Integrating Molecular Dynamics, Molecular Docking, and Machine Learning for Predicting SARS-CoV-2 Papain-like Protease Binders
Abstract
1. Introduction
2. Results and Discussion
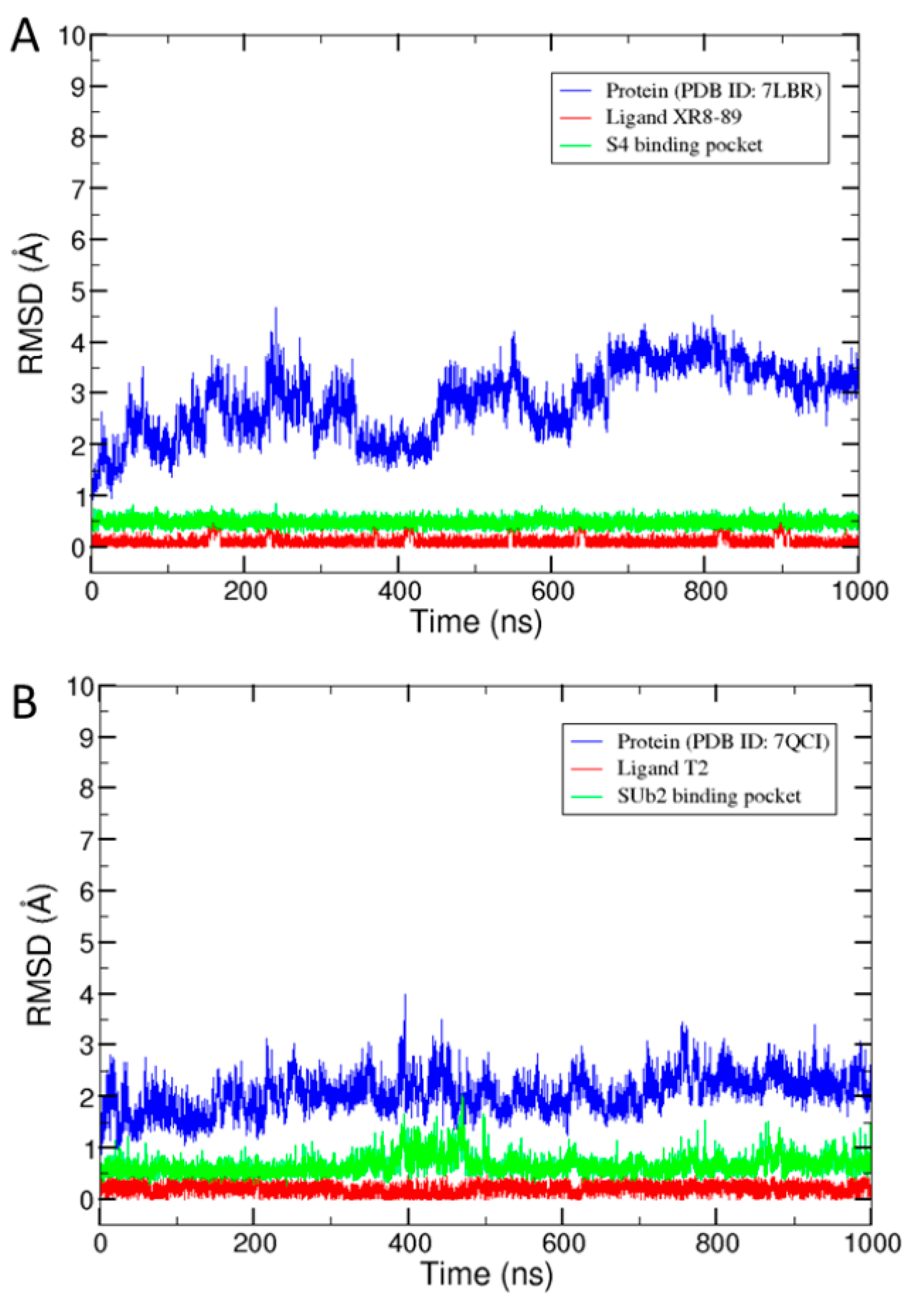
2.1. Conformation Dynamics
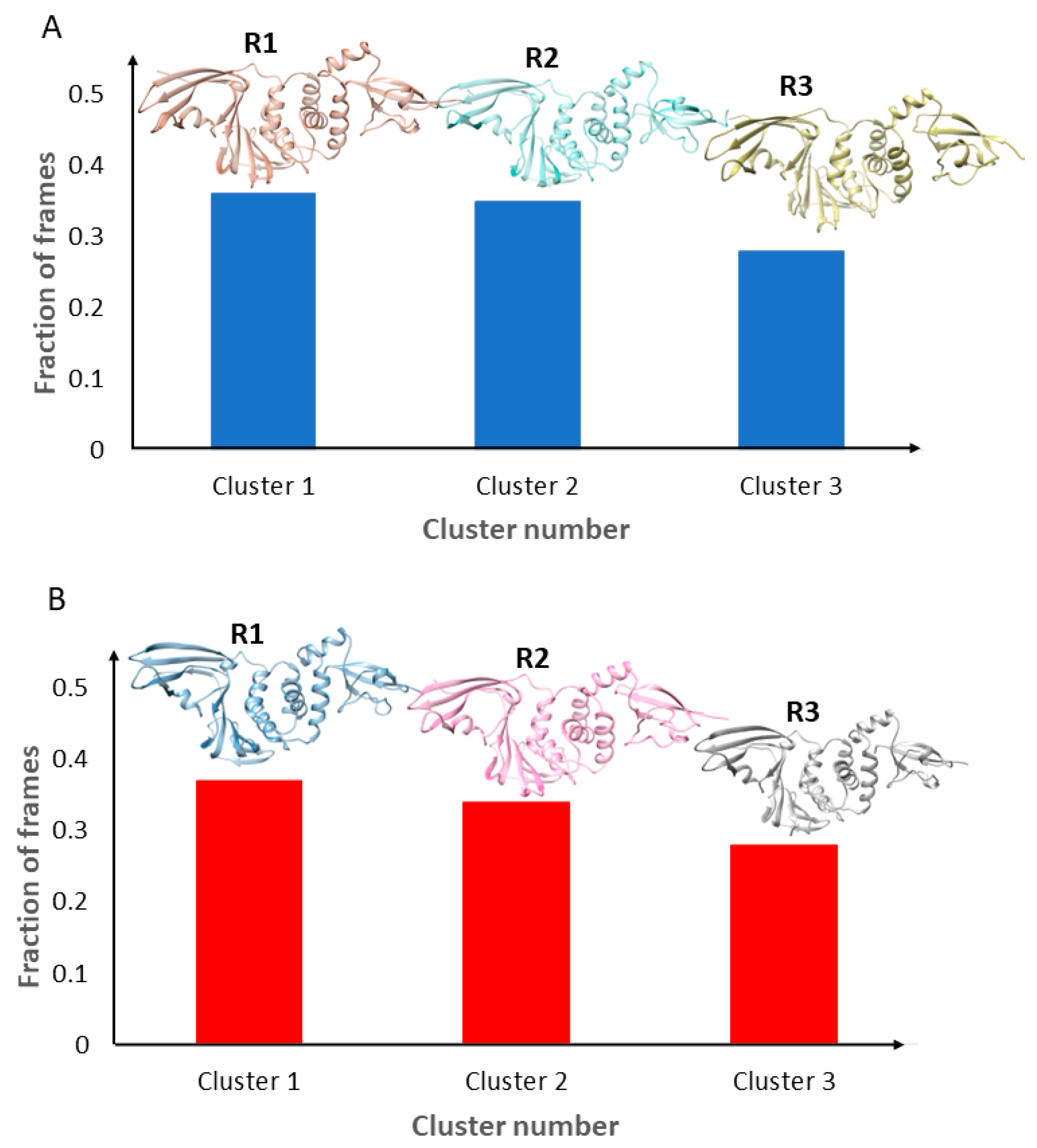
2.2. Representative Structures
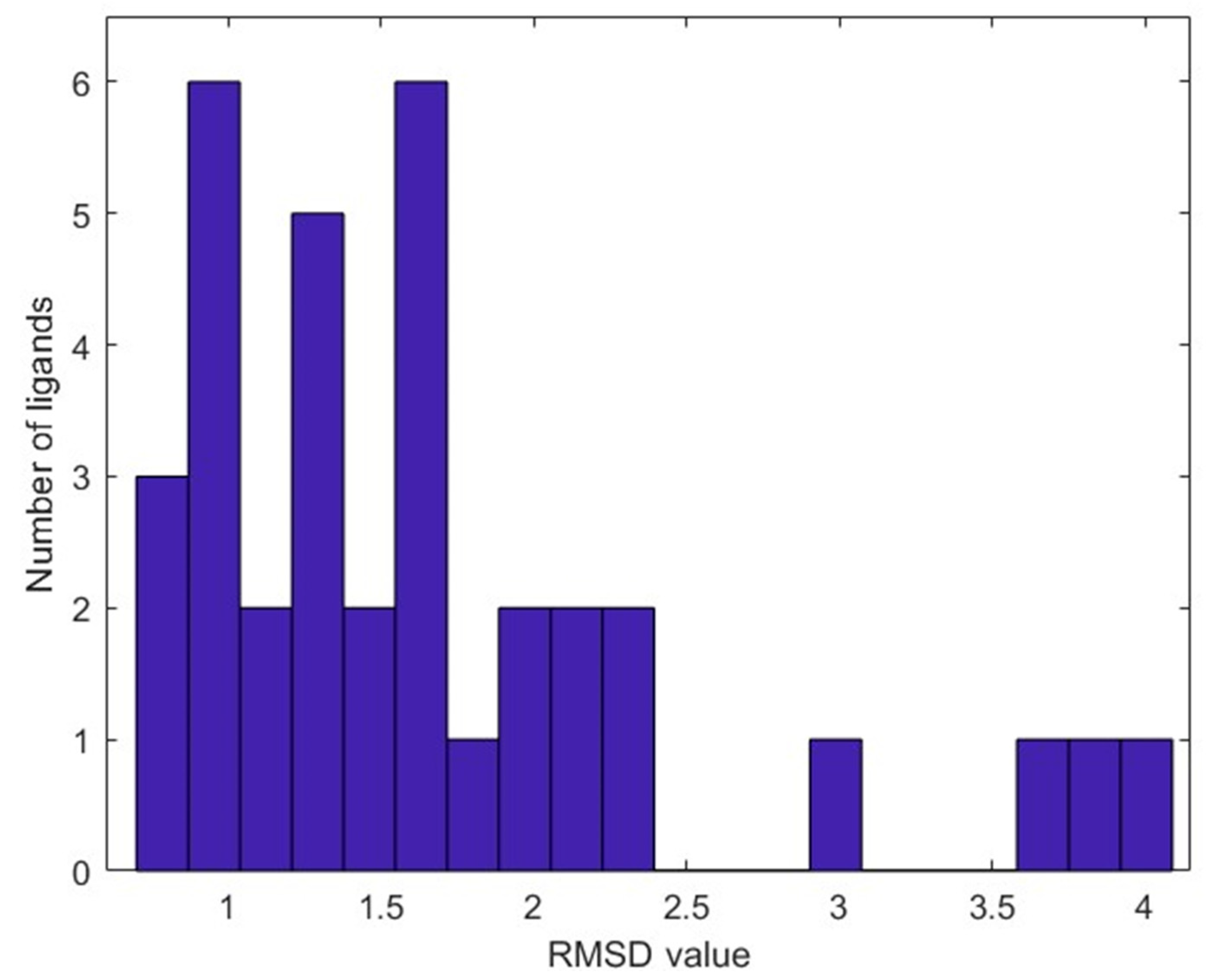
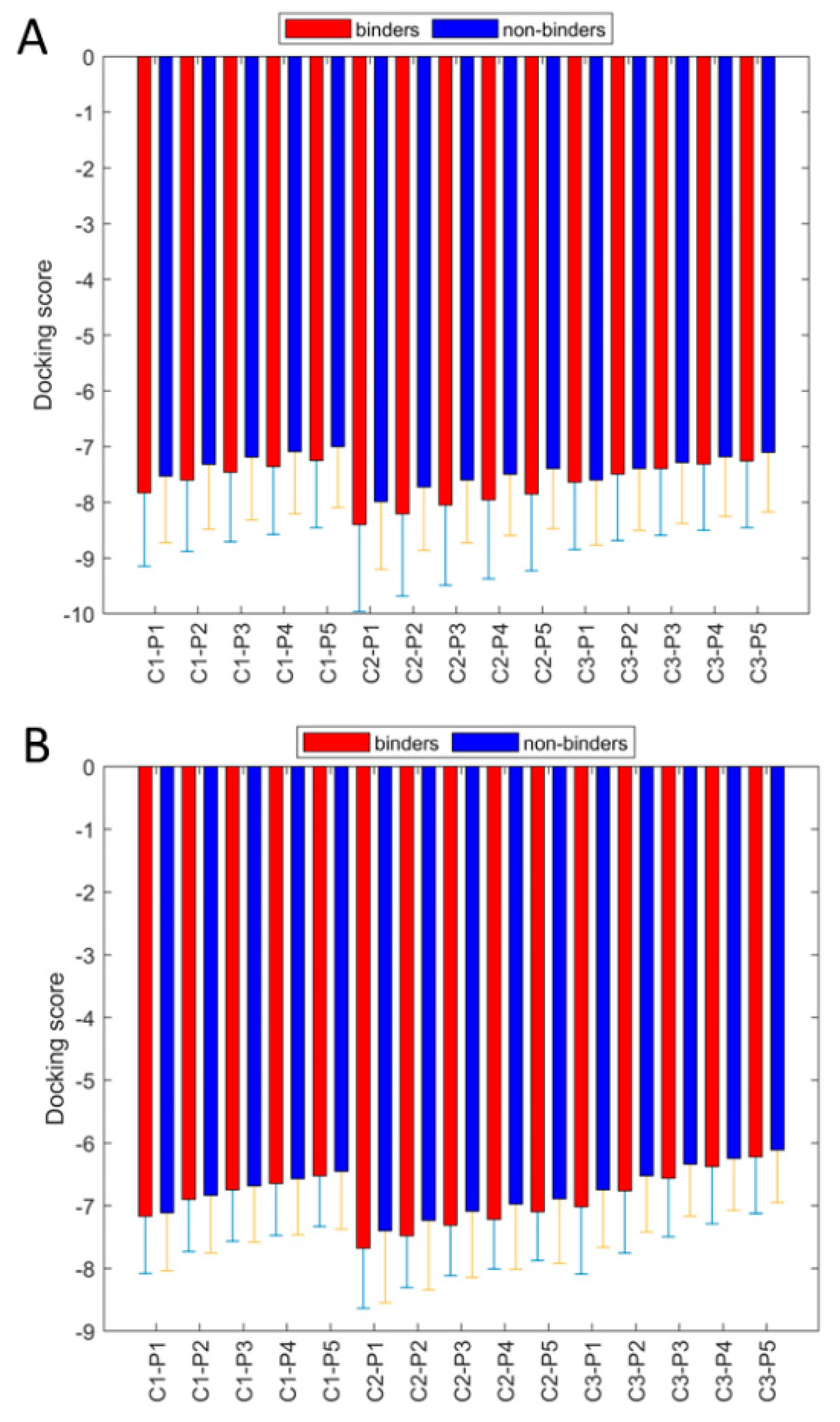
2.3. Molecular Docking
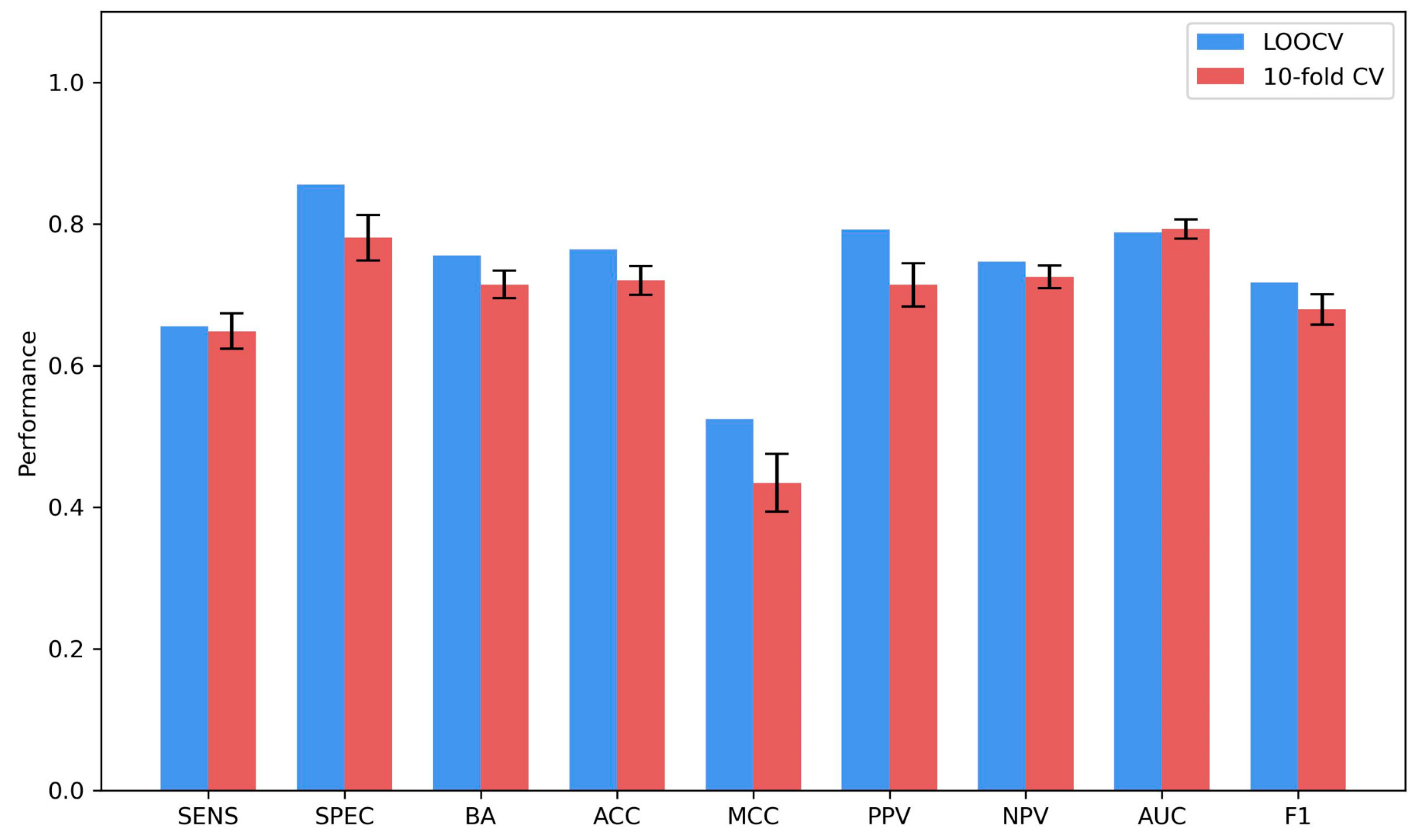
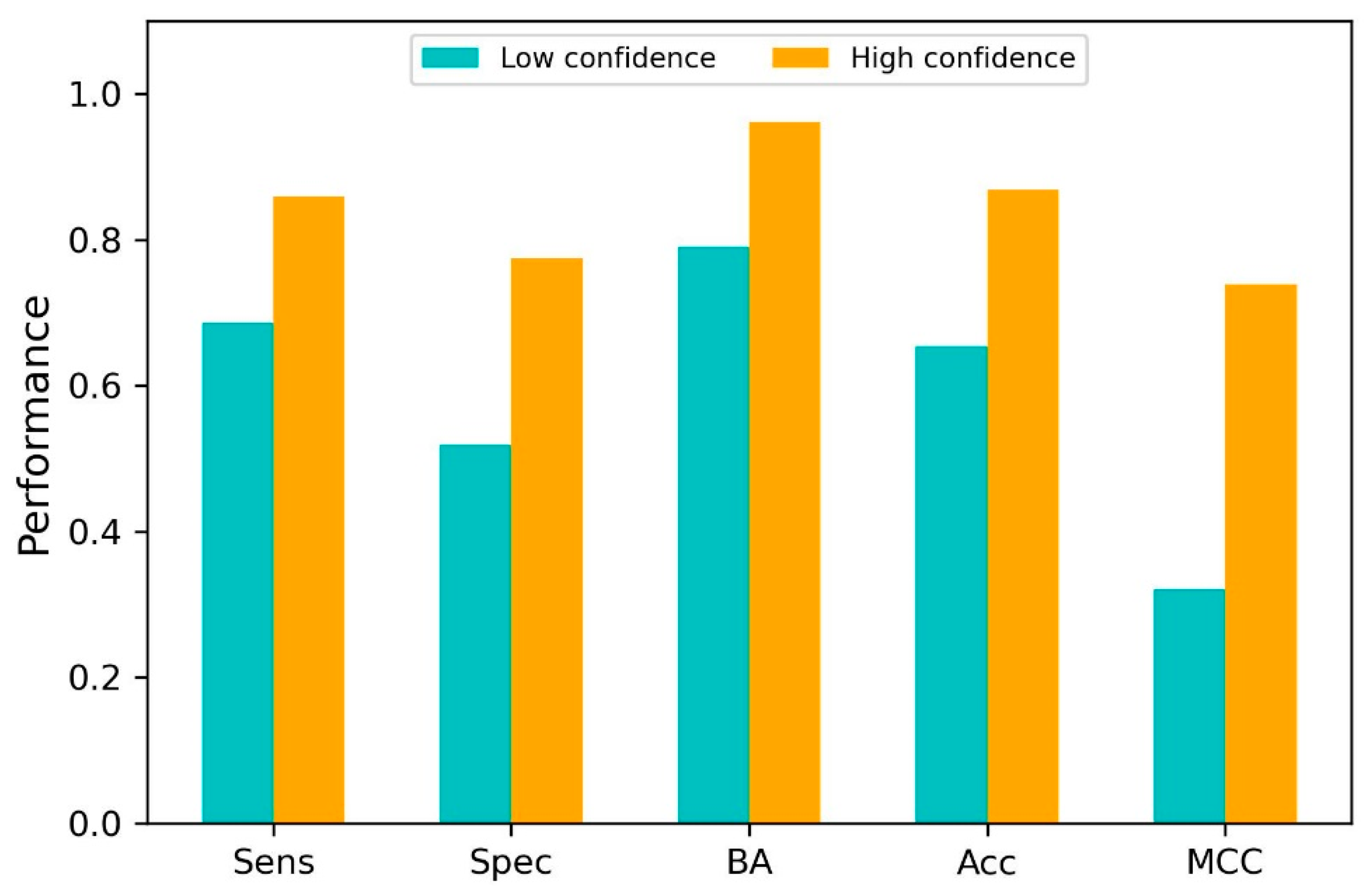
2.4. Model Performance
2.5. Identification of Drugs as Candidates for Repurposing
3. Materials and Methods
3.1. Study Design
3.2. MD Simulation System Preparation
3.3. MD Simulations
3.4. Prepare Protein and Ligand Structures for Molecular Docking
3.5. Molecular Docking
3.6. RF Model Development
3.7. Model Performance Measurement
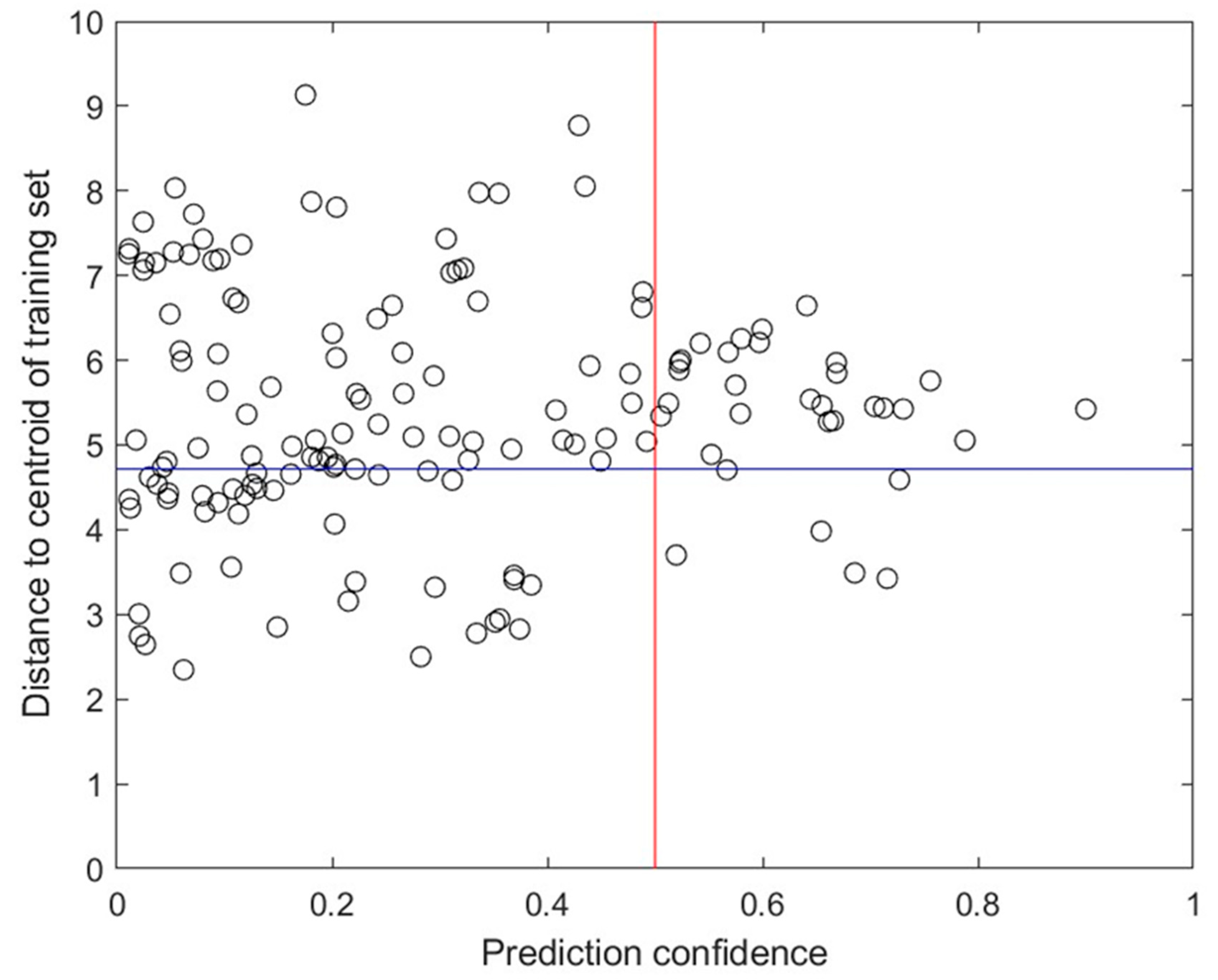
3.8. Prediction Confidence
3.9. Identification of Important Docking Scores
3.10. Applicability Domain Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Author Disclaimer
Conflicts of Interest
References
