Nitrate–Nitrite Interplay in the Nitrogen Biocycle
Abstract
1. Introduction
2. Redox Chemistry of N-Cycle
3. Oxygen Atom Transfer in the Nitrogen Cycle: Control of Active and Inactive Sites of Enzymes Involved—A General Mechanism
4. Nitrate Reduction and Nitrite Oxidation
4.1. Nitrate Reduction (NO3− → NO2−)
4.1.1. Enzymatic Machinery
4.1.2. Enzymatic Mechanism
4.2. Nitrite Oxidation (NO2− → NO3−)
4.2.1. Enzymatic Machinery
4.2.2. Enzymatic Mechanism
4.3. How Similar Are Nitrate Reductases and Nitrite Oxidoreductases
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Anammox | Anammox: Anaerobic Ammonium Oxidation |
| AO | Aldehyde Oxidase |
| Comammox | Complete Ammonia Oxidation |
| cd1NiR | cytochrome cd1 Nitrite reductase |
| ccNiR: | cytochrome c Nitrite reductase |
| Cu-NiR | Copper-containing Nitrite reductase; |
| C. necator | Cupriavidus necator |
| DNRA | Dissimilatory nitrate reduction to ammonium |
| DMSOR | Dimethyl Sulfoxide Reductase |
| D. desulfuricans | Desulfovibrio desulfuricans |
| E. coli | Escherichia coli |
| FDH | formate dehydrogenase |
| Moco | molybdenum cofactors |
| MGD | molybdopterin guanine dinucleotide |
| N-cycle | Nitrogen cycle |
| NXR | Nitrite oxidoreductase |
| NAP | periplasmic Nitrate reductase |
| NAR | respiratory Nitrate reductase |
| NAS | assimilatory Nitrate reductase |
| NOR | Nitric oxide reductase |
| N2OR | Nitrous oxide reductase |
| NOB | Nitrite-oxidizing bacteria |
| OAT | oxygen atom transfer |
| PDB | protein data bank |
| SO | Sulfite oxidase |
| XO/XD | Xanthine Oxidase/Xanthine Dehydrogenase |
References
- Hutchins, D.A.; Capone, D.G. The Marine Nitrogen Cycle: New Developments and Global Change. Nat. Rev. Microbiol. 2022, 20, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zheng, T.; Li, Y.; Zheng, X. A Critical Review of the Central Role of Microbial Regulation in the Nitrogen Biogeochemical Process: New Insights for Controlling Groundwater Nitrogen Contamination. J. Environ. Manag. 2023, 328, 116959. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ward, B.B.; Sigman, D.M. Global Nitrogen Cycle: Critical Enzymes, Organisms, and Processes for Nitrogen Budgets and Dynamics. Chem. Rev. 2020, 120, 5308–5351. [Google Scholar] [CrossRef] [PubMed]
- Kuypers, M.M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Gruber, N.; Galloway, J.N. An Earth-system perspective of the global nitrogen cycle. Nature 2008, 451, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Maia, L.B.; Moura, J.J.G. How Biology Handles Nitrite. Chem. Rev. 2014, 114, 5273–5357. [Google Scholar] [CrossRef] [PubMed]
- Canfield, D.E.; Glazer, A.N.; Falkowski, P.G. The evolution and future of Earth’s nitrogen cycle. Science 2010, 330, 192–196. [Google Scholar] [CrossRef] [PubMed]
- Moura, I.; Maia, L.; Pauleta, S.R.; Moura, J.J.G. A bird’s-eye view of denitrification in relation to the nitrogen cycle. In Metalloenzymes in Denitrification: Applications and Environmental Impacts; Moura, I., Moura, J.J.G., Pauleta, S.R., Maia, L., Eds.; RSC Metallobiology Series No. 9; Royal Society of Chemistry: Cambridge, UK, 2017; pp. 1–10. [Google Scholar]
- Lehnert, N.; Musselman, B.W.; Seefeldt, L.C. Grand challenges in the nitrogen cycle. Chem. Soc. Rev. 2021, 50, 3640–3646. [Google Scholar] [CrossRef] [PubMed]
- Fowler, D.; Coyle, M.; Skiba, U.; Sutton, M.A.; Cape, J.N.; Reis, S.; Sheppard, L.J.; Jenkins, A.; Grizzetti, B.; Galloway, J.N.; et al. The global nitrogen cycle in the twenty-first century. Trans. R. Soc. B 2013, 368, 20130164. [Google Scholar] [CrossRef] [PubMed]
