Theoretical Investigation of the Material Usage During On-Bead Enrichment of Post-Translationally Modified Peptides in Suspension Systems
Abstract
1. Introduction
2. Results
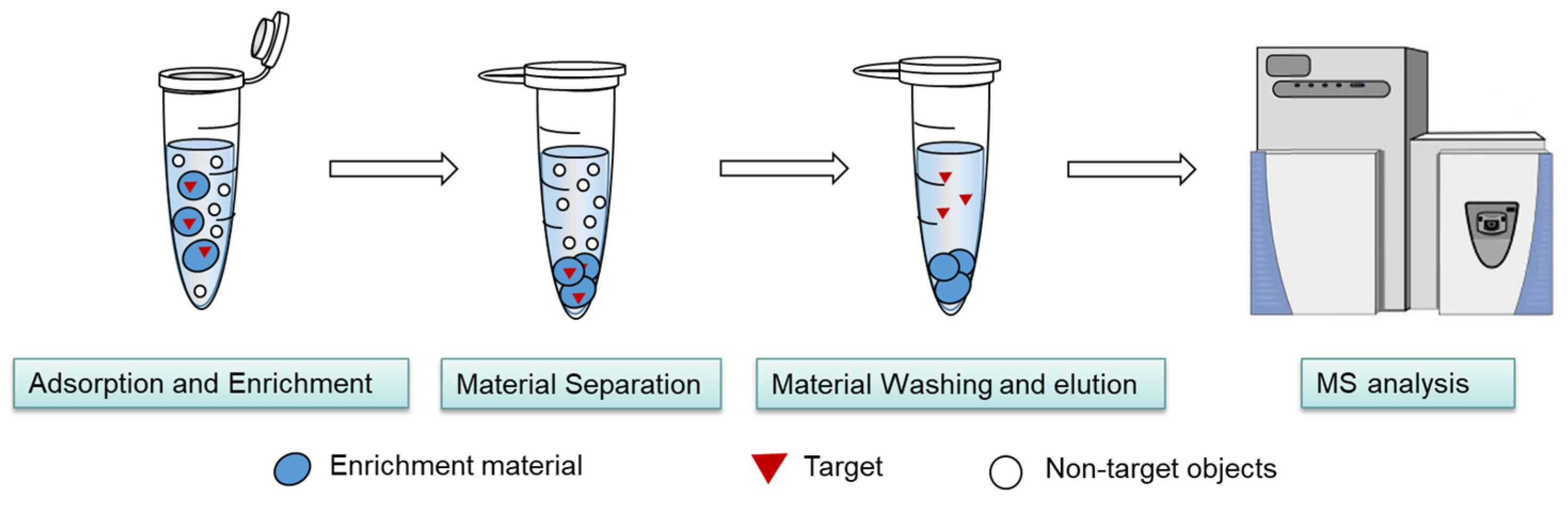
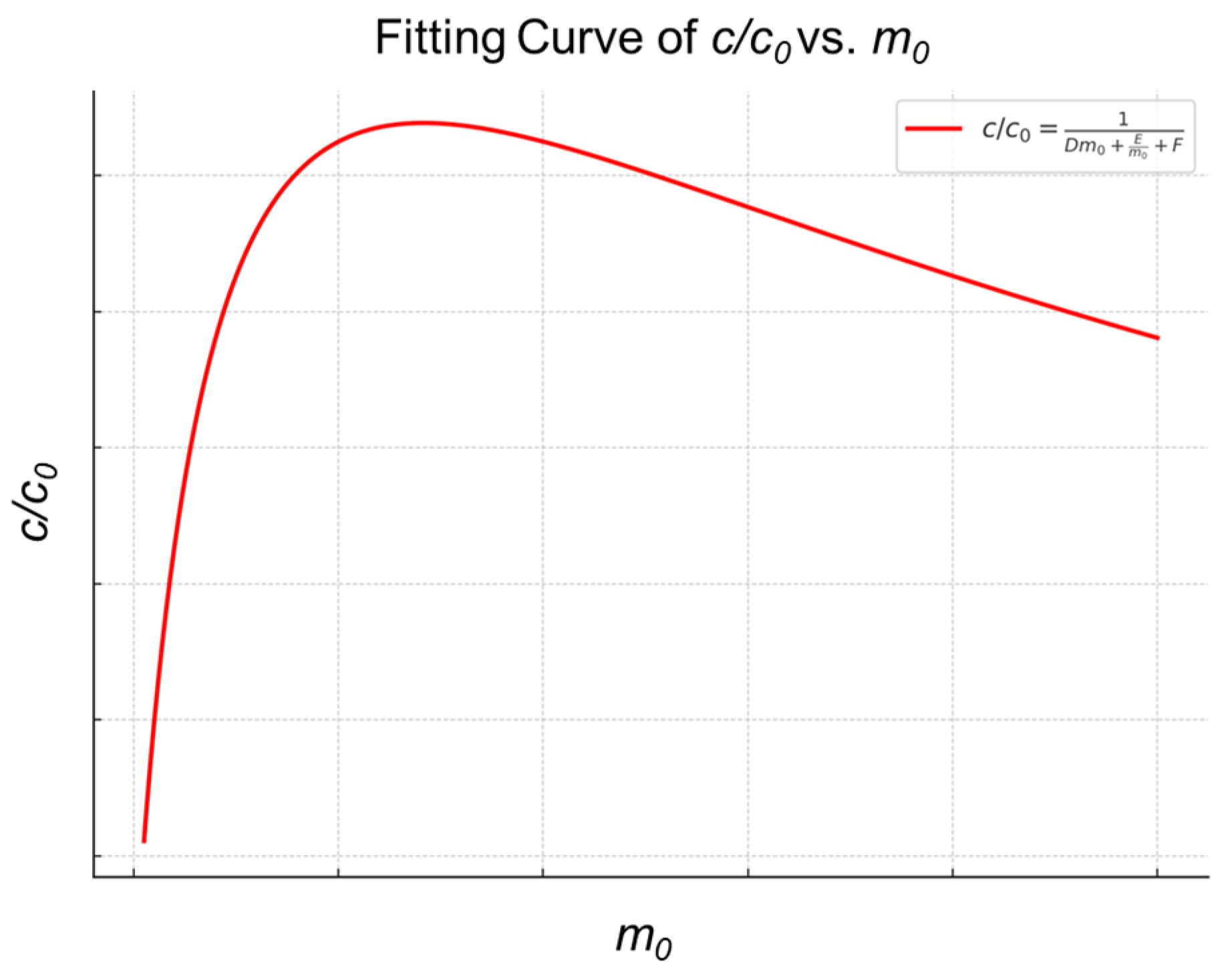
2.1. Thermodynamic Characteristics of the MSPE Separation and Enrichment Process
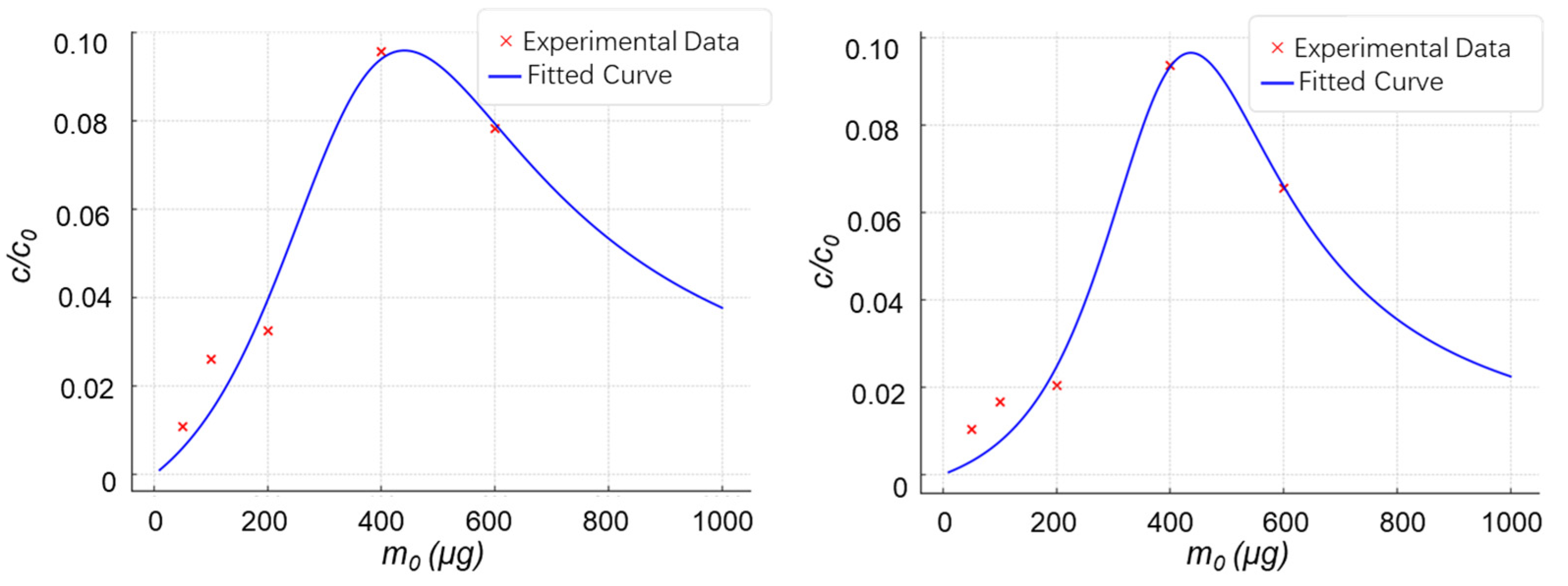
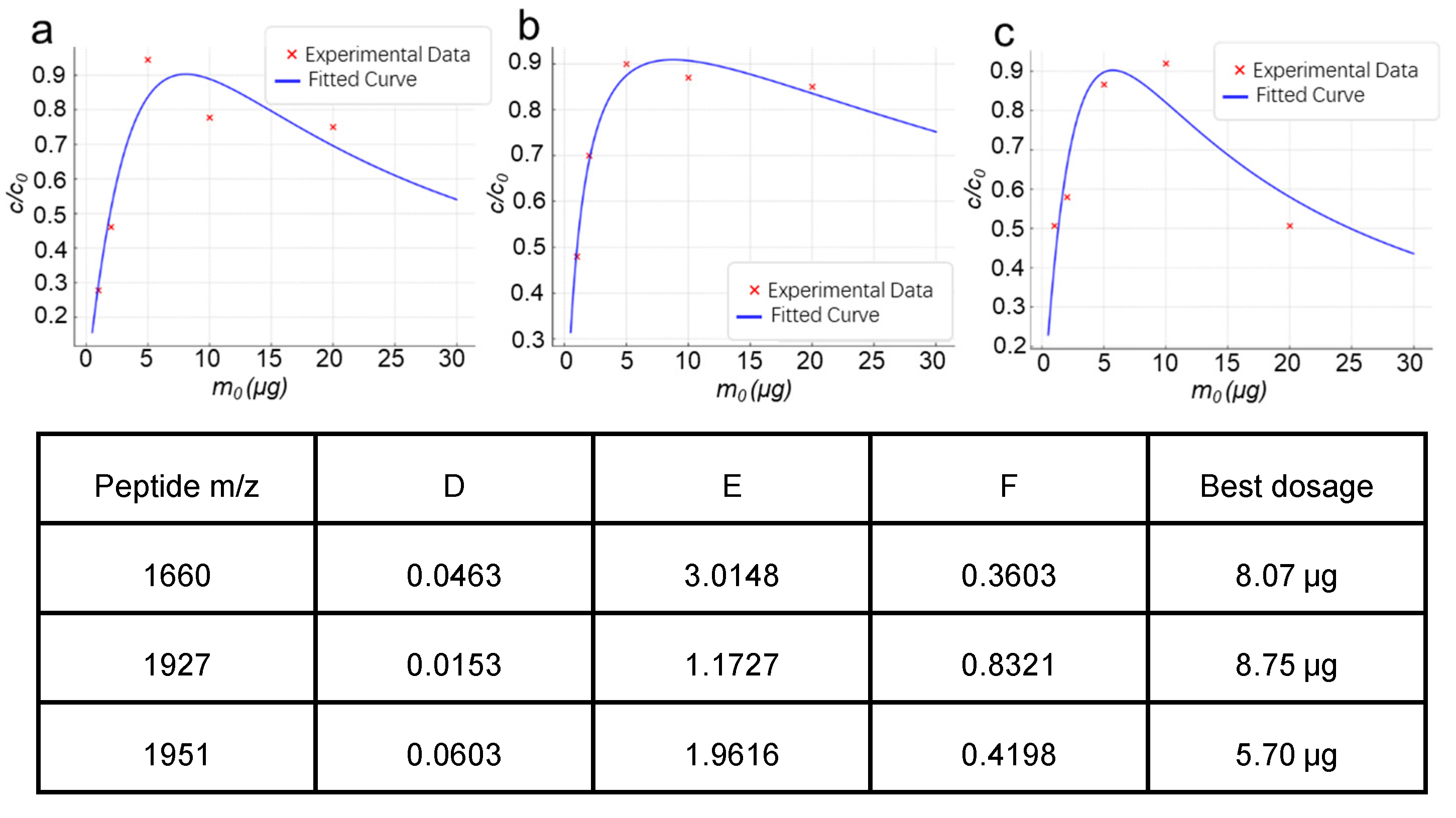
2.2. Experimental Validation of the Material Dosage-Analyte Relationship Trend
2.2.1. Highly Selective Enrichment of N-Glycopeptides Using Multilayered Mesoporous Covalent Organic Polymers (Centrifuge-Based)
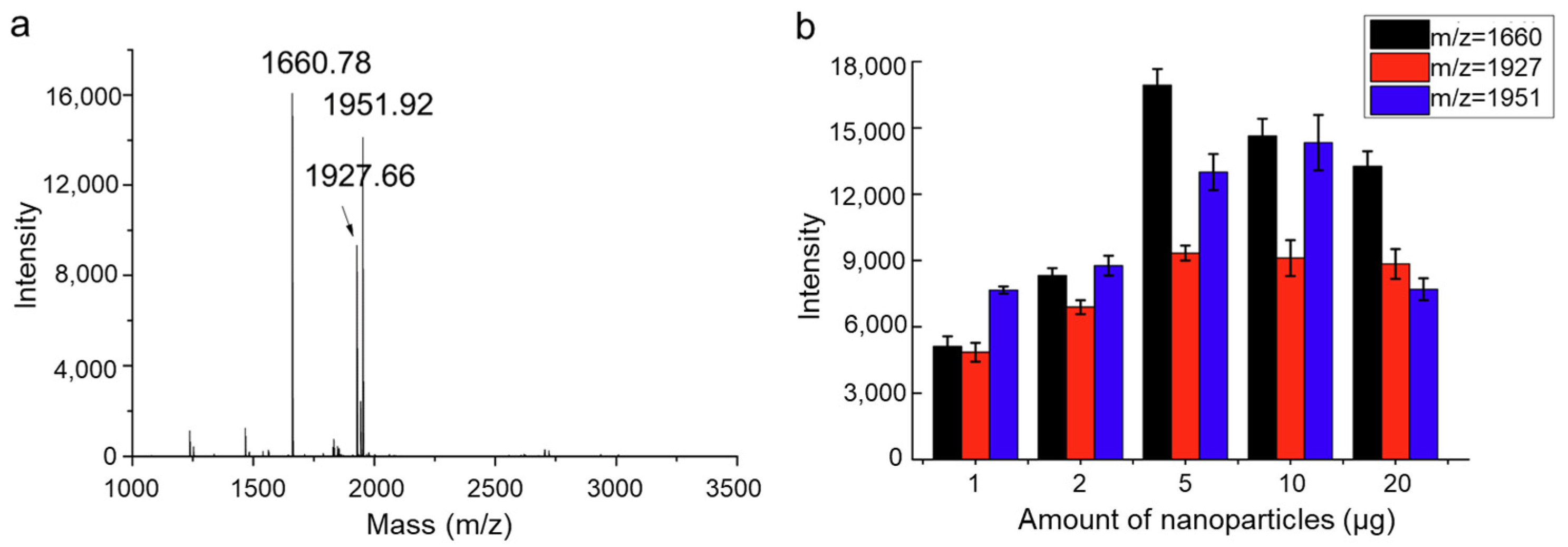
2.2.2. Functionalized Magnetic Covalent Organic Framework Materials for the Selective Enrichment of Phosphopeptides (Magnetic-Based)
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Synthesis of Multilayered p-TpBDH and p-TpBDH-OH Materials
4.3. Synthesis of Magnetic Fe3O4@SiO2@TpPa-Ti4+ Nanoparticles
4.4. Enzymatic Digestion of Standard Proteins
4.5. Enrichment of N-Glycopeptides
4.6. Enrichment of Phosphopeptides
4.7. MALDI-TOF/TOF Mass Spectrometry Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mann, M.; Jensen, O.N. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 2003, 21, 255–261. [Google Scholar] [CrossRef]
- Geffen, Y.; Anand, S.; Akiyama, Y.; Yaron, T.M.; Song, Y.; Johnson, J.L.; Govindan, A.; Babur, O.; Li, Y.; Huntsman, E.; et al. Pan-cancer analysis of post-translational modifications reveals shared patterns of protein regulation. Cell 2023, 186, 3945–3967. [Google Scholar] [CrossRef]