- Parvathaneni, V.; Kulkarni, N.S.; Muth, A.; Gupta, V. Drug repurposing: A promising tool to accelerate the drug discovery process. Drug Discov. Today 2019, 24, 2076–2085. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, V.S.; Alagarsamy, V.; Solomon, V.R.; Jose, P.A.; Murugesan, S. Drug Repurposing: An Effective Tool in Modern Drug Discovery. Russ. J. Bioorg. Chem. 2023, 49, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Novac, N. Challenges and opportunities of drug repositioning. Trends Pharmacol. Sci. 2013, 34, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Rudrapal, M.; Khairnar, S.J.; Jadhav, A.G. Drug Repurposing (DR): An Emerging Approach in Drug Discovery. In Drug Repurposing—Hypothesis, Molecular Aspects and Therapeutic Applications; IntechOpen: London, UK, 2020. [Google Scholar]
- Bakshi, A.; Gangopadhyay, K.; Basak, S.; De, R.K.; Sengupta, S.; Dasgupta, A. Integrating state-space modeling, parameter estimation, deep learning, and docking techniques in drug repurposing: A case study on COVID-19 cytokine storm. J. Am. Med. Inform. Assn 2025, ocaf035. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Yang, B.; Wang, S.; Yuan, S.; Zhu, W.J.; Zhou, Z.Y.; Zhang, Y.J.; Hu, G. Drug Repurposing for COVID-19 by Constructing a Comorbidity Network with Central Nervous System Disorders. Int. J. Mol. Sci. 2024, 25, 8917. [Google Scholar] [CrossRef] [PubMed]