- Lehnert, N.; Dong, H.T.; Harland, J.B.; Hunt, A.P.; White, C.J. Reversing nitrogen fixation. Nat. Rev. Chem. 2018, 2, 278. [Google Scholar] [CrossRef]
- Shimizu, R.; Dempo, Y.; Nakayama, Y.; Nakamura, S.; Bamba, T.; Fukusaki, E.; Fukui, T. New Insight into the Role of the Calvin Cycle: Reutilization of CO2 Emitted through Sugar Degradation. Sci. Rep. 2015, 5, 11617. [Google Scholar] [CrossRef] [PubMed]
- Kroneck, P.M.H. Nature’s nitrite-to-ammonia expressway, with no stop at dinitrogen. J. Biol. Inorg. Chem. 2022, 27, 1–21. [Google Scholar] [CrossRef] [PubMed]
- van Kessel, M.A.; Speth, D.R.; Albertsen, M.; Nielsen, P.H.; Op den Camp, H.J.; Kartal, B.; Jetten, M.S.; Lücker, S. Complete nitrification by a single microorganism. Nature 2015, 528, 555–559. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Cruz, S.; Stultiens, K.; van Kessel, M.A.H.J.; Versantvoort, W.; Jetten, M.S.M.; Op den Camp, H.J.M.; Kartal, B. Key physiology of a nitrite-dependent methane-oxidizing enrichment culture. Appl. Environ. Microbiol. 2019, 85, e00124-19. [Google Scholar] [CrossRef] [PubMed]
- Versantvoort, W.; Guerrero-Castillo, S.; Wessels, H.J.C.T.; van Niftrik, L.; Jetten, M.S.M.; Brandt, U.; Reimann, J.; Kartal, B. Complexome analysis of the nitrite-dependent methanotroph Methylomirabilis lanthanidiphila. Biochim. Biophys. Acta Bioenerg. 2019, 1860, 734–744. [Google Scholar] [CrossRef] [PubMed]
- Kartal, B.; Maalcke, W.J.; de Almeida, N.M.; Cirpus, I.; Gloerich, J.; Geerts, W.; Op den Camp, H.J.; Harhangi, H.R.; Janssen-Megens, E.M.; Francoijs, K.J.; et al. Molecular mechanism of anaerobic ammonium oxidation. Nature 2011, 479, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Kartal, B.; Kuenen, J.G.; van Loosdrecht, M.C.M. Sewage treatment with anammox. Science 2010, 328, 702–703. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.; Zhang, L.; Zipfel, S.; Topitsch, A.; Lutz, M.; Eckert, J.; Prasser, B.; Chami, M.; Lü, W.; Du, J.; et al. Molecular interplay of an assembly machinery for nitrous oxide reductase. Nature 2022, 608, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xie, P.; Huang, Y.; Gao, H. Complex Interplay of Heme-Copper Oxidases with Nitrite and Nitric Oxide. Int. J. Mol. Sci. 2022, 23, 979. [Google Scholar] [CrossRef] [PubMed]
- Maia, L.B. Bringing Nitric Oxide to the Molybdenum World-A Personal Perspective. Molecules 2023, 28, 5819. [Google Scholar] [CrossRef] [PubMed]
- Pauleta, S.R.; Carepo, M.S.P.; Moura, I. Source and reduction of nitrous oxide. Coord. Chem. Rev. 2019, 387, 436–449. [Google Scholar] [CrossRef]
- Sparacino-Watkins, C.; Stolz, J.F.; Basu, P. Nitrate and periplasmic nitrate reductases. Chem. Soc. Rev. 2014, 43, 676–706. [Google Scholar] [CrossRef] [PubMed]
- Maia, L.B.; Pereira, V.; Mira, L.; Moura, J.J.G. Nitrite reductase activity of rat and human xanthine oxidase, xanthine dehydrogenase, and aldehyde oxidase: Evaluation of their contribution to NO formation in vivo. Biochemistry 2015, 54, 685–710. [Google Scholar] [CrossRef] [PubMed]
- Hille, R.; Hall, J.; Basu, P. The Mononuclear Molybdenum Enzymes. Chem. Rev. 2014, 114, 3963–4038. [Google Scholar] [CrossRef] [PubMed]
- Maia, L.; Moura, J.J.G. Chapter 1: Molybdenum and tungsten-containing enzymes: An overview. In Molybdenum and Tungsten Enzymes; Hille, R., Schulzke, C., Kirk, M.L., Eds.; Royal Society of Chemistry: Cambridge, UK, 2017; pp. 1–80. [Google Scholar]