- Huang, K.Y.; Su, M.G.; Kao, H.J.; Hsieh, Y.C.; Jhong, J.H.; Cheng, K.H.; Huang, H.D.; Lee, T.Y. dbPTM 2016: 10-year anniversary of a resource for post-translational modification of proteins. Nucleic Acids Res. 2016, 44, 435–446. [Google Scholar] [CrossRef]
- Lee, J.M.; Hammaren, H.M.; Savitski, M.M.; Baek, S.H. Control of protein stability by post-translational modifications. Nat. Commun. 2023, 14, 201. [Google Scholar] [CrossRef]
- Guccione, E.; Richard, S. The regulation, functions and clinical relevance of arginine methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 642–657. [Google Scholar] [CrossRef] [PubMed]
- Madahar, V.; Dang, R.; Zhang, Q.; Liu, C.; Rodgers, V.G.J.; Liao, J. Human Post-Translational SUMOylation Modification of SARS-CoV-2 Nucleocapsid Protein Enhances Its Interaction Affinity with Itself and Plays a Critical Role in Its Nuclear Translocation. Viruses 2023, 15, 1600. [Google Scholar] [CrossRef] [PubMed]
- Orth, K.; Xu, Z.; Mudgett, M.B.; Bao, Z.Q.; Palmer, L.E.; Bliska, J.B.; Mangel, W.F.; Staskawicz, B.; Dixon, J.E. Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science 2000, 290, 1594–1597. [Google Scholar] [CrossRef] [PubMed]
- Witze, E.S.; Old, W.M.; Resing, K.A.; Ahn, N.G. Mapping protein post-translational modifications with mass spectrometry. Nat. Methods 2007, 4, 798–806. [Google Scholar] [CrossRef]
- Guo, T.; Steen, J.A.; Mann, M. Mass-spectrometry-based proteomics: From single cells to clinical applications. Nature 2025, 638, 901–911. [Google Scholar] [CrossRef]