- Raghav, P.K.; Mann, Z.; Ahluwalia, S.K.; Rajalingam, R. Potential treatments of COVID-19: Drug repurposing and therapeutic interventions. J. Pharmacol. Sci. 2023, 152, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Nishiyama, K.; Nishimura, A.; Noda, T.; Okabe, K.; Kusakabe, T.; Kanda, Y.; Nishida, M. Drug repurposing for the treatment of COVID-19. J. Pharmacol. Sci. 2022, 149, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Kumari, P.; Pradhan, B.; Koromina, M.; Patrinos, G.P.; Van Steen, K. Discovery of new drug indications for COVID-19: A drug repurposing approach. PLoS ONE 2022, 17, e0267095. [Google Scholar] [CrossRef] [PubMed]
- Mule, S.; Singh, A.; Greish, K.; Sahebkar, A.; Kesharwani, P.; Shukla, R. Drug repurposing strategies and key challenges for COVID-19 management. J. Drug Target. 2022, 30, 413–429. [Google Scholar] [CrossRef] [PubMed]
- Ciliberto, G.; Cardone, L. Boosting the arsenal against COVID-19 through computational drug repurposing. Drug Discov. Today 2020, 25, 946–948. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.X.; Hong, H.X.; Yang, S.Y.; Wei, Y.Q. Individualized network-based drug repositioning infrastructure for precision oncology in the panomics era. Brief. Bioinform. 2017, 18, 682–697. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Wei, J.; Tang, K.L.; Feuers, R.; Hong, H.X. Drug Repositioning Through Network Pharmacology. Curr. Top. Med. Chem. 2016, 16, 3646–3656. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Mattes, W.; Mendrick, D.L.; Hong, H.X. Molecular Docking for Identification of Potential Targets for Drug Repurposing. Curr. Top. Med. Chem. 2016, 16, 3636–3645. [Google Scholar] [CrossRef] [PubMed]
- Wan, Z.M.; Sun, X.R.; Li, Y.; Chu, T.Y.; Hao, X.Y.; Cao, Y.; Zhang, P. Applications of Artificial Intelligence in Drug Repurposing. Adv. Sci. 2025, 12, e2411325. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, F.; Tang, J.; Nussinov, R.; Cheng, F. Artificial intelligence in COVID-19 drug repurposing. Lancet Digit. Health 2020, 2, e667–e676. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, F.; Soomro, A.M.; Chethikkattuveli Salih, A.R.; Samantasinghar, A.; Asif, A.; Kang, I.S.; Choi, K.H. A comprehensive review of artificial intelligence and network based approaches to drug repurposing in COVID-19. Biomed. Pharmacother. 2022, 153, 113350. [Google Scholar] [CrossRef] [PubMed]
- Kowshik, A.V.; Manoj, M.; Sowmyanarayan, S.; Chatterjee, J. Drug repurposing: Databases and pipelines. Cns Spectrums 2024, 29, 6–9. [Google Scholar] [CrossRef] [PubMed]