- Cantu-Medellin, N.; Kelley, E.E. Xanthine oxidoreductase-catalyzed reduction of nitrite to nitric oxide: Insights regarding where, when and how. Nitric Oxide 2013, 34, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Bender, D.; Schwarz, G. Nitrite-dependent nitric oxide synthesis by molybdenum enzymes. FEBS Lett. 2018, 592, 2126–2139. [Google Scholar] [CrossRef] [PubMed]
- Omar, S.A.; Webb, A.J. Nitrite reduction and cardiovascular protection. J. Mol. Cell. Cardiol. 2014, 73, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Castiglione, N.; Rinaldo, S.; Giardina, G.; Stelitano, V.; Cutruzzolà, F. Nitrite and nitrite reductases: From molecular mechanisms to significance in human health and disease. Antioxid Redox Signal. 2012, 17, 684–716. [Google Scholar] [CrossRef] [PubMed]
- Fraústo da Silva, J.J.R.; Williams, R.J.P. The Biological Chemistry of the Elements, The Inorganic Chemistry of Life; Oxford University Press: Oxford, UK, 2001. [Google Scholar]
- Stein, L.Y.; Klotz, M.G. The nitrogen cycle. Curr. Biol. 2016, 26, R94–R98. [Google Scholar] [CrossRef] [PubMed]
- Maiti, B.K.; Maia, L.B.; Moura, J.J.G. Sulfide and transition metals-A partnership for life. J. Inorg. Biochem. 2021, 227, 111687. [Google Scholar] [CrossRef] [PubMed]
- Koch, H.; Lücker, S.; Albertsen, M.; Kitzinger, K.; Herbold, C.; Spieck, E.; Nielsen, P.H.; Wagner, M.; Daims, H. Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc. Natl. Acad. Sci. USA 2015, 112, 11371–11376. [Google Scholar] [CrossRef] [PubMed]
- Frost, A.A. Oxidation Potential-Free Energy Diagrams. J. Am. Chem. Soc. 1951, 73, 2680–2682. [Google Scholar] [CrossRef]
- Mou, T.; Long, J.; Frauenheim, T.; Xiao, J. Advances in Electrochemical Ammonia Synthesis Beyond the Use of Nitrogen Gas as a Source. ChemPlusChem 2021, 86, 1211–1224. [Google Scholar] [CrossRef] [PubMed]
- Latimer, W.M. The Oxidation States of the Elements and Their Potentials in Aqueous Solutions, 1st ed.; Prentice-Hall: New York, NY, USA, 1938; pp. 1–352. [Google Scholar]
- Dutton, K.G.; Lipke, M.C. Correcting Frost Diagram Misconceptions Using Interactive Frost Diagrams. J. Chem. Educ. 2021, 98, 2578–2583. [Google Scholar] [CrossRef]
- Hawley, A.K.; Nobu, M.K.; Wright, J.J.; Durno, W.E.; Morgan-Lang, C.; Sage, B.; Schwientek, P.; Swan, B.K.; Rinke, C.; Torres-Beltrán, M.; et al. Diverse Marinimicrobia bacteria may mediate coupled biogeochemical cycles along eco-thermodynamic gradients. Nat. Commun. 2017, 8, 1507. [Google Scholar] [CrossRef] [PubMed]
- Hawley, A.K.; Brewer, H.M.; Norbeck, A.D.; Paša-Tolić, L.; Hallam, S.J. Metaproteomics reveals differential modes of metabolic coupling among ubiquitous oxygen minimum zone microbes. Proc. Natl. Acad. Sci. USA 2014, 111, 11395–11400. [Google Scholar] [CrossRef] [PubMed]
- Oren, A. Anammox revisited: Thermodynamic considerations in early studies of the microbial nitrogen cycle. FEMS Microbiol. Lett. 2015, 362, fnv114. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Wessels, H.J.; van Alen, T.; Jetten, M.S.; Kartal, B. Nitric oxide-dependent anaerobic ammonium oxidation. Nat. Commun. 2019, 10, 1244. [Google Scholar] [CrossRef] [PubMed]
- Broda, E. Two kinds of lithotrophs missing in nature. Z. Allg. Mikrobiol. 1977, 17, 491–493. [Google Scholar] [CrossRef] [PubMed]
- Mulder, A.; Graaf, A.A.; Robertson, L.A.; Kuenen, J.G. Anaerobic ammonium oxidation discovered in a denitrifying fluidized bed reactor. FEMS Microbiol. Ecol. 1995, 16, 177–184. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, M.; Ran, X.; Wang, W.; Wang, H. A powerful but frequently overlooked role of thermodynamics in environmental microbiology: Inspirations from anammox. Appl. Environ. Microbiol. 2025, 91, e01668-24. [Google Scholar] [CrossRef] [PubMed]