- Fang, F.; Xu, T.; Chien Hagar, H.T.; Hovde, S.; Kuo, M.H.; Sun, L. Pilot Study for Deciphering Post-Translational Modifications and Proteoforms of Tau Protein by Capillary Electrophoresis-Mass Spectrometry. J. Proteome Res. 2024, 23, 5085–5095. [Google Scholar] [CrossRef]
- Wenger, K.; Viode, A.; Kumar, M.; Steen, H.; Steen, J.A. Quantitative profiling of posttranslational modifications of pathological tau via sarkosyl fractionation and mass spectrometry. Nat. Protoc. 2024, 19, 1235–1251. [Google Scholar] [CrossRef]
- Movassaghi, C.S.; Sun, J.; Jiang, Y.; Turner, N.; Chang, V.; Chung, N.; Chen, R.J.; Browne, E.N.; Lin, C.; Schweppe, D.K.; et al. Recent Advances in Mass Spectrometry-Based Bottom-Up Proteomics. Anal. Chem. 2025, 97, 4728–4749. [Google Scholar] [CrossRef]
- Dens, C.; Adams, C.; Laukens, K.; Bittremieux, W. Machine Learning Strategies to Tackle Data Challenges in Mass Spectrometry-Based Proteomics. J. Am. Soc. Mass. Spectrom. 2024, 35, 2143–2155. [Google Scholar] [CrossRef] [PubMed]
- Geiszler, D.J.; Kong, A.T.; Avtonomov, D.M.; Yu, F.; Leprevost, F.D.V.; Nesvizhskii, A.I. PTM-Shepherd: Analysis and Summarization of Post-Translational and Chemical Modifications From Open Search Results. Mol. Cell. Proteom. 2021, 20. [Google Scholar] [CrossRef] [PubMed]
- Mun, D.G.; Bhat, F.A.; Joshi, N.; Sandoval, L.; Ding, H.; Jain, A.; Peterson, J.A.; Kang, T.; Pujari, G.P.; Tomlinson, J.L.; et al. Diversity of post-translational modifications and cell signaling revealed by single cell and single organelle mass spectrometry. Commun. Biol. 2024, 7, 884. [Google Scholar] [CrossRef] [PubMed]