- Ng, H.W.; Leggett, C.; Sakkiah, S.; Pan, B.; Ye, H.; Wu, L.; Selvaraj, C.; Tong, W.; Hong, H. Competitive docking model for prediction of the human nicotinic acetylcholine receptor alpha7 binding of tobacco constituents. Oncotarget 2018, 9, 16899–16916. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ji, Z.; Guo, W.; Wood, E.L.; Liu, J.; Sakkiah, S.; Xu, X.; Patterson, T.A.; Hong, H. Machine Learning Models for Predicting Cytotoxicity of Nanomaterials. Chem. Res. Toxicol. 2022, 35, 125–139. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.J.; Liu, J.; Dong, F.; Hong, H.X. Unlocking the potential of AI: Machine learning and deep learning models for predicting carcinogenicity of chemicals. J. Environ. Sci. Health C-Tox 2025, 43, 23–50. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.K.H.; Guo, W.; Liu, J.; Dong, F.; Li, Z.; Patterson, T.A.; Hong, H. Machine learning and deep learning for brain tumor MRI image segmentation. Exp. Biol. Med. 2023, 248, 1974–1992. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Liu, J.; Dong, F.; Song, M.; Li, Z.; Khan, M.K.H.; Patterson, T.A.; Hong, H. Review of machine learning and deep learning models for toxicity prediction. Exp. Biol. Med. 2023, 248, 1952–1973. [Google Scholar] [CrossRef] [PubMed]