- Giri, N.C.; Wedasingha, L.; Manicke, N.; Szaleniec, M.; Basu, P. Reversible Interconversion of Nitrate and Nitrite Catalyzed by Periplasmic Nitrate Reductase from Campylobacter jejuni. J. Am. Chem. Soc. 2025, 147, 13243–13250. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hodak, M.; Bernholc, J. Enzymatic Mechanism of Copper-Containing Nitrite Reductase. Biochemistry 2015, 54, 1233–1242. [Google Scholar] [CrossRef] [PubMed]
- Rinaldo, S.; Cutruzzolà, F. Nitrite Reductases in Denitrification. In Biology of the Nitrogen Cycle; Elsevier: Amsterdam, The Netherlands, 2007; pp. 37–55. [Google Scholar]
- Averill, B.A. Dissimilatory Nitrite and Nitric Oxide Reductases. Chem. Rev. 1996, 96, 2951–2964. [Google Scholar] [CrossRef] [PubMed]
- Caranto, J.D.; Weitz, A.; Hendrich, M.P.; Kurtz, D.M. The Nitric Oxide Reductase Mechanism of a Flavo-Diiron Protein: Identification of Active-Site Intermediates and Products. J. Am. Chem. Soc. 2014, 136, 7981–7992. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira, N.M.F.S.A.; Fernandes, P.A.; Gonzalez, P.J.; Moura, J.J.G.; Ramos, M.J. The sulfur shift: An activation mechanism for periplasmic nitrate reductase and formate dehydrogenase. Inorg. Chem. 2013, 52, 10766–10772. [Google Scholar] [CrossRef] [PubMed]
- Dias, J.M.; Bursakov, S.; Carneiro, C.; Moura, J.J.; Moura, I.; Romão, M.J. Crystallization and preliminary X-ray analysis of a nitrate reductase from Desulfovibrio desulfuricans ATCC 27774. Acta Crystallogr. D Biol. Crystallogr. 1999, 55, 877–879. [Google Scholar] [CrossRef] [PubMed]
- Coelho, C.; Gonzalez, P.J.; Moura, J.G.; Moura, I.; Trincão, J.; Romão, M.J. The crystal structure of Cupriavidus necator nitrate reductase in oxidized and partially reduced states. J. Mol. Biol. 2011, 408, 932–948. [Google Scholar] [CrossRef] [PubMed]
- Kukimoto, M.; Nishiyama, M.; Murphy, M.E.P.; Turley, S.; Adman, E.T.; Horinouchi, S.; Beppu, T. X-ray Structure and Site-Directed Mutagenesis of a Nitrite Reductase from Alcaligenes Faecalis S-6: Roles of Two Copper Atoms in Nitrite Reduction. Biochemistry 1994, 33, 5246–5252. [Google Scholar] [CrossRef] [PubMed]
- Horrell, S.; Kekilli, D.; Strange, R.W.; Hough, M.A. Recent structural insights into the function of copper nitrite reductases. Metallomics 2017, 9, 1470–1482. [Google Scholar] [CrossRef] [PubMed]
- Albertsson, I.; Sjöholm, J.; Beek, J.T.; Watmough, N.J.; Widengren, J.; Ädelroth, P. Functional interactions between nitrite reductase and nitric oxide reductase from Paracoccus denitrificans. Sci. Rep. 2019, 9, 17234. [Google Scholar] [CrossRef] [PubMed]
- Alfaro, V.S.; Campeciño, J.; Tracy, M.; Elliott, S.J.; Hegg, E.L.; Lehnert, N. Elucidating Electron Storage and Distribution within the Pentaheme Scaffold of Cytochrome c Nitrite Reductase (NrfA). Biochemistry 2021, 60, 1853–1867. [Google Scholar] [CrossRef] [PubMed]
- Siegbahn, P.E.M. The Mechanism of Nitrite Reductase. J. Comput. Chem. 2025, 46, e70088. [Google Scholar] [CrossRef] [PubMed]
- Moura, I.; Pauleta, S.R.; Moura, J.J.G. Enzymatic activity mastered by altering metal coordination spheres. J. Biol. Inorg. Chem. 2008, 13, 1185–1195. [Google Scholar] [CrossRef] [PubMed]
- Mendel, R.R.; Bittner, F. Cell biology of molybdenum. Biochim. Biophys. Acta 2006, 1763, 621–635. [Google Scholar] [CrossRef] [PubMed]