- Orsburn, B.C. Single cell proteomics by mass spectrometry reveals deep epigenetic insight into the actions of an orphan histone deacetylase inhibitor. bioRxiv 2024. [Google Scholar] [CrossRef]
- Maier, T.; Guell, M.; Serrano, L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009, 583, 3966–3973. [Google Scholar] [CrossRef]
- Ellis, R.J. Most Abundant Protein in the World. Trends. Biochem. Sci. 1979, 4, 241–244. [Google Scholar] [CrossRef]
- Koonin, E.V.; Wolf, Y.I.; Karev, G.P. The structure of the protein universe and genome evolution. Nature 2002, 420, 218–223. [Google Scholar] [CrossRef]
- Csizmok, V.; Forman-Kay, J.D. Complex regulatory mechanisms mediated by the interplay of multiple post-translational modifications. Curr. Opin. Struct. Biol. 2018, 48, 58–67. [Google Scholar] [CrossRef]
- Huang, Q.; Chang, J.; Cheung, M.K.; Nong, W.; Li, L.; Lee, M.T.; Kwan, H.S. Human proteins with target sites of multiple post-translational modification types are more prone to be involved in disease. J. Proteome Res. 2014, 13, 2735–2748. [Google Scholar] [CrossRef] [PubMed]
- Tokmakov, A.A.; Kurotani, A.; Takagi, T.; Toyama, M.; Shirouzu, M.; Fukami, Y.; Yokoyama, S. Multiple post-translational modifications affect heterologous protein synthesis. J. Biol. Chem. 2012, 287, 27106–27116. [Google Scholar] [CrossRef] [PubMed]