- March-Vila, E.; Pinzi, L.; Sturm, N.; Tinivella, A.; Engkvist, O.; Chen, H.; Rastelli, G. On the Integration of In Silico Drug Design Methods for Drug Repurposing. Front. Pharmacol. 2017, 8, 298. [Google Scholar] [CrossRef] [PubMed]
- Jourdan, J.P.; Bureau, R.; Rochais, C.; Dallemagne, P. Drug repositioning: A brief overview. J. Pharm. Pharmacol. 2020, 72, 1145–1151. [Google Scholar] [CrossRef] [PubMed]
- Ciotti, M.; Ciccozzi, M.; Terrinoni, A.; Jiang, W.C.; Wang, C.B.; Bernardini, S. The COVID-19 pandemic. Crit. Rev. Clin. Lab. Sci. 2020, 57, 365–388. [Google Scholar] [CrossRef] [PubMed]
- Hardenbrook, N.J.; Zhang, P. A structural view of the SARS-CoV-2 virus and its assembly. Curr. Opin. Virol. 2022, 52, 123–134. [Google Scholar] [CrossRef] [PubMed]
- van de Leemput, J.; Han, Z. Understanding Individual SARS-CoV-2 Proteins for Targeted Drug Development against COVID-19. Mol. Cell Biol. 2021, 41, e0018521. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Lacko, L.A.; Chen, S. Druggable targets and therapeutic development for COVID-19. Front. Chem. 2022, 10, 963701. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Chen, R.; Liu, J.; Patterson, T.A.; Hong, H. Analyzing 3D structures of the SARS-CoV-2 main protease reveals structural features of ligand binding for COVID-19 drug discovery. Drug Discov. Today 2023, 28, 103727. [Google Scholar] [CrossRef] [PubMed]