- Hille, R.; Nishino, T.; Bittner, F. Molybdenum enzymes in higher organisms. Coord. Chem. Rev. 2011, 255, 1179–1205. [Google Scholar] [CrossRef] [PubMed]
- Hille, R. Molybdenum enzymes containing the pyranopterin cofactor: An overview. Met. Ions Biol. Syst. 2002, 39, 187–226. [Google Scholar] [PubMed]
- Schwarz, G.; Mendel, R.R.; Ribbe, M.W. Molybdenum cofactors, enzymes and pathways. Nature 2009, 460, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Maiti, B.K. Cross-talk Between (Hydrogen) Sulfite and Metalloproteins: Impact on Human Health. Chem. Eur. J. 2022, 28, e202104342. [Google Scholar] [CrossRef] [PubMed]
- Maiti, B.K.; Maia, L.B.; Silveira, C.M.; Todorovic, S.; Carreira, C.; Carepo, M.S.P.; Grazina, R.; Moura, I.; Pauleta, S.R.; Moura, J.J.G. Incorporation of molybdenum in rubredoxin: Models for mononuclear molybdenum enzymes. JBIC J. Biol. Inorg. Chem. 2015, 20, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Magalon, A.; Ceccaldi, P.; Schoepp-Cothenet, B. The prokaryotic Mo/W-bisPGD family. In Molybdenum and Tungsten Enzymes; Hille, R., Schulzke, C., Kirk, M.L., Eds.; Royal Society of Chemistry: Cambridge, UK, 2017; pp. 143–191. [Google Scholar]
- Wells, M.; Kim, M.; Akob Denise, M.; Basu, P.; Stolz John, F. Impact of the Dimethyl Sulfoxide Reductase Superfamily on the Evolution of Biogeochemical Cycles. Microbiol. Spectr. 2023, 11, e04145-22. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, F.V.; Schulz, L.; Zarzycki, J.; Prinz, S.; Oehlmann, N.N.; Erb, T.J.; Rebelein, J.G. Structural insights into the iron nitrogenase complex. Nat. Struct. Mol. Biol. 2024, 31, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Sippel, D.; Einsle, O. The structure of vanadium nitrogenase reveals an unusual bridging ligand. Nat. Chem. Biol. 2017, 13, 956–960. [Google Scholar] [CrossRef] [PubMed]
- Trncik, C.; Detemple, F.; Einsle, O. Iron-only Fe-nitrogenase underscores common catalytic principles in biological nitrogen fixation. Nat. Catal. 2023, 6, 415–424. [Google Scholar] [CrossRef]
- Besson, S.; Almeida, M.G.; Silveira, C.M. Nitrite reduction in bacteria: A comprehensive view of nitrite reductases. Coord. Chem. Rev. 2022, 464, 214560. [Google Scholar] [CrossRef]
- Bertero, M.G.; Rothery, R.A.; Palak, M.; Hou, C.; Lim, D.; Blasco, F.; Weiner, J.H.; Strynadka, N.C.J. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat. Struct. Mol. Biol. 2003, 10, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Chicano, T.M.; Dietrich, L.; de Almeida, N.M.; Akram, M.; Hartmann, E.; Leidreiter, F.; Leopoldus, D.; Mueller, M.; Sánchez, R.; Nuijten, G.H.L.; et al. Structural and functional characterization of the intracellular filament-forming nitrite oxidoreductase multiprotein complex. Nat. Microbiol. 2021, 6, 1129–1139. [Google Scholar] [CrossRef] [PubMed]
- Sundermeyer-Klinger, H.; Meyer, W.; Warninghoff, B.; Bock, E. Membrane-bound nitrite oxidoreductase of Nitrobacter: Evidence for a nitrate reductase system. Arch. Microbiol. 1984, 140, 153–158. [Google Scholar] [CrossRef]
- Meincke, M.; Bock, E.; Kastrau, D.; Kroneck, P.M.H. Nitrite oxidoreductase from Nitrobacter hamburgensis: Redox centers and their catalytic role. Arch. Microbiol. 1992, 158, 127–131. [Google Scholar] [CrossRef]
- Su, Z.; Liu, T.; Guo, J.; Zheng, M. Nitrite Oxidation in Wastewater Treatment: Microbial Adaptation and Suppression Challenges. Environ. Sci. Technol. 2023, 57, 12557–12570. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Jia, C.; Huang, S.; Yu, K.; Luo, A.; Peng, Y. Nitrite oxidation in oxygen-deficient conditions during landfill leachate treatment. Environ. Res. 2022, 214, 114090. [Google Scholar] [CrossRef] [PubMed]