- Kanaujia, P.K.; Sharma, Y.K.; Garg, M.O.; Tripathi, D.; Singh, R. Review of analytical strategies in the production and upgrading of bio-oils derived from lignocellulosic biomass. J. Anal. Appl. Pyrolysis 2014, 105, 55–74. [Google Scholar] [CrossRef]
- Lu, Y.; Li, G.S.; Lu, Y.C.; Fan, X.; Wei, X.Y. Analytical Strategies Involved in the Detailed Componential Characterization of Biooil Produced from Lignocellulosic Biomass. Int. J. Anal. Chem. 2017, 1, 9298523. [Google Scholar] [CrossRef]
- Herrero, M.; Ibanez, E.; Cifuentes, A.; Bernal, J. Multidimensional chromatography in food analysis. J. Chromatogr. A 2009, 1216, 7110–7129. [Google Scholar] [CrossRef]
- Le Masle, A.; Angot, D.; Gouin, C.; D’Attoma, A.; Ponthus, J.; Quignard, A.; Heinisch, S. Development of on-line comprehensive two-dimensional liquid chromatography method for the separation of biomass compounds. J. Chromatogr. A. 2014, 1340, 90–98. [Google Scholar] [CrossRef]
- Chen, Z.; Li, Z.; Chen, J.; Kallem, P.; Banat, F.; Qiu, H. Recent advances in selective separation technologies of rare earth elements: A review. J. Environ. Chem. Eng. 2022, 10, 107104. [Google Scholar] [CrossRef]
- Li, J.R.; Kuppler, R.J.; Zhou, H.C. Selective gas adsorption and separation in metal-organic frameworks. Chem. Soc. Rev. 2009, 38, 1477–1504. [Google Scholar] [CrossRef]
- Ritter, J.A.; Ebner, A.D. State of-the Art Adsorption and Membrane Separation Processes for Hydrogen Production in the Chemical and Petrochemical Industries. Sep. Sci. Technol. 2007, 42, 1123–1193. [Google Scholar] [CrossRef]