- Sternberg, A.; Naujokat, C. Structural features of coronavirus SARS-CoV-2 spike protein: Targets for vaccination. Life Sci. 2020, 257, 118056. [Google Scholar] [CrossRef] [PubMed]
- Sakkiah, S.; Guo, W.J.; Pan, B.H.; Ji, Z.W.; Yavas, G.; Azevedo, M.; Hawes, J.; Patterson, T.A.; Hong, H.X. Elucidating Interactions Between SARS-CoV-2 Trimeric Spike Protein and ACE2 Using Homology Modeling and Molecular Dynamics Simulations. Front. Chem. 2021, 8, 622632. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Anirudhan, V.; Du, R.K.; Cui, Q.H.; Rong, L.J. RNA-dependent RNA polymerase of SARS-CoV-2 as a therapeutic target. J. Med. Virol. 2021, 93, 300–310. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wu, J.Q.; Wang, H.F.; Gao, Y.; Liu, Q.J.; Mu, A.; Ji, W.X.; Yan, L.M.; Zhu, Y.; Zhu, C.; et al. Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase. Cell 2020, 182, 417–428. [Google Scholar] [CrossRef] [PubMed]
- Vangeel, L.; Chiu, W.; De Jonghe, S.; Maes, P.; Slechten, B.; Raymenants, J.; Andre, E.; Leyssen, P.; Neyts, J.; Jochmans, D. Remdesivir, Molnupiravir and Nirmatrelvir remain active against SARS-CoV-2 Omicron and other variants of concern. Antivir. Res. 2022, 198, 105252. [Google Scholar] [CrossRef] [PubMed]