- Moura, J.J.; Brondino, C.D.; Trincão, J.; Romão, M.J. Mo and W bis-MGD enzymes: Nitrate reductases and formate dehydrogenases. J. Biol. Inorg. Chem. 2004, 9, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, P.J.; Rivas, M.G.; Mota, C.; Brondino, C.D.; Moura, I.; Moura, J.J.G. Periplasmic nitrate reductases and formate dehydrogenases: Biological control of the chemical properties of Mo and W for fine tuning of reactivity, substrate specificity, and metabolic role. Coord. Chem. Rev. 2013, 257, 315–331. [Google Scholar] [CrossRef]
- Pinho, D.; Besson, S.; Silva, P.J.; de Castro, B.; Moura, I. Isolation and Spectroscopic Characterization of the Membrane-Bound Nitrate Reductase from Pseudomonas chlororaphis DSM 50135. Biochim. Biophys. Acta 2005, 1723, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Najmudin, S.; González, P.J.; Trincão, J.; Coelho, C.; Mukhopadhyay, A.; Cerqueira, N.M.F.S.A.; Romão, C.C.; Moura, I.; Moura, J.J.G.; Brondino, C.D.; et al. Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum. J. Biol. Inorg. Chem. 2008, 13, 737–753. [Google Scholar] [CrossRef] [PubMed]
- Asamoto, C.K.; Rempfert, K.R.; Luu, V.H.; Younkin, A.D.; Kopf, S.H. Enzyme-Specific Coupling of Oxygen and Nitrogen Isotope Fractionation of the Nap and Nar Nitrate Reductases. Environ. Sci. Technol. 2021, 55, 5537–5546. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, S.; Schoepp-Cothenet, B.; Ceccaldi, P.; Guigliarelli, B.; Magalon, A. The prokaryotic Mo/W bis-PGD enzymes family: A catalytic workhorse in bioenergetics. Biochim. Biophys. Acta. 2013, 1827, 1048–1085. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.; Barbier, G.G.; Hecht, H.J.; Mendel, R.R.; Campbell, W.H.; Schwarz, G. Structural basis of eukaryotic nitrate reduction: Crystal structures of the nitrate reductase active site. Plant Cell. 2005, 17, 1167–1179. [Google Scholar] [CrossRef] [PubMed]
- Stolz, J.F.; Basu, P. Evolution of nitrate reductase: Molecular and structural variations on a common function. ChemBioChem. 2002, 3, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Correia, C.; Besson, S.; Brondino, C.D.; Gonzalez, P.J.; Fauque, G.; Lampreia, J.; Moura, I.; Moura, J.J.G. Biochemical and spectroscopic characterization of the membrane-bound nitrate reductase from Marinobacter hydrocarbonoclasticus 617. J. Biol. Inorg. Chem. 2018, 13, 1321–1333. [Google Scholar] [CrossRef] [PubMed]
- Jormakka, M.; Richardson, D.; Byrne, B.; Iwata, S. Architecture of NarGH reveals a structural classification of Mo-bisMGD enzymes. Structure 2004, 12, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Dias, J.M.; Than, M.E.; Humm, A.; Huber, R.; Bourenkov, G.P.; Bartunik, H.D.; Bursakov, S.; Calvete, J.; Caldeira, J.; Carneiro, C.; et al. Crystal structure of the first dissimilatory nitrate reductase at 1.9 Å solved by MAD methods. Structure 1999, 7, 65. [Google Scholar] [PubMed]
- Kumar, H.; Khosraneh, M.; Bandaru, S.S.M.; Schulzke, C.; Leimkühler, S. The Mechanism of Metal-Containing Formate Dehydrogenases Revisited: The Formation of Bicarbonate as Product Intermediate Provides Evidence for an Oxygen Atom Transfer Mechanism. Molecules 2023, 28, 1537. [Google Scholar] [CrossRef] [PubMed]
- Coelho, C.; Romão, M.J. Structural and mechanistic insights on nitrate reductases. Protein Sci. 2015, 24, 1901–1911. [Google Scholar] [CrossRef] [PubMed]