- Mamanova, L.; Coffey, A.J.; Scott, C.E.; Kozarewa, I.; Turner, E.H.; Kumar, A.; Howard, E.; Shendure, J.; Turner, D.J. Target-enrichment strategies for next-generation sequencing. Nat. Methods. 2010, 7, 111–118. [Google Scholar] [CrossRef]
- Mertes, F.; Elsharawy, A.; Sauer, S.; van Helvoort, J.M.; van der Zaag, P.J.; Franke, A.; Nilsson, M.; Lehrach, H.; Brookes, A.J. Targeted enrichment of genomic DNA regions for next-generation sequencing. Brief. Funct. Genomics. 2011, 10, 374–386. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Chu, Z.; Zhang, Q.; Liu, H.; Zhang, W. Facile synthesis of layered mesoporous covalent organic polymers for highly selective enrichment of N-glycopeptides. Anal. Chim. Acta 2019, 1057, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Zhao, Y.; Liu, H.; Zhang, W. Core-shell magnetic microporous covalent organic framework with functionalized Ti (iv) for selective enrichment of phosphopeptides. Analyst 2020, 145, 4341–4351. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Huang, Y.; Jiang, B.; Hu, Y.; Xie, J.; Gao, X.; Jia, B.; Shen, H.; Zhang, W.; Yang, P. In Situ Synthesis of Magnetic Mesoporous Phenolic Resin for the Selective Enrichment of Glycopeptides. Anal. Chem. 2018, 90, 7357–7363. [Google Scholar] [CrossRef]
- Jiang, H.-L.; Li, N.; Cui, L.; Wang, X.; Zhao, R.-S. Recent application of magnetic solid phase extraction for food safety analysis. TrAC Trends Anal. Chem. 2019, 120, 115632. [Google Scholar] [CrossRef]
- Wierucka, M.; Biziuk, M. Application of magnetic nanoparticles for magnetic solid-phase extraction in preparing biological, environmental and food samples. TrAC Trends Anal. Chem. 2014, 59, 50–58. [Google Scholar] [CrossRef]
