfaces 2020, 12, 1533–1538. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.H.; Yang, X.; Mo, Z.L.; Pan, Z.; Liu, Z.Y.; Shuai, C.; Gao, Q.Q.; Liu, N.J.; Guo, R.B. Electrochemical chiral recognition at molecular imprinting Au surfaces. J. Electrochem. Soc. 2019, 166, B1126–B1130. [Google Scholar] [CrossRef]
- Deng, Y.A.; Zhang, Z.H.; Zhou, X.Y.; Wang, Y.; Zhang, Y.; Yuan, Y.L. Common materials, extraordinary behavior: An ultrasensitive and enantioselective strategy for D-tryptophan recognition based on electrochemical Au@p-L-cysteine chiral interface. Anal. Chim. Acta 2022, 1227, 340331. [Google Scholar] [CrossRef]
- Chen, H.B.; Zhao, C.Q.; Li, Y.; Li, J.Y.; Cai, W.R.; Kong, Y.; Yin, Z.Z. A chiral sensing platform with reversible chirality based on Au nanoparticles-D-methionine/chitosan. J. Electroanal. Chem. 2023, 942, 117562. [Google Scholar] [CrossRef]
- Wei, X.F.; Chen, Y.; He, S.; Lian, H.T.; Cao, X.G.; Liu, B. L-histidine-regulated zeolitic imidazolate framework modified electrochemical interface for enantioselective determination of L-glutamate. Electrochim. Acta 2021, 400, 139464. [Google Scholar] [CrossRef]
- Gong, T.; Zhu, S.; Huang, S.Q.; Gu, P.C.; Xiong, Y.; Zhang, J.; Jiang, X.H. A renewable electrochemical sensor based on a self-assembled framework of chiral molecules for efficient identification of tryptophan isomers. Anal. Chim. Acta 2022, 1191, 339276. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.; He, J.H.; Wang, S.J.; Xu, D.F.; Zhang, R.; Bradley, M.; Sun, Y.X. A signal amplification for Trp isomers electrochemical recognition based on PEDOT:PSS and CS/PAA multilayers. Talanta 2023, 265, 124885. [Google Scholar] [CrossRef]
- Hou, H.P.; Tang, S.S.; Liu, Y.Q.; Wang, W.; Liang, A.X.; Sun, L.Q.; Luo, A.Q. Drop-coated molybdenum disulfide-ionic liquid for improving the electrochemical chiral recognition ability of chitosan. J. Electroanal. Chem. 2022, 904, 115921. [Google Scholar] [CrossRef]
- Gou, H.; He, J.X.; Nie, R.; Xu, D.Q.; Rao, H.H.; Zhao, G.H. A stable electrochemical chiral interface based on graphene-chitosan composites for tyrosine enantiomers recognition. Microchem. J. 2023, 190, 108712. [Google Scholar] [CrossRef]
- Han, S.; Wang, L.H.; Li, H.X.; Niu, X.H.; Li, Y.N.; Liu, X.Y.; Wang, K.J. Chiral electrochemical recognition of tryptophan enantiomers by polysaccharide modified MOFs. J. Electroanal. Chem. 2023, 935, 117326. [Google Scholar] [CrossRef]
- Du, Y.X.; Mo, Z.L.; Wang, J.; Shuai, C.; Pei, H.B.; Chen, Y.; Yue, R.M. A novel chiral carbon nanocomposite based on cellulose gum modifying chiral tri-electrode system for the enantiorecognition of tryptophan. J. Electroanal. Chem. 2021, 895, 115390. [Google Scholar] [CrossRef]
- Du, Y.X.; Mo, Z.L.; Pei, H.B.; Liu, W.T.; Yue, R.M.; Wang, X.R. The fabrication of a highly electroactive chiral-interface self-assembled Cu(ii)-coordinated binary-polysaccharide composite for the differential pulse voltammetry (DPV) detection of tryptophan isomers. New J. Chem. 2022, 46, 9811–9818. [Google Scholar] [CrossRef]
- Niu, X.H.; Yang, X.; Li, H.X.; Shi, Q.Y.; Wang, K.J. Chiral voltammetric sensor for tryptophan enantiomers by using a self-assembled multiwalled carbon nanotubes/polyaniline/sodium alginate composite. Chirality 2021, 33, 248–260. [Google Scholar] [CrossRef]
- Pei, H.B.; Chen, F.; Niu, X.H.; Jia, Q.Q.; Guo, R.B.; Liu, N.J.; Mo, Z.L. Self-assembled chitosan-sodium alginate composite material for electrochemical recognition of tyrosine isomers. J. Electroanal. Chem. 2021, 895, 115525. [Google Scholar] [CrossRef]
- Niu, X.H.; Yang, X.; Mo, Z.L.; Wang, J.; Pan, Z.; Liu, Z.Y.; Shuai, C.; Liu, G.G.; Liu, N.J.; Guo, R.B. Fabrication of an electrochemical chiral sensor via an integrated polysaccharides/3D nitrogen-doped graphene-CNT frame. Bioelectrochemistry 2020, 131, 107396. [Google Scholar] [CrossRef]
- Ji, J.; Qu, L.H.; Wang, Z.R.; Li, G.Y.; Feng, W.; Yang, G. A facile electrochemical chiral sensor for tryptophan enantiomers based on multiwalled carbon nanotube/hydroxypropyl-β-cyclodextrin functionalized carboxymethyl cellulose. Microchem. J. 2022, 175, 107133. [Google Scholar] [CrossRef]
- Yi, Y.H.; Zhang, D.P.; Ma, Y.Z.; Wu, X.Y.; Zhu, G.B. Dual-signal electrochemical enantiospecific recognition system via competitive supramolecular host-guest interactions: The case of phenylalanine. Anal. Chem. 2019, 91, 2908–2915. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, F.; Wang, Z.X.; Wang, J.P. Electrochemical sensor based on sodium alginate as a chiral selective agent for the detection of tryptophan isomers in food. Microchem. J. 2025, 211, 113126. [Google Scholar] [CrossRef]