- Jepson, B.J.; Mohan, S.; Clarke, T.A.; Gates, A.J.; Cole, J.A.; Butler, C.S.; Butt, J.N.; Hemmings, A.M.; Richardson, D.J. Spectropotentiometric and structural analysis of the periplasmic nitrate reductase from Escherichia coli. J. Biol. Chem. 2007, 282, 6425–6437. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Mintmier, B.; Kc, K.; Metzger, M.C.; Radhakrishnan, M.; McGarry, J.; Wilcoxen, J.; Basu, P.; Kirk, M.L. Active Site Characterization of a Campylobacter jejuni Nitrate Reductase Variant Provides Insight into the Enzyme Mechanism. Inorg. Chem. 2024, 63, 13191–13196. [Google Scholar] [CrossRef] [PubMed]
- McGarry, J.; Mintmier, B.; Metzger, M.C.; Giri, N.C.; Britt, N.; Basu, P.; Wilcoxen, J. Insights into periplasmic nitrate reductase function under single turnover. J. Biol. Inorg. Chem. 2024, 29, 811–819. [Google Scholar] [CrossRef] [PubMed]
- Rendon, J.; Biaso, F.; Ceccaldi, P.; Toci, R.; Seduk, F.; Magalon, A.; Guigliarelli, B.; Grimaldi, S. Elucidating the structures of the low- and high-pH Mo(V) species in respiratory nitrate reductase: A combined EPR, 14,15N HYSCORE, and DFT study. Inorg. Chem. 2017, 56, 4422–4434. [Google Scholar] [CrossRef] [PubMed]
- Marangon, J.; Paes de Sousa, P.M.; Moura, I.; Brondino, C.D.; Moura, J.J.G. The effect of the sixth sulfur ligand in the catalytic mechanism of periplasmic nitrate reductase. J. Comput. Chem. 2009, 30, 2466–2484. [Google Scholar] [CrossRef] [PubMed]
- Sousa, S.F.; Cerqueira, N.M.F.S.A.; Bras, N.F.; Fernandes, P.A.; Ramos, M.J. Enzymatic “tricks”: Carboxylate shift and sulfur shift. Int. J. Quantum Chem. 2014, 114, 1253–1256. [Google Scholar] [CrossRef]
- Lücker, S.; Wagner, M.; Maixner, F.; Pelletier, E.; Koch, H.; Vacherie, B.; Rattei, T.; Damsté, J.S.; Spieck, E.; Paslier, D.L.; et al. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl. Acad. Sci. USA 2010, 107, 13479–13484. [Google Scholar] [CrossRef]
- Hemp, J.; Lücker, S.; Schott, J.; Pace, L.A.; Johnson, J.E.; Schink, B.; Daims, H.; Fischer, W.W. Genomics of a phototrophic nitrite oxidizer: Insights into the evolution of photosynthesis and nitrification. ISME J. 2016, 10, 2669–2678. [Google Scholar] [CrossRef] [PubMed]
- Daims, H.; Lucker, S.; Wagner, M.A. new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends. Microbiol. 2016, 24, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, K.M.; Caranto, J.D.; Majer, S.H.; Smith, M.A. Alternative Bioenergy: Updates to and Challenges in Nitrification Metalloenzymology. Joule 2018, 2, 421–441. [Google Scholar] [CrossRef]
- Spieck, E.; Muller, S.; Engel, A.; Mandelkow, E.; Patel, H.; Bock, E. Two-dimensional structure of membrane-bound nitrite oxidoreductase from Nitrobacter hamburgensis. J. Struct. Biol. 1996, 117, 117–123. [Google Scholar] [CrossRef]
- de Almeida, N.M.; Neumann, S.; Mesman, R.J.; Ferousi, C.; Keltjens, J.T.; Jetten, M.S.M.; Kartal, B.; van Niftrik, L. Immunogold localization of key metabolic enzymes in the anammoxosome and on the tubule-like structures of Kuenenia stuttgartiensis. J. Bacteriol. 2015, 197, 2432–2441. [Google Scholar] [CrossRef] [PubMed]
- Kartal, B.; Keltjens, J.T. Anammox Biochemistry: A Tale of Heme c Proteins. Trends Biochem. Sci. 2016, 41, 998–1011. [Google Scholar] [CrossRef] [PubMed]
- Park, C.K.; Horton, N.C. Structures, functions, and mechanisms of filament forming enzymes: A renaissance of enzyme filamentation. Biophys. Rev. 2019, 11, 927–994. [Google Scholar] [CrossRef] [PubMed]