- Xu, X.; Deng, C.; Gao, M.; Yu, W.; Yang, P.; Zhang, X. Synthesis of Magnetic Microspheres with Immobilized Metal Ions for Enrichment and Direct Determination of Phosphopeptides by Matrix Assisted Laser Desorption Ionization Mass Spectrometry. Adv. Mater. 2006, 18, 3289–3293. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, K.; Huang, Y.; Huang, T.; Yang, P.; Kong, J.; Shen, H.; Zhang, Q. Theoretical Investigation of the Material Usage During On-Bead Enrichment of Post-Translationally Modified Peptides in Suspension Systems. Molecules 2025, 30, 3245. https://doi.org/10.3390/molecules30153245
Liu K, Huang Y, Huang T, Yang P, Kong J, Shen H, Zhang Q. Theoretical Investigation of the Material Usage During On-Bead Enrichment of Post-Translationally Modified Peptides in Suspension Systems. Molecules. 2025; 30(15):3245. https://doi.org/10.3390/molecules30153245
Chicago/Turabian StyleLiu, Kai, Yuanyu Huang, Thomas Huang, Pengyuan Yang, Jilie Kong, Huali Shen, and Quanqing Zhang. 2025. "Theoretical Investigation of the Material Usage During On-Bead Enrichment of Post-Translationally Modified Peptides in Suspension Systems" Molecules 30, no. 15: 3245. https://doi.org/10.3390/molecules30153245
APA StyleLiu, K., Huang, Y., Huang, T., Yang, P., Kong, J., Shen, H., & Zhang, Q. (2025). Theoretical Investigation of the Material Usage During On-Bead Enrichment of Post-Translationally Modified Peptides in Suspension Systems. Molecules, 30(15), 3245. https://doi.org/10.3390/molecules30153245