- Sun, X.Z.; Fu, Z.B.; Zhang, M.; Fu, H.; Lin, C.H.; Kuang, J.J.; Zhang, H.Y.; Hu, P. An electrochemical sensor employing β-cyclodextrin chiral cross-linked metal organic framework and graphene oxide for chiral enantiomer recognition. Microchem. J. 2022, 183, 108074. [Google Scholar] [CrossRef]
- Zou, J.; Zhao, G.Q.; Guan, J.F.; Jiang, X.Y.; Yu, J.G. Single-layer graphene oxide-amino-β-cyclodextrin/black phosphorus nanosheet composites for recognition of tyrosine enantiomers. ACS Appl. Nano Mater. 2021, 4, 13329–13338. [Google Scholar] [CrossRef]
- Ye, Q.M.; Guo, L.L.; Wu, D.T.; Yang, B.Z.; Tao, Y.X.; Deng, L.H.; Kong, Y. Covalent functionalization of bovine serum albumin with graphene quantum dots for stereospecific molecular recognition. Anal. Chem. 2019, 91, 11864–11871. [Google Scholar] [CrossRef]
- Tortolini, C.; Gigli, V.; Rizzo, F.; Lenzi, A.; Bizzarri, M.; Angeloni, A.; Antiochia, R. Stereoselective voltammetric biosensor for myo-inositol and D-chiro-inositol recognition. Sensors 2023, 23, 9211. [Google Scholar] [CrossRef]
- Wang, L.J.; Gao, W.L.; Ng, S.; Pumera, M. Chiral protein-covalent organic framework 3D-printed structures as chiral biosensors. Anal. Chem. 2021, 93, 5277–5283. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.F.; Chen, Y.; He, S.; Lian, H.T.; Cao, X.G.; Liu, B. Bovine serum albumin-coated titanium dioxide modified electrochemical interface for enantioselective discrimination of D/L-aspartic acid. Chirality 2021, 33, 731–744. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.M.; Gao, J.X.; Luo, K.; Li, J.P.; Zeng, Y. Protein synergistic action-based development and application of a molecularly imprinted chiral sensor for highly stereoselective recognition of S-fluoxetine. Biosens. Bioelectron. 2023, 223, 115027. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.X.; He, J.H.; Huang, J.W.; Sheng, Y.; Xu, D.F.; Bradley, M.; Zhang, R. Electrochemical recognition of tryptophan enantiomers based on the self-assembly of polyethyleneimine and chiral peptides. J. Electroanal. Chem. 2020, 865, 114130. [Google Scholar] [CrossRef]
- Ye, Q.M.; Hu, J.H.; Wu, D.T.; Yang, B.Z.; Tao, Y.X.; Qin, Y.; Kong, Y. Smart construction of an efficient enantioselective sensing device based on bioactive tripeptide. Anal. Methods 2019, 11, 1951–1957. [Google Scholar] [CrossRef]
- Ye, Q.M.; Yin, Z.Z.; Wu, H.W.; Wu, D.T.; Tao, Y.X.; Kong, Y. Decoration of glutathione with copper-platinum nanoparticles for chirality sensing of tyrosine enantiomers. Electrochem. Commun. 2020, 110, 106638. [Google Scholar] [CrossRef]
- Yao, W.Y.; Li, S.; Xie, L.C.; Jiang, Y. Chiral recognition of tryptophan enantiomer based on the electrode modified by polyaniline adsorption bovine serum albumin complex. Chirality 2023, 35, 129–144. [Google Scholar] [CrossRef]
- Zhu, G.B.; Zhang, D.P.; Ma, Y.Z.; Yi, Y.H. Electrochemical chiral recognition for a complex system based on specific enzymatic reactions. J. Electrochem. Soc. 2020, 167, 027523. [Google Scholar] [CrossRef]
- Liu, H.; Shao, J.J.; Shi, L.X.; Ke, W.; Zheng, F.J.; Zhao, Y. Electroactive NPs and D-amino acids oxidase engineered electrochemical chiral sensor for D-alanine detection. Sens. Actuators B-Chem. 2020, 304, 127333. [Google Scholar] [CrossRef]
- Tian, T.T.; Liu, M.X.; Chen, L.X.; Zhang, F.J.; Yao, X.; Zhao, H.; Li, X.J. D-amino acid electrochemical biosensor based on D-amino acid oxidase: Mechanism and high performance against enantiomer interference. Biosens. Bioelectron. 2020, 151, 111971. [Google Scholar] [CrossRef]
- Muñoz, J.; Redondo, E.; Pumera, M. Chiral 3D-printed bioelectrodes. Adv. Funct. Mater. 2021, 31, 2010608. [Google Scholar] [CrossRef]
- Zhang, L.M.; Luo, K.; Gao, J.X.; Li, J.P. DNA-immobilized special conformation recognition of L-penicillamine using a chiral molecular imprinting technique. Polymers 2022, 14, 4133. [Google Scholar] [CrossRef]
- Zhang, L.M.; Luo, K.; Li, D.; Zhang, Y.F.; Zeng, Y.; Li, J.P. Chiral molecular imprinted sensor for highly selective determination of D-carnitine in enantiomers via dsDNA-assisted conformation immobilization. Anal. Chim. Acta 2020, 1136, 82–90. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Zhou, X.Q.; Hu, Z.H.; Lu, D.C.; Chen, Y.Y.; Feng, L.Y. Stereoselective signal amplification for multiplex detection of tyrosinamide. J. Electroanal. Chem. 2024, 967, 118480. [Google Scholar] [CrossRef]