- Park, C.K.; Horton, N.C. Novel insights into filament-forming enzymes. Nat. Rev. Mol. Cell Biol. 2020, 21, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Kartal, B.; Kuypers, M.M.M.; Lavik, G.; Schalk, J.; den Camp, H.J.O.; Jetten, M.S.M.; Strous, M. Anammox bacteria disguised as denitrifiers: Nitrate reduction to dinitrogen gas via nitrite and ammonium. Environ. Microbiol. 2007, 9, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Giri, N.C.; Mintmier, B.; Radhakrishnan, M.; Mielke, J.W.; Wilcoxen, J.; Basu, P. The critical role of a conserved lysine residue in periplasmic nitrate reductase catalyzed reactions. J. Biol. Inorg. Chem. 2024, 29, 395–405. [Google Scholar] [CrossRef] [PubMed]
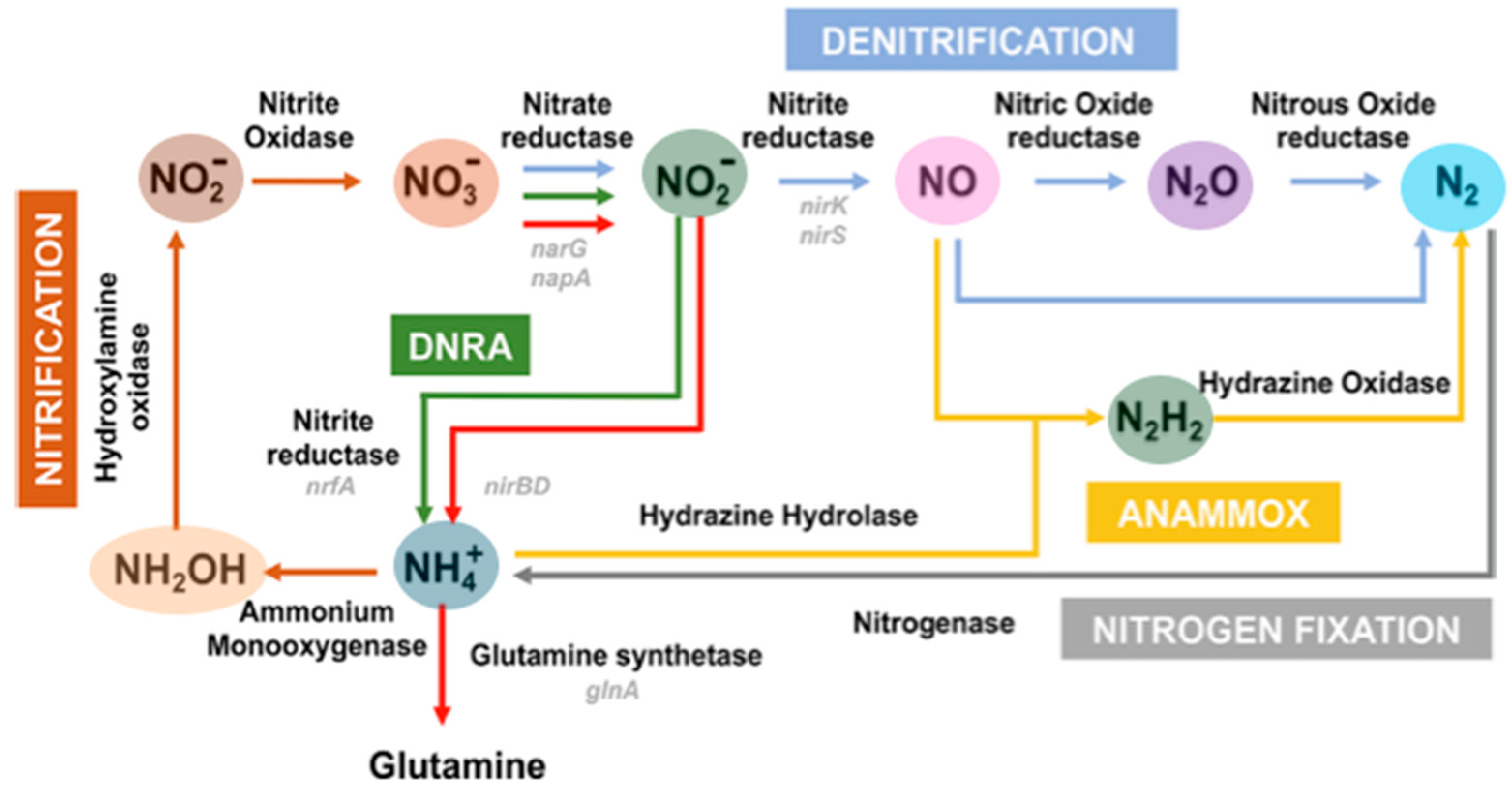
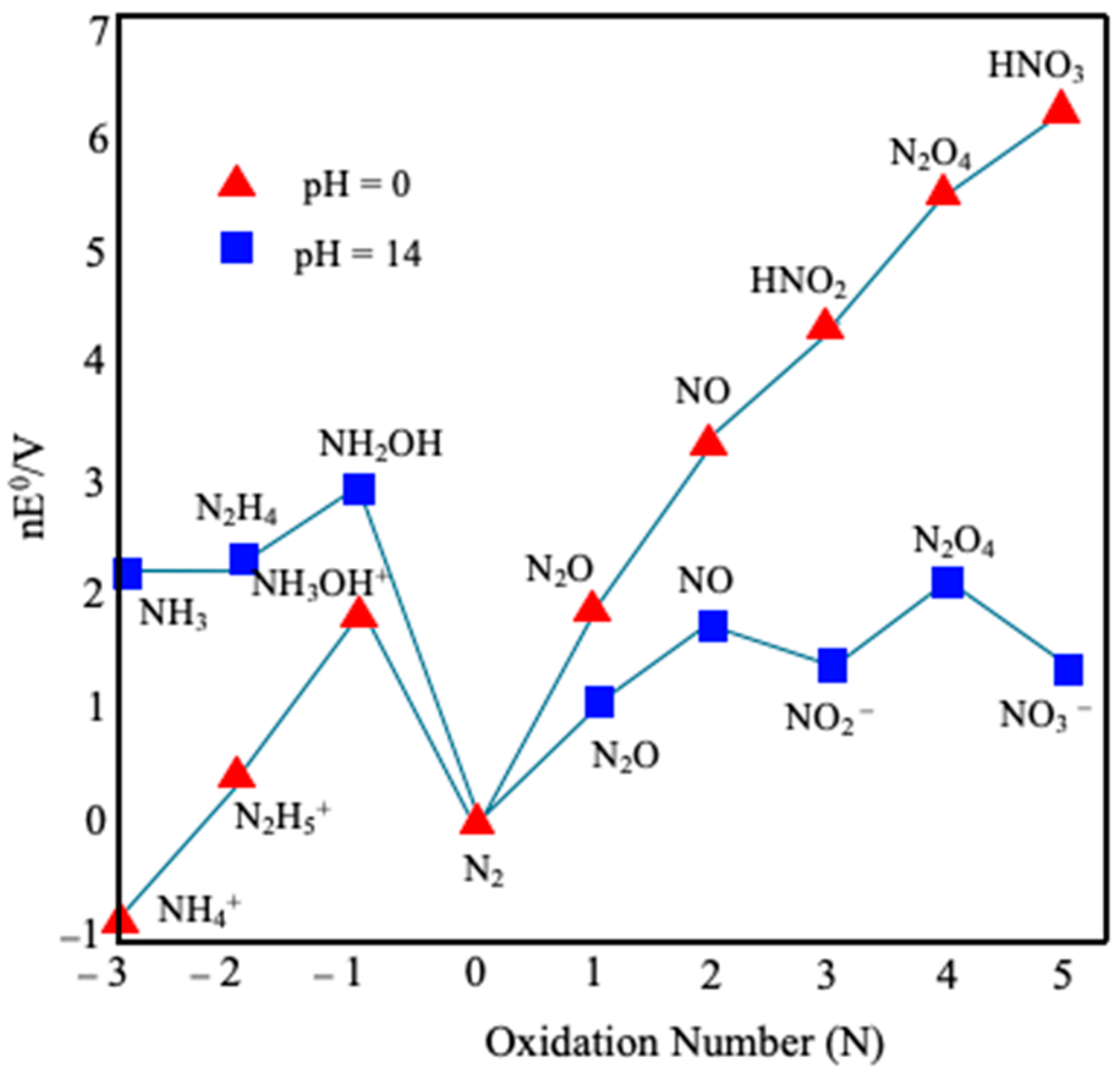
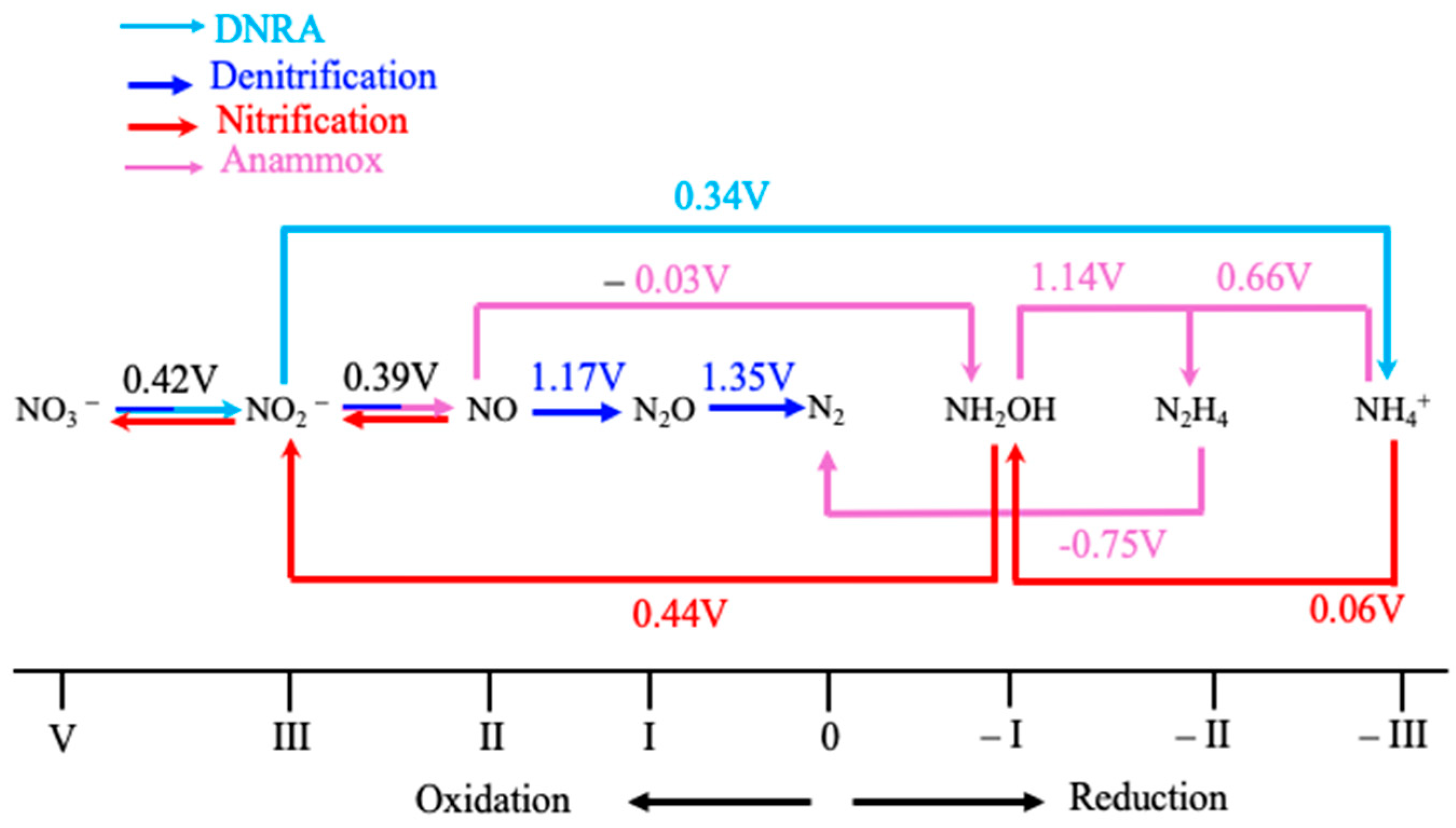
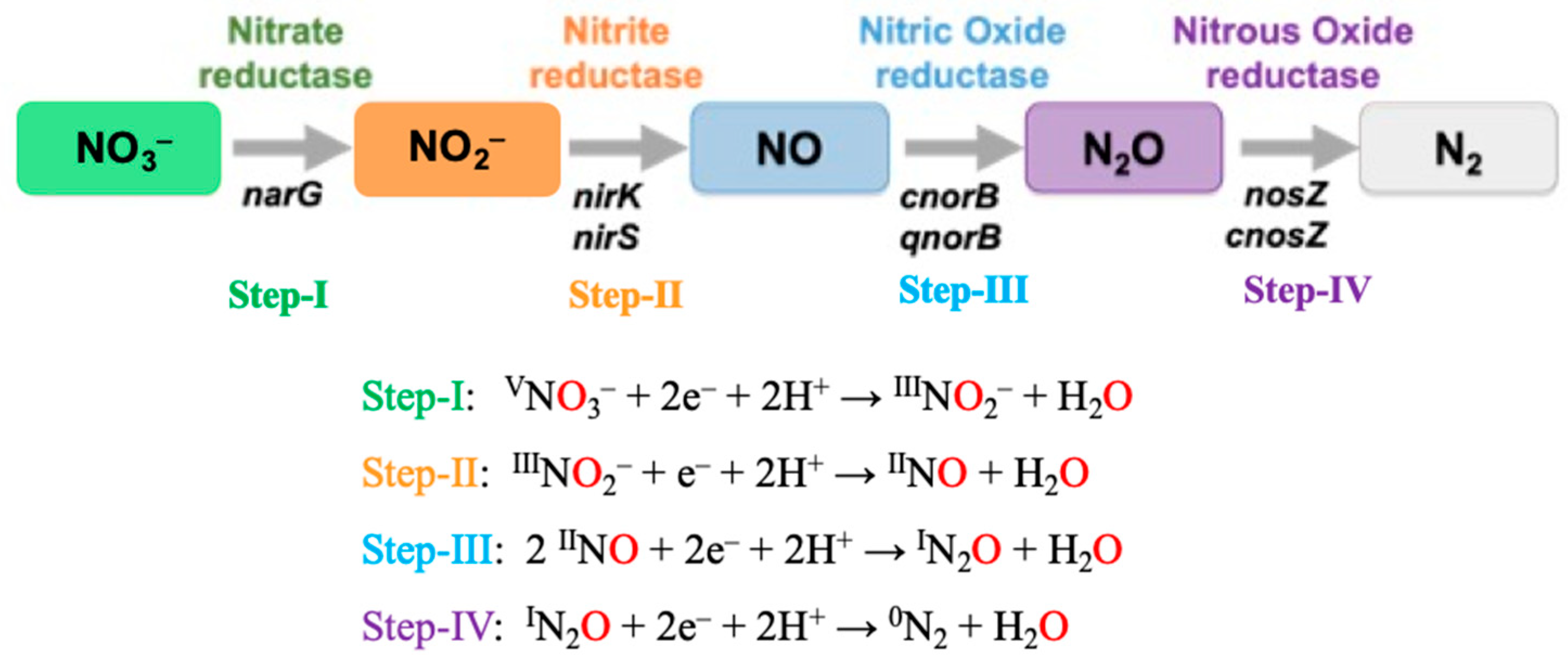
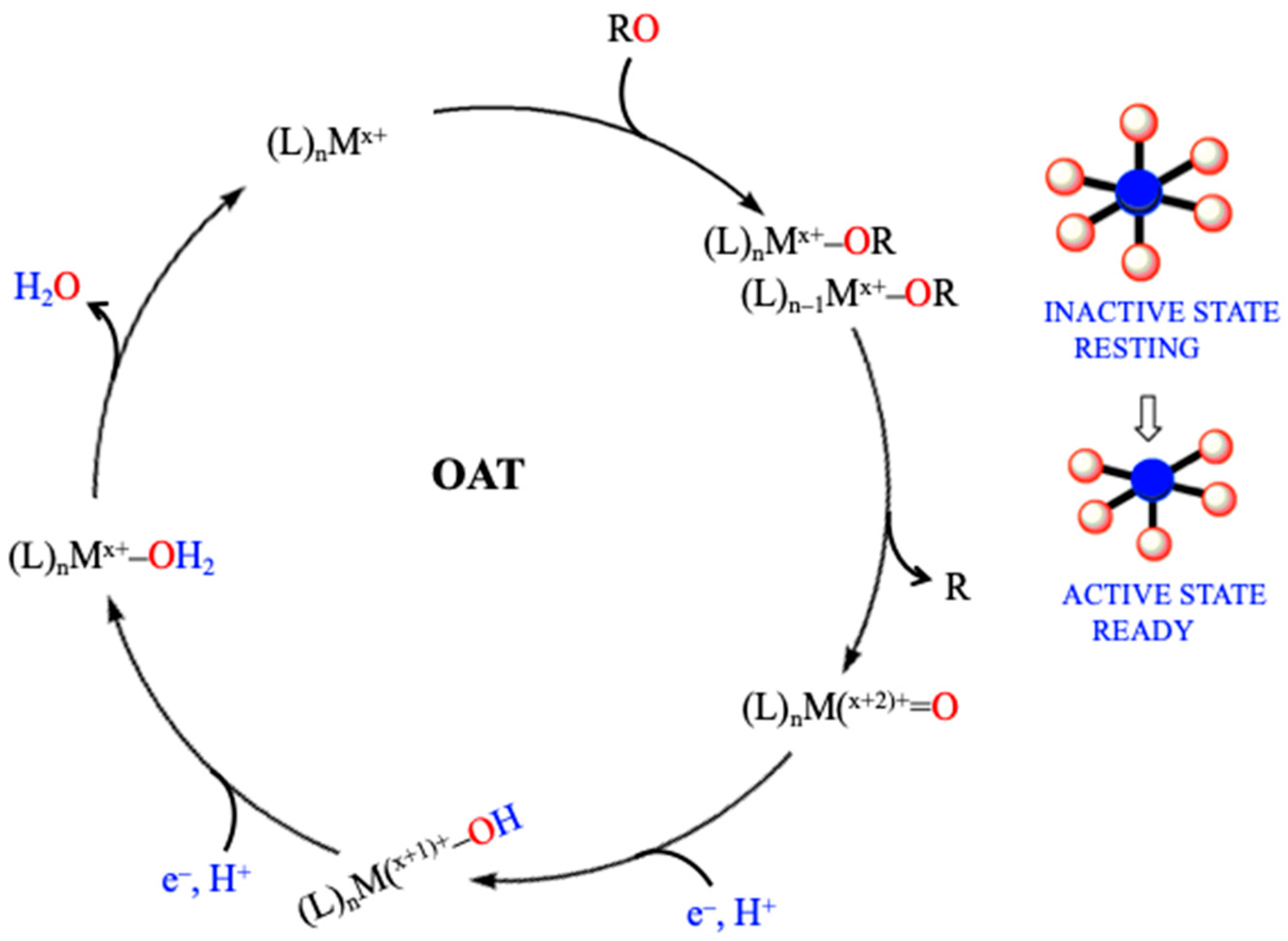
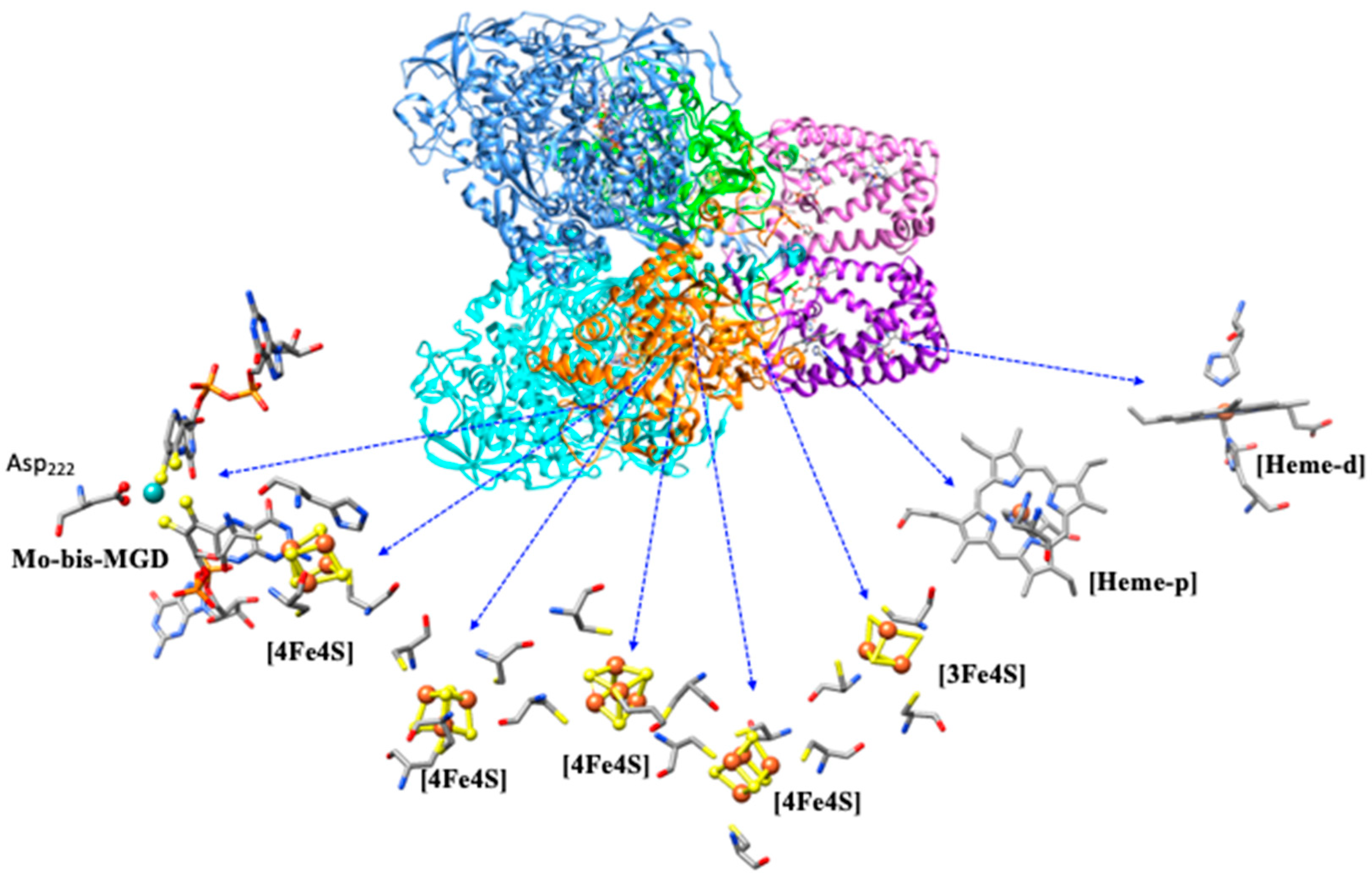



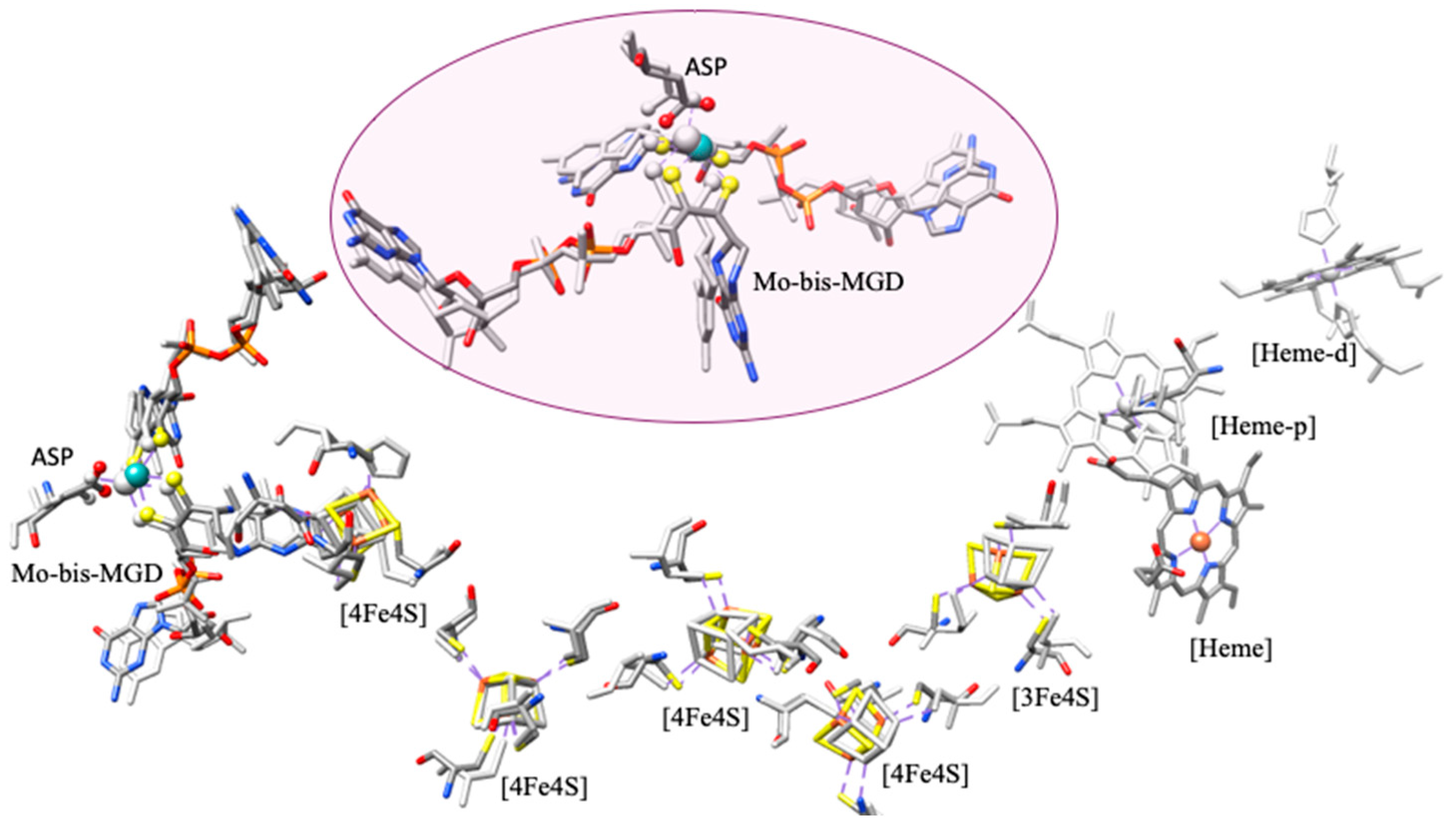
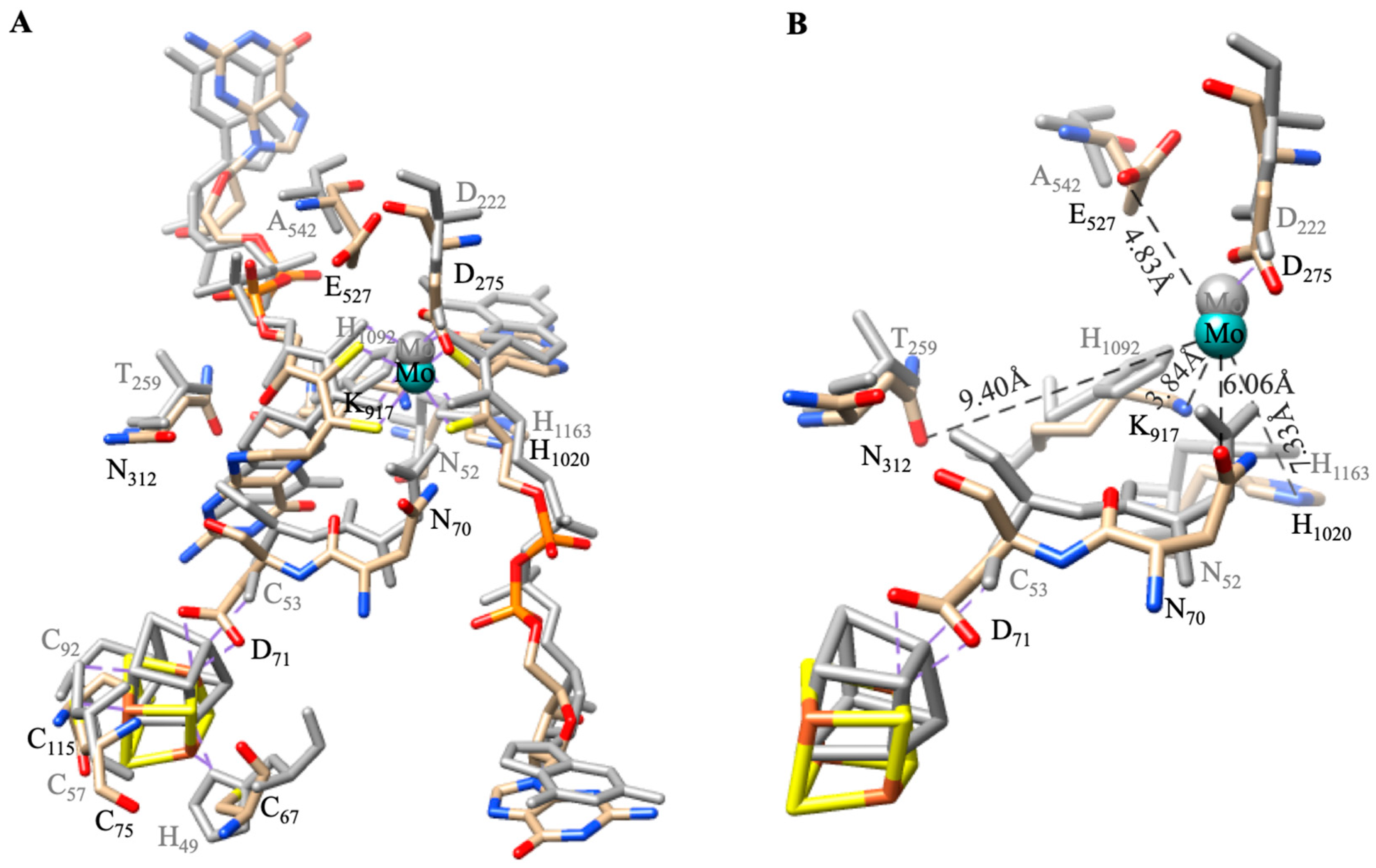
| Pathways | Reactions | ReductionPotentials (E0) (at pH 7) |
|---|---|---|
| Assimilatory and dissimilatory nitrate reduction | Nitrate Reduction (NO3− → NO2−) VNO3− + 2H+ + 2e− → IIINO2− + H2O | +0.42 V |
| Dissimilatory nitrate reduction to Ammonium (DNRA) | Nitrate to Ammonium (NO3− → NO2−→ NH3) IIINO2− + 8H+ + 6e− → -IIINH4+ + 2H2O | +0.34 V |
| Denitrification | Nitrite Reduction (NO2−→ NO, N2O, or N2) Several steps occur in denitrification: Nitrite to Nitric Oxide (NO2− → NO) IIINO2− + 2H+ + e− → IINO + H2O | +0.39 V |
| Nitric Oxide to Nitrous Oxide (NO →N2O) 2 IINO + 2H+ + 2e−→ IN2O + H2O | +1.17 V | |
| Nitrous Oxide to Dinitrogen Gas (N2O → N2) IN2O + 2H+ + 2e− → 0N2 + H2O | +1.35 V | |
| Nitrification | Nitrite Oxidation (NO2− → NO3−) IIINO2− + H2O → VNO3− + 2H+ + 2e− | +0.42 V |
| Ammonia Oxidation (NH4+ → NO2−) Ammonia to Hydroxylamine -IIINH3 + 1/2O2 + H+ + 2e− → -INH2OH | +0.06 V | |
| Hydroxylamine to Nitrite (NH2OH → NO2−) -INH2OH + H2O → IIINO2− + 5H+ + 4e− | +0.44 V | |
| The overall oxidation of ammonia to nitrite: -IIINH3 + 1/2O2 → IIINO2− + H2O + H+ | +0.34 V | |
| Anammox | Hydrazine to Nitrogen (N2H4 → N2) -IIN2H4 → 0N2 + 4H+ + 4e−. IINO + −IIINH4+ + 2H+ + 3e− → −IIN2H4 + H2O | −0.75 V +0.126 |
| Nitrogen fixation | Nitrogen to Ammonium (N2 → NH3) 0N2 + 6H+ + 6e− → 2 -IIINH3 | +0.09 V |
| Reactions | ΔGo (kJ/mol) | Equation |
|---|---|---|
| -IIINH4+ + IIINO2− → 0N2 + 2H2O | −358 | (1) |
| 2 -IIINH3 + 2O2 → IVNO3− + 3H2O | −349 | (2) |
| 2 IVNO3− + 2H+ + 5H2 → 0N2 + 6H2O | −1121 | (3) |
| 2 IVNO3− + 2H+ + 4H2 → -IIINH4+ + 3H2O | −591 | (4) |
| IIINO2− + 2H+ + e− → IINO + H2O | −113.38 | (5) |
| IIINO2− + −IIINH4+ → N2 + 2H2O | −357 | (6) |
| IINO + −IIINH4+ + 2H+ + 3e− → −IIN2H4 + H2O | −116.27 | (7) |
| -IIN2H4 → 0N2 + 4H+ + 4e− | −128.10 | (8) |
| 0N2 + 2H+ + 3H2 → 2 -IIINH3 | −39 | (9) |
| -IIINH4+ + NO2− → N2 + 2H2O | −119 | (10) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maiti, B.K.; Moura, I.; Moura, J.J.G. Nitrate–Nitrite Interplay in the Nitrogen Biocycle. Molecules 2025, 30, 3023. https://doi.org/10.3390/molecules30143023
Maiti BK, Moura I, Moura JJG. Nitrate–Nitrite Interplay in the Nitrogen Biocycle. Molecules. 2025; 30(14):3023. https://doi.org/10.3390/molecules30143023
Chicago/Turabian StyleMaiti, Biplab K., Isabel Moura, and José J. G. Moura. 2025. "Nitrate–Nitrite Interplay in the Nitrogen Biocycle" Molecules 30, no. 14: 3023. https://doi.org/10.3390/molecules30143023
APA StyleMaiti, B. K., Moura, I., & Moura, J. J. G. (2025). Nitrate–Nitrite Interplay in the Nitrogen Biocycle. Molecules, 30(14), 3023. https://doi.org/10.3390/molecules30143023