- Sun, Y.X.; He, J.H.; Zhang, D.D.; Sheng, Y.; Xu, D.F.; Zhang, R.; Bradley, M. Synergistic effects of chitosan and DNA self-assembly films on the chiral discrimination of tryptophan enantiomers. Microchem. J. 2021, 165, 106118. [Google Scholar] [CrossRef]
- Zhai, F.F.; Yu, Q.; Zhou, H.; Liu, J.; Yang, W.R.; You, J.M. Electrochemical selective detection of carnitine enantiomers coupling copper ion dependent DNAzyme with DNA assistant hybridization chain reaction. J. Electroanal. Chem. 2019, 837, 137–142. [Google Scholar] [CrossRef]
- Han, Q.; Mo, F.J.; Wu, J.L.; Wang, C.; Chen, M.; Fu, Y.Z. Engineering DNAzyme cyclic amplification integrated dual-signal chiral sensing system for specific recognition of histidine enantiomers. Sens. Actuators B-Chem. 2020, 302, 127191. [Google Scholar] [CrossRef]
- Peluso, P.; Chankvetadze, B. Recognition in the Domain of Molecular Chirality: From Noncovalent Interactions to Separation of Enantiomers. Chem. Rev. 2022, 122, 13235–13400. [Google Scholar] [CrossRef]
- Ogston, A.G. Interpretation of experiments on metabolic processes, using isotopic tracer elements. Nature 1948, 162, 963. [Google Scholar] [CrossRef]
- Pirkle, W.H.; Pochapsky, T.C. Considerations of chiral recognition relevant to the liquid chromatography separation of enantiomers. Chem. Rev. 1989, 89, 347–362. [Google Scholar] [CrossRef]
- Pirkle, W.H.; Pochapsky, T.C.; Mahler, G.S.; Corey, D.E.; Reno, D.S.; Alessi, D.M. Useful and easily prepared chiral stationary phases for the direct chromatographic separation of the enantiomers of a variety of derivatized amines, amino acids, alcohols, and related compounds. J. Org. Chem. 1986, 51, 4991–5000. [Google Scholar] [CrossRef]
- Davankov, V.A.; Meyer, V.R.; Rais, M. A vivid model illustrating chiral recognition induced by achiral structures. Chirality 1990, 2, 208–210. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhu, X.C.; Jiang, W.; Liu, H.L.; Sun, B.G. Chiral recognition for chromatography and membrane-based separations: Recent developments and future prospects. Molecules 2021, 26, 1145. [Google Scholar] [CrossRef] [PubMed]
- Scriba, G.K.E. Update on chiral recognition mechanisms in separation science. J. Sep. Sci. 2024, 47, 35. [Google Scholar] [CrossRef]
- Takahashi, M.; Takani, D.; Haba, M.; Hosokawa, M. Investigation of the chiral recognition ability of human carboxylesterase 1 using indomethacin esters. Chirality 2020, 32, 73–80. [Google Scholar] [CrossRef]
- Ostovan, A.; Arabi, M.; Wang, Y.Q.; Li, J.H.; Li, B.W.; Wang, X.Y.; Chen, L.X. Greenificated molecularly imprinted materials for advanced applications. Adv. Mater. 2022, 34, 32. [Google Scholar] [CrossRef]
- Li, S.H.; Wu, Y.W.; Ma, X.H.; Pang, C.H.; Wang, M.Y.; Xu, Z.; Li, B. Monitoring levamisole in food and the environment with high selectivity using an electrochemical chiral sensor comprising an MOF and molecularly imprinted polymer. Food Chem. 2024, 430, 137105. [Google Scholar] [CrossRef]
- Yang, Y.; Li, M.X.; Zhu, Z.W. A disposable dual-signal enantioselective electrochemical sensor based on stereogenic porous chiral carbon nanotubes hydrogel. Talanta 2021, 232, 122445. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Liu, Z.X.; Tong, L.; Zhao, L.; Kuang, X.; Kuang, R.; Ju, H.X. One-step electrodeposition of the MOF@CCQDs/NiF electrode for chiral recognition of tyrosine isomers. Dalton Trans. 2020, 49, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chen, R.B.; Jia, L. A colorimetric sensor for chiral recognition and detection of mandelic acid based on the peroxidase-like activity of chiral copper oxide. Microchem. J. 2024, 200, 110187. [Google Scholar] [CrossRef]
- Hoyos-Arbeláez, J.; Vázquez, M.; Contreras-Calderón, J. Electrochemical methods as a tool for determining the antioxidant capacity of food and beverages: A review. Food Chem. 2017, 221, 1371–1381. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.X.; Kou, M.C.; Quan, K.J.; Wang, J.J.; Zhang, H.X.; Ihara, H.; Takafuji, M.; Qiu, H.D. Enantioselective glutamic acid discrimination and nanobiological Imaging by chiral fluorescent silicon nanoparticles. Anal. Chem. 2024, 96, 2173–2182. [Google Scholar] [CrossRef]
- Kelly, B.; Pearce, E.L. Amino assests: How amino acids support Immunity. Cell Metab. 2020, 32, 154–175. [Google Scholar] [CrossRef]
- Liao, X.; Wu, B.Y.; Li, H.X.; Zhang, M.T.; Cai, M.Z.; Lang, B.Z.; Wu, Z.Z.; Wang, F.L.; Sun, J.N.; Zhou, P.P.; et al. Fluorescent/colorimetric dual-mode discriminating Gln and Val enantiomers based on carbon dots. Anal. Chem. 2023, 95, 14573–14581. [Google Scholar] [CrossRef]
- Gou, H.; He, J.X.; Mo, Z.L.; Wei, X.J.; Hu, R.R.; Wang, Y.W.; Guo, R.B. A highly effective electrochemical chiral sensor of tryptophan enantiomers based on covalently functionalize reduced graphene oxide with L-Lysine. J. Electrochem. Soc. 2016, 163, B272–B279. [Google Scholar] [CrossRef]
- Lourenço, A.S.; Nascimento, R.F.; Silva, A.C.; Ribeiro, W.F.; Araujo, M.C.U.; Oliveira, S.C.B.; Nascimento, V.B. Voltammetric determination of tartaric acid in wines by electrocatalytic oxidation on a cobalt(II)-phthalocyanine-modified electrode associated with multiway calibration. Anal. Chim. Acta 2018, 1008, 29–37. [Google Scholar] [CrossRef]
- Heng, H.; Gu, Q.Y.; Jin, H.B.; Shen, P.; Wei, J.X.; Er, X.; Sun, J. Fabrication of a ratiometric fluorescence nanoprobe for detecting tryptophan enantiomers. Talanta 2024, 268, 125291. [Google Scholar] [CrossRef]
- Gu, W.W.; Yang, M.W.; Chen, Z.; Cao, T.; Zhang, Y.M.; Li, Y.F.; Zhang, R.R. New insights into enhanced electrochemical advanced oxidation mechanism of B-doped graphene aerogel: Experiments, molecular dynamics simulations and DFT. J. Hazard. Mater. 2023, 443, 130331. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.L.; Cao, Y.Y.; Diao, D.F. N-doped graphene sheets induced high electrochemical activity in carbon film. Appl. Surf. Sci. 2019, 470, 205–211. [Google Scholar] [CrossRef]
- Zhang, Y.F.; Wang, H.Z.; Yu, H.; Sun, X.X. Chiral fluorescent sensor based on H8-BINOL for the high enantioselective recognition of d- and l-phenylalanine. Rsc Adv. 2022, 12, 11967–11973. [Google Scholar] [CrossRef]
- Xu, H.J.; Guo, J.L.; Zhao, J.J.; Gao, Z.D.; Song, Y.Y. Enantioselective target transport-mediated nanozyme decomposition for the identification of reducing enantiomers in asymmetric nanochannel arrays. Anal. Chem. 2023, 95, 14465–14474. [Google Scholar] [CrossRef]
- Tan, X.P.; Zhang, Y.H.; Mao, H.P.; Yang, J.D. Recognition of chiral propranolol by fluorescent aptamerlight switch based on GO. Spectrochim. Acta Part A-Mol. Biomol. Spectrosc. 2024, 305, 123436. [Google Scholar] [CrossRef]
- Chen, X.X.; Yang, J.; Shen, M.Y.; Chen, Y.; Yu, Q.; Xie, J.H. Structure, function and advance application of microwave-treated polysaccharide: A review. Trends Food Sci. Technol. 2022, 123, 198–209. [Google Scholar] [CrossRef]
- Zagitova, L.; Yarkaeva, Y.; Zagitov, V.; Nazyrov, M.; Gainanova, S.; Maistrenko, V. Voltammetric chiral recognition of naproxen enantiomers by N-tosylproline functionalized chitosan and reduced graphene oxide based sensor. J. Electroanal. Chem. 2022, 922, 116744. [Google Scholar] [CrossRef]
- Bao, L.P.; Dai, J.Y.; Yang, L.; Ma, J.F.; Tao, Y.X.; Deng, L.H.; Kong, Y. Electrochemical recognition of tyrosine enantiomers based on chiral ligand exchange with sodium alginate as the chiral selector. J. Electrochem. Soc. 2015, 162, H486–H491. [Google Scholar] [CrossRef]
- Salehpour, N.; Nojavan, S. Maltodextrin-starch chiral composite membranes: Preparation, enantioselective recognition of phenylalanine and ibuprofen enantiomers, and molecular docking simulation. J. Membr. Sci. 2025, 717, 123660. [Google Scholar] [CrossRef]
- Jafari Sanjari, A.; Asghari, M. A Review on chitosan utilization in membrane synthesis. ChemBioEng Rev. 2016, 3, 134–158. [Google Scholar] [CrossRef]
- Wang, J.R.; Wang, L.H.; Zhao, H.F.; Li, H.X.; Niu, X.H.; Wang, Y.; Wang, K.J. Chiral electrochemical sensing platform constructed by chiral chitosan materials for enhanced electrochemical chiral analysis. Electroanalysis 2025, 37, e202400303. [Google Scholar] [CrossRef]
- Luo, H.; Li, H.; Ge, Q.M.; Cong, H.; Tao, Z.; Liu, M. An electrochemical sensor for enantiorecognition of tyrosine based on a chiral macrocycle functionalized rGO. Microchem. J. 2021, 164, 105949. [Google Scholar] [CrossRef]
- Ye, D.; Rongpipi, S.; Kiemle, S.N.; Barnes, W.J.; Chaves, A.M.; Zhu, C.H.; Norman, V.A.; Liebman-Peláez, A.; Hexemer, A.; Toney, M.F.; et al. Preferred crystallographic orientation of cellulose in plant primary cell walls. Nat. Commun. 2020, 11, 4720. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, H.; Quan, K.; Zhao, L.; Qiu, H.; Li, Z. Preparation and applications of cellulose-functionalized chiral stationary phases: A review. Talanta 2021, 225, 121987. [Google Scholar] [CrossRef] [PubMed]
- Hecht, H.; Srebnik, S. Structural characterization of sodium alginate and calcium alginate. Biomacromolecules 2016, 17, 2160–2167. [Google Scholar] [CrossRef]
- Wang, M.Q.; Nie, Z.W.; He, Y.H.; Vasilakos, A.V.; Cheng, Q.; Ren, Z.X. Deep learning methods for protein representation and function prediction: A comprehensive overview. Eng. Appl. Artif. Intell. 2025, 155, 110977. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, X.; Ji, Y. Monoliths with proteins as chiral selectors for enantiomer separation. Talanta 2012, 91, 7–17. [Google Scholar] [CrossRef]
- Haginaka, J. Enantiomer separation of drugs by capillary electrophoresis using proteins as chiral selectors. J. Chromatogr. A 2000, 875, 235–254. [Google Scholar] [CrossRef]
- Lin, X.; Zhu, S.; Wang, Q.H.; Xia, Q.; Ran, P.Y.; Fu, Y.Z. Chiral recognition of penicillamine enantiomers using hemoglobin and gold nanoparticles functionalized graphite-like carbon nitride nanosheets via electrochemiluminescence. Colloids Surf. B-Biointerfaces 2016, 148, 371–376. [Google Scholar] [CrossRef]
- Zor, E.; Saf, A.O.; Bingol, H.; Ersoz, M. Voltammetric discrimination of mandelic acid enantiomers. Anal. Biochem. 2014, 449, 83–89. [Google Scholar] [CrossRef]
- Richerson, H.B.; Seebohm, P.M. Anaphylactoid reaction to human gamma globulin. Arch. Intern. Med. 1966, 117, 568–572. [Google Scholar] [CrossRef]
- Fu, Y.Z.; Chen, Q.; Zhou, J.; Han, Q.; Wang, Y.H. Enantioselective recognition of mandelic acid based on γ-globulin modified glassy carbon electrode. Anal. Biochem. 2012, 421, 103–107. [Google Scholar] [CrossRef]
- Han, Q.; Wang, Y.H.; Huang, Y.H.; Guo, L.J.; Fu, Y.Z. Reagentless biosensor using for stereospecific interaction between γ-globulin and N-isobutyryl-cysteine. J. Electrochem. Soc. 2013, 160, B102–B106. [Google Scholar] [CrossRef]
- Xu, X.H.; Hu, J.Y.; Xue, H.Q.; Hu, Y.Y.; Liu, Y.N.; Lin, G.Y.; Liu, L.L.; Xu, R.A. Applications of human and bovine serum albumins in biomedical engineering: A review. Int. J. Biol. Macromol. 2023, 253, 126914. [Google Scholar] [CrossRef]
- Lete, I.; Martínez, A.; Lasaga, I.; Centurión, E.; Vesga, A. Update on the combination of myo-inositol/d-chiro-inositol for the treatment of polycystic ovary syndrome. Gynecol. Endocrinol. 2024, 40, 2301554. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, P.D., Jr.; Ellena, J.; Diniz, L.F.; Cardoso, T.F.M.; da Silva, C.C.P. Spontaneous resolution of RS-fluoxetine to a racemic conglomerate upon salt formation with oxalic acid. Cryst. Growth Des. 2022, 22, 5966–5973. [Google Scholar] [CrossRef]
- Misra, R.; Rudnick-Glick, S.; Adler-Abramovich, L. From folding to assembly: Functional supramolecular architectures of peptides comprised of non-canonical amino acids. Macromol. Biosci. 2021, 21, 2100090. [Google Scholar] [CrossRef]
- Li, W.H.; Li, M.H.; Qi, J. Nano-drug design based on the physiological properties of glutathione. Molecules 2021, 26, 5567. [Google Scholar] [CrossRef] [PubMed]
- Robinson, P.K. Enzymes: Principles and biotechnological applications. Essays Biochem. 2015, 59, 1–41. [Google Scholar] [CrossRef]
- Bauer, J.A.; Zámocká, M.; Majtán, J.; Bauerová-Hlinková, V. Glucose oxidase, an enzyme “ferrari”: Its structure, function, production and properties in the light of various industrial and biotechnological applications. Biomolecules 2022, 12, 472. [Google Scholar] [CrossRef]
- Marcone, G.L.; Binda, E.; Rosini, E.; Abbondi, M.; Pollegioni, L. Antibacterial properties of D-Amino acid oxidase: Impact on the food industry. Front. Microbiol. 2019, 10, 2786. [Google Scholar] [CrossRef]
- Cho, N.H.; Shaw, J.E.; Karuranga, S.; Huang, Y.; Fernandes, J.D.D.; Ohlrogge, A.W.; Malanda, B. IDF diabetes atlas: Global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res. Clin. Pract. 2018, 138, 271–281. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, J.H.; Wu, D.T.; Ma, J.F.; Tao, Y.X.; Qin, Y.; Kong, Y. Multi-templates based molecularly imprinted sodium alginate/MnO2 for simultaneous enantiorecognition of lysine, alanine and cysteine isomers. Int. J. Biol. Macromol. 2019, 129, 786–791. [Google Scholar] [CrossRef]
- Duan, J.W.; Wang, X.; Kizer, M.E. Biotechnological and therapeutic applications of natural nucleic acid structural motifs. Top. Curr. Chem. 2020, 378, 26. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Huang, Y.H.; Guo, L.J.; Chen, C.; Guo, D.M.; Chen, Y.; Fu, Y.Z. DNA-based nanocomposite as electrochemical chiral sensing platform for the enantioselective interaction with quinine and quinidine. New J. Chem. 2014, 38, 4600–4606. [Google Scholar] [CrossRef]
- Rehman, S.U.; Sarwar, T.; Husain, M.A.; Ishqi, H.M.; Tabish, M. Studying non-covalent drug-DNA interactions. Arch. Biochem. Biophys. 2015, 576, 49–60. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.; Wu, L.; Ding, L.; Effah, C.Y.; Wu, Y.; Xiong, Y.; He, L. Construction and bioapplications of aptamer-based dual recognition strategy. Biosens. Bioelectron. 2022, 195, 113661. [Google Scholar] [CrossRef] [PubMed]
- Farzin, M.A.; Naghib, S.M. A Cu foil/CuO nanosheets-integrated redox-free electrochemical biosensor based on aptananozymes with improved enzyme-mimetic activity toward non-electroactive L-tyrosinamide substrate. Microchem. J. 2025, 215, 114282. [Google Scholar] [CrossRef]
- Wang, M.; Liu, Z.; Liu, C.; He, W.H.; Qin, D.; You, M.L. DNAzyme-based ultrasensitive immunoassay: Recent advances and emerging trends. Biosens. Bioelectron. 2024, 251, 116122. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.R.; Li, X.L.; Xu, J.J.; Chen, H.Y. Acid-switchable DNAzyme nanodevice for imaging multiple metal ions in living cells. ACS Appl. Mater. Interfaces 2020, 12, 13005–13012. [Google Scholar] [CrossRef] [PubMed]
- McConnell, E.M.; Cozma, I.; Mou, Q.B.; Brennan, J.D.; Lu, Y.; Li, Y.F. Biosensing with DNAzymes. Chem. Soc. Rev. 2021, 50, 8954–8994. [Google Scholar] [CrossRef]
- Bai, R.N.; Ma, Y.C.; Gao, X.Y.; Obaid, E.; Yang, J.; Ouyang, H.; Guo, T.; Fu, Z.F. Chiral cobalt nanocluster-driven chemiluminescent system for the facile discrimination of carnitine enantiomers with a hydrogel-assisted strategy. Anal. Chem. 2024, 96, 20139–20146. [Google Scholar] [CrossRef]
- Zhang, L.J.; Liu, Z.; Xiong, C.; Zheng, L.; Ding, Y.S.; Lu, H.B.; Zhang, G.B.; Qiu, L.Z. Selective recognition of Histidine enantiomers using novel molecularly imprinted organic transistor sensor. Org. Electron. 2018, 61, 254–260. [Google Scholar] [CrossRef]
- Zhang, Q.; Guo, L.J.; Huang, Y.H.; Chen, Y.; Guo, D.M.; Chen, C.; Fu, Y.Z. An electrochemical chiral sensing platform for propranolol enantiomers based on size-controlled gold nanocomposite. Sens. Actuators B-Chem. 2014, 199, 239–246. [Google Scholar] [CrossRef]
- Guo, L.J.; Zhang, Q.; Huang, Y.H.; Han, Q.; Wang, Y.H.; Fu, Y.Z. The application of thionine-graphene nanocomposite in chiral sensing for Tryptophan enantiomers. Bioelectrochemistry 2013, 94, 87–93. [Google Scholar] [CrossRef]
- Wang, J.; Mo, Z.L.; Liu, N.J.; Guo, R.B.; Shuai, C.; Chen, F.; Du, Y.X.; Liu, J.J.; Liu, G.G.; Dong, Q.B.; et al. Construction of electrochemical chiral interface of C3N4/Ppy/ self-assembled polysaccharide. J. Electroanal. Chem. 2021, 886, 115118. [Google Scholar] [CrossRef]
- Yao, W.Y.; Li, S.; Kong, Y.; Xie, L.C.; Jiang, Y. Chiral recognition of phenylglycinamide enantiomer based on electrode modified by silver-ammonia ion-functionalized carbon nanotubes complex. Chemosensors 2023, 11, 86. [Google Scholar] [CrossRef]
- Niu, X.H.; Yang, X.; Mo, Z.L.; Liu, N.J.; Guo, R.B.; Pan, Z.; Liu, Z.Y. Electrochemical chiral sensing of tryptophan enantiomers by using 3D nitrogen-doped reduced graphene oxide and self-assembled polysaccharides. Microchim. Acta 2019, 186, 557. [Google Scholar] [CrossRef]
- Chen, M.T.; Lan, X.Y.; Zhu, L.J.; Ru, P.; Liu, H.Y.; Xu, W.T. Nucleic acid-aided molecular amplification techniques for food microorganism detection. TrAC-Trends Anal. Chem. 2023, 165, 117116. [Google Scholar] [CrossRef]
- Qiu, X.; Ke, J.; Chen, W.B.; Liu, H.X.; Bai, X.P.; Ji, Y.B.; Chen, J.Q. β-Cyclodextrin-ionic liquid functionalized chiral composite membrane for enantioseparation of drugs and molecular simulation. J. Membr. Sci. 2022, 660, 120870. [Google Scholar] [CrossRef]
- Huggins, D.J.; Biggin, P.C.; Dämgen, M.A.; Essex, J.W.; Harris, S.A.; Henchman, R.H.; Khalid, S.; Kuzmanic, A.; Laughton, C.A.; Michel, J.; et al. Biomolecular simulations: From dynamics and mechanisms to computational assays of biological activity. Wiley Interdiscip. Rev.-Comput. Mol. Sci. 2019, 9, e1393. [Google Scholar] [CrossRef]
- Kaptan, S.; Vattulainen, I. Machine learning in the analysis of biomolecular simulations. Adv. Phys. X 2022, 7, 2006080. [Google Scholar] [CrossRef]
- Semenova, E.V.; Belova, E.V.; Sulimov, A.V.; Sulimov, V.B. Molecular docking of chiral drug enantiomers with different bioactivities. Chirality 2024, 36, e23712. [Google Scholar] [CrossRef] [PubMed]

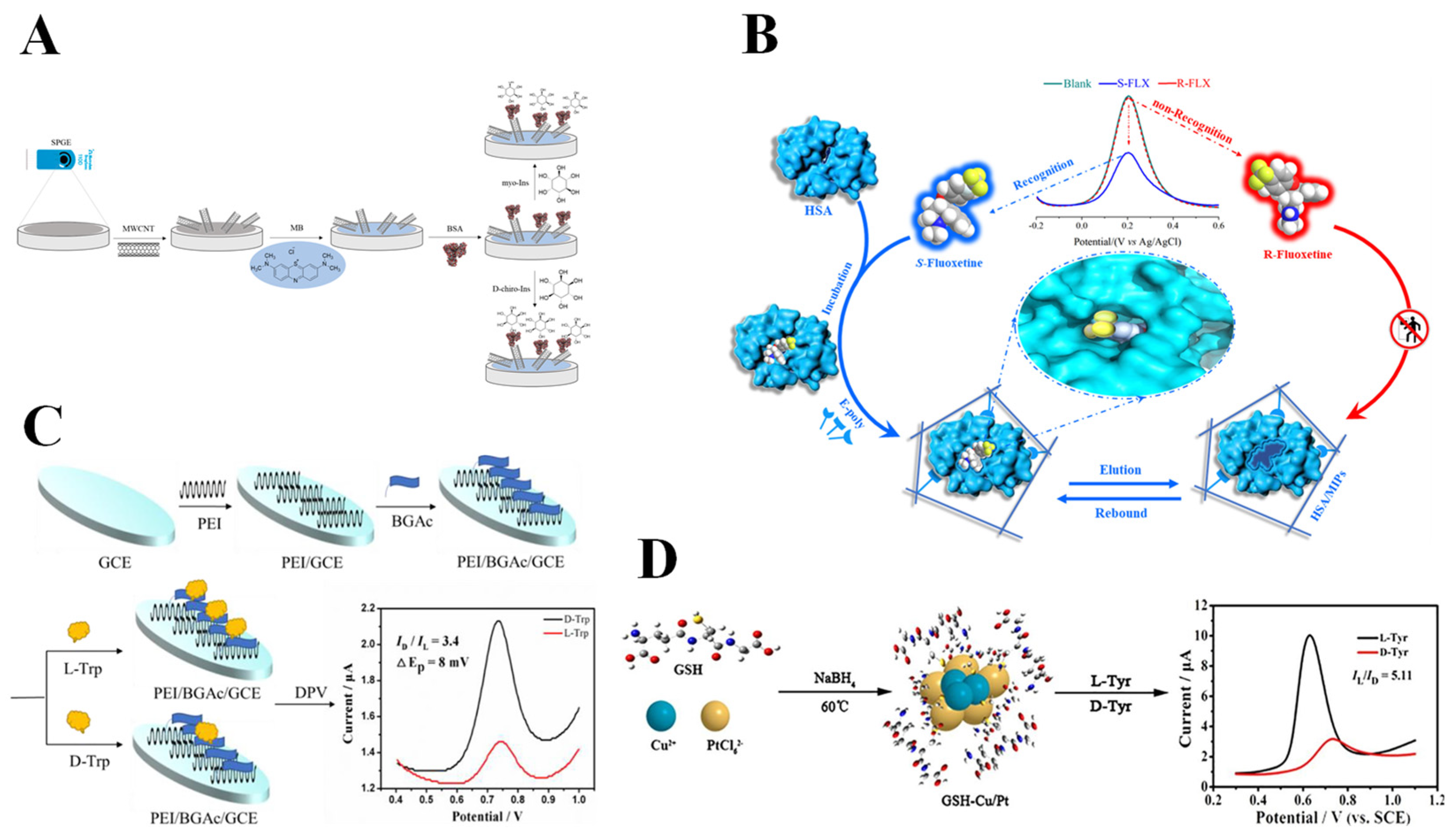
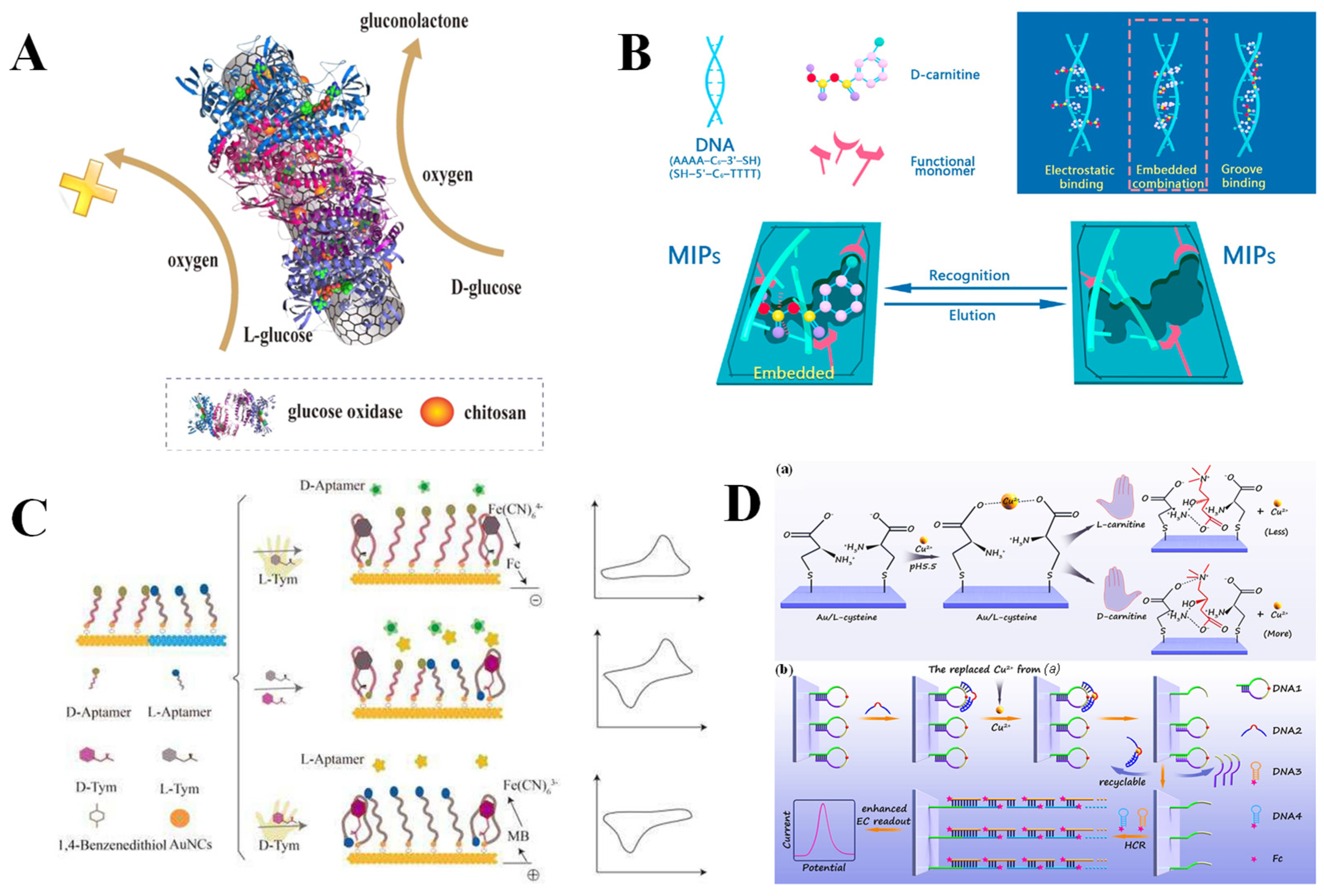
| Biomaterial | Modifier | Method | Mechanism | Analyte | Recognition Ability | LOD | Real Sample | Ref. |
|---|---|---|---|---|---|---|---|---|
| Amino acids and their derivatives | L-GO D-GO | EIS and LSV | Three-point interaction | L-Trp D-Trp | IL/ID = 1.4, ΔEP = 10 mV ID/IL = 1.53, ΔEP = 22 mV | – | – | [34] |
| RGO-Au/L-Glu RGO-Au/D-Glu | DPV | Three-point interaction | L-Trp D-Trp | IL/ID = 2.56 IL/ID = 2.05 | 0.28 mM 0.86 mM | – | [35] | |
| L-NGQDs D-NGQDs | EIS and LSV | Three-point interaction | L-Tart D-Tart | – | – | – | [36] | |
| L-CQDs/TCPP D-CQDs/TCPP | DPV | Three-point interaction | L-Phe D-Phe | IL/ID = 2.3 ID/IL = 2.7 | – | Human serum | [37] | |
| KB/D-His-ZIF-8 | DPV | Three-point interaction | L-Trp D-Trp | ID/IL = 5.69 | 0.51 μM 0.23 μM | Human serum and human urine | [32] | |
| D-His-ZIF-8@Au@ZIF-8 | DPV | Three-point interaction | L-Phe D-Phe | IL/ID = 1.97 | 0.195 mM 0.195 mM 0.588 mM | Human serum | [38] | |
| D-His-ZIF-8@CoFe-PDA | DPV | Three-point interaction | L-Trp D-Trp | IL/ID = 2.01 | 0.066 mM 0.15 mM | Human serum and human urine | [39] | |
| L-PCN-224 D-PCN-224 | DPV | Three-point interaction | L-Trp D-Trp | ID/IL = 2.723 IL/ID = 2.682 | – | – | [40] | |
| MOF-Glu MOF-His MOF-Lys | DPV | Three-point interaction | L-Trp, D-Trp L-Phe, D-Phe L-Glu, D-Glu | IL/ID = 2.69 IL/ID = 2.58 IL/ID = 2.00 | – | – | [41] | |
| h-HDGA@ZIF-67 | CV | Three-point interaction | L-Pen D-Pen | ΔEP = 272 mV | 0.015 μM 0.022 μM | – | [42] | |
| LCMS DCMS | DPV | Molecular imprinting recognition | L-Trp D-Trp | IL/ID = 2.788 ID/IL = 3.186 | 0.012 μM 0.009 μM | Human serum and human urine | [43] | |
| Au@p-L-cysteine | DPV | Chiral ligand exchange recognition | L-Trp D-Trp | ΔID = Iblank − ID > 0 ΔIL = Iblank − IL < 0 | 75 nM | Human serum | [44] | |
| Au-D-Met/CS | DPV | Three-point interaction | L-Trp D-Trp | IL/ID = 1.54 | 42.16 pM 7.75 pM | Nutritional supplement | [45] | |
| L-Cys-AuNPs D-Cys-AuNPs | DPV and EIS | Three-point interaction | R(+)-PRNL S(-)-PRNL | ΔEP = 40 mV | – | – | [33] | |
| L-His-ZIF | EIS | Three-point interaction | L-Glu | – | 0.06 nM | Soy sauce | [46] | |
| Polysaccharides and their derivatives | Cu2-β-CD/NH2-CS-MWCNTs | DPV | Host–guest recognition | L-Trp D-Trp | IL/ID = 1.64 ΔEP = 40 mV | 13.4 μM 18.5 μM | Rat serum | [47] |
| (CS/PAA)n@PEDOT:PSS | DPV | Three-point interaction | L-Trp D-Trp | ID/IL = 6.71 ΔEP = 64 mV | 0.33 μM 0.67 μM | – | [48] | |
| MoS2-IL/CS | DPV | Three-point interaction | L-Trp D-Trp | ΔEP = 53.3 mV | – | – | [49] | |
| RGO/CS | CV and DPV | Three-point interaction | L-Tyr D-Tyr | ID/IL = 1.39 ΔI = 5.0 μA | – | – | [50] | |
| CMC-MIL-88(Fe) | DPV | Three-point interaction | L-Trp D-Trp | IL/ID = 2.04 | – | Human serum and human urine | [51] | |
| rGO-PhenCu-CMC | DPV | Chiral ligand exchange recognition | L-Trp D-Trp | IL/ID = 2.41 | – | – | [52] | |
| rGO-PANI/CD-Cu-CMC | DPV | – | L-Trp D-Trp | IL/ID =3.58 | – | – | [53] | |
| CNT/PANI/SA | DPV | Three-point interaction | L-Trp D-Trp | ID/IL = 2.1 | – | Human serum and human urine | [54] | |
| CS-SA | SWV | Three-point interaction | L-Tyr D-Tyr | IL/ID = 1.63 | 29 nM 107 nM | – | [55] | |
| SA-CS-NGC | DPV | Three-point interaction | L-Trp D-Trp | IL/ID = 4.52 | – | Human serum and human urine | [56] | |
| MWCNTs/CMC-CD-Cu | DPV | Chiral ligand exchange recognition | L-Trp D-Trp | IL/ID = 2.2 | 0.81 μM 1.9 μM | Human urine | [57] | |
| β-CD/CNTs@rGO | DPV | Host–guest recognition | L-Phe D-Phe | – | 0.08 μM | – | [58] | |
| SA/CuNPs/rGO | CV and DPV | Three-point interaction | L-Trp D-Trp | IL/ID = 2.11 ΔEP = 36 mV | 0.205 μM 0.319 μM | Cow milk, goat milk, beef, and millet | [59] | |
| GO-CLMOF | DPV | Host–guest recognition | L-MA D-MA | IL/ID = 1.8 ΔEP = 40 mV | 0.09 mM 0.15 mM | Human urine | [60] | |
| SGO-NH2-βCD/BPNs | SWV | Host–guest recognition | L-Tyr D-Tyr | ID/IL = 1.94 ΔI = 0.89 μA | 1.74 μM 1.02 μM | – | [61] | |
| Proteins | BSA-GQD | DPV | Biological macromolecule recognition | L-Trp D-Trp | IL/ID = 3.67 ΔEP = 96 mV | – | – | [62] |
| BSA/MB/MWCNT | DPV | Biological macromolecule recognition | Myo-Ins D-chiro-Ins | – | 0.5 μM 1 μM | Commercial pharmaceutical preparation | [63] | |
| Fe3O4@COF@BSA | LSV | Biological macromolecule recognition | L-Trp D-Trp | IL/ID = 1.45 ΔEP = 23 mV | – | – | [64] | |
| BSA/TiO2 | EIS | Biological macromolecule recognition | L-Asp D-Asp | ΔR = 5584.3 Ω | 9.39 nM 8.34 nM | – | [65] | |
| MIPs/HSA | DPV | Molecular imprinting recognition | S-FLX | – | 6.43 × 10−17 M | – | [66] | |
| PEI/D-BGAc | DPV | Three-point interaction | L-Trp D-Trp | ID/IL = 3.4 ΔEP = 8 mV | 0.67 μM 0.33 μM | – | [67] | |
| α-CD/GSH | DPV | Three-point interaction | L-Trp D-Trp | IL/ID = 3.88 | – | – | [68] | |
| GSH-Cu/Pt | DPV | Three-point interaction | L-Tyr D-Tyr | IL/ID = 5.11 ΔEP = 104 mV | – | – | [69] | |
| APS-DPAN-BSA | DPV | Biological macromolecule recognition | L-Trp D-Trp | ID/IL = 1.95 | 0.071 mM 0.0478 mM | – | [70] | |
| Enzymes | β-D-GOD@CS-CNTs | CV | Biological macromolecule recognition | L-Glc D-Glc | – | 0.085 mM | Human urine | [71] |
| Fe3O4@Au@Ag@CuxO NPs | DPV | Biological macromolecule recognition | D-Ala | – | 52 pM | Human urine | [72] | |
| DAAO/CNTs | CV | Biological macromolecule recognition | D-Ala | – | 7.91 μM | Milk and human urine | [73] | |
| 3D-nCE | EIS | Biological macromolecule recognition | L-Ala | – | 10 × 10−15 M | - | [74] | |
| Nucleic acids | MIP/dsDNA | DPV | Molecular imprinting recognition | L-Pen | – | 2.48 × 10−16 M | Penicillamine tablets | [75] |
| MIPs/dsDNA | DPV | Molecular imprinting recognition | D-carnitine | – | 2.24 × 10−16 M | Baby milk powder and weight loss capsules | [76] | |
| L-Apt@AuNCs D-Apt@AuNCs | DPV | Biological macromolecule recognition | L-Tym D-Tym | ID/IL = 4.0 IL/ID = 3.0 | – | – | [77] | |
| DNA/CS | DPV | Biological macromolecule recognition | L-Trp D-Trp | ID/IL = 4.02 | 1.67 μM 1.33 μM | – | [78] | |
| DNA3/4/Cu2+/MCH/DNA1/2/Au | DPV | Biological macromolecule recognition | L-carnitine D-carnitine | – | – | – | [79] | |
| MCH/HP/Au/Fe3O4@rGO | DPV | Biological macromolecule recognition | L-His D-His | IL/ID = 2.61 IL/ID = 2.68 | 0.28 pM | Human serum | [80] |
| Biomaterials | Advantages | Disadvantages | Detection Objects | Lowest LOD | Highest Recognition Efficiency |
|---|---|---|---|---|---|
| Amino acids | Clear structures, appropriate molecular sizes, and easy to form complexes | Provides fewer chiral sites and chiral recognition ability is weak | Trp, Tart, Phe, Glu, Pen, and PRNL | 42.16 pM (L-Trp) 7.75 pM (D-Trp) [45] | ID-Trp/IL-Trp = 5.69 [38] |
| Polysaccharides | Good biocompatibility, low toxicity, good hydrophilicity, and easy modification | Easy agglomeration and poor conductivity | Trp, Tyr, Phe, and MA | 29 nM (L-Tyr) 107 nM (D-Tyr) [55] | ID-Trp/IL-Trp = 6.71 [48] |
| Proteins | Rich functional groups, easily accessible, and good solubility | Adsorption occurs on the electrode surface and covers the redox sites | Trp, Inositol, Asp, FLX, and Tyr | 6.43 × 10−17 M (S-FLX) [66] | IL-Tyr/ID-Tyr = 5.11 [69] |
| Enzymes | High specificity, sensitivity, and interference-resistance ability | Easily affected by pH, temperature, and solvent | Glc and Ala | 10 × 10−15 M (L-Ala) [74] | – |
| Nucleic acid | Biocompatibility, controllable self-assembly, and stable chemical properties | Limited recognition ability for natural DNA and uncertainty of design | Pen, carnitine, Tym, Trp, and His | 2.24 × 10−15 M (D-carnitine) [76] | ID-Trp/IL-Trp = 4.02 [78] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiu, S.; Chen, G.-Y.; Qin, Y.-D.; Li, T.-T.; Yang, F.-Q. Recent Advance in Electrochemical Chiral Recognition Based on Biomaterials (2019–2024). Molecules 2025, 30, 3386. https://doi.org/10.3390/molecules30163386
Qiu S, Chen G-Y, Qin Y-D, Li T-T, Yang F-Q. Recent Advance in Electrochemical Chiral Recognition Based on Biomaterials (2019–2024). Molecules. 2025; 30(16):3386. https://doi.org/10.3390/molecules30163386
Chicago/Turabian StyleQiu, Shan, Guo-Ying Chen, Yi-Dan Qin, Ting-Ting Li, and Feng-Qing Yang. 2025. "Recent Advance in Electrochemical Chiral Recognition Based on Biomaterials (2019–2024)" Molecules 30, no. 16: 3386. https://doi.org/10.3390/molecules30163386
APA StyleQiu, S., Chen, G.-Y., Qin, Y.-D., Li, T.-T., & Yang, F.-Q. (2025). Recent Advance in Electrochemical Chiral Recognition Based on Biomaterials (2019–2024). Molecules, 30(16), 3386. https://doi.org/10.3390/molecules30163386