- Citarella, A.; Scala, A.; Piperno, A.; Micale, N. SARS-CoV-2 Mpro: A Potential Target for Peptidomimetics and Small-Molecule Inhibitors. Biomolecules 2021, 11, 607. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Q.; Zhu, G.H.; Zhu, R.; Lei, J.X.; Liu, S.Y.; Tu, D.Z.; Zhang, Y.N.; Song, Y.Q.; Hou, X.D.; Zhuang, X.Y.; et al. Discovery of baicalein derivatives as novel covalent inhibitors of SARS CoV-2 Mpro: Structure-activity relationships and inhibitory mechanisms. Bioorg. Chem. 2025, 161, 108560. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Zhang, B.; Jiang, X.M.; Su, H.; Li, J.; Zhao, Y.; Xie, X.; Jin, Z.; Peng, J.; Liu, F.; et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 2020, 368, 1331–1335. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Xu, F. PLpro Inhibitors as a Potential Treatment for COVID-19. Biomedicines 2025, 13, 1417. [Google Scholar] [CrossRef] [PubMed]
- Bader, S.M.; Calleja, D.J.; Devine, S.M.; Kuchel, N.W.; Lu, B.G.C.; Wu, X.; Birkinshaw, R.W.; Bhandari, R.; Loi, K.; Volpe, R.; et al. A novel PLpro inhibitor improves outcomes in a pre-clinical model of long COVID. Nat. Commun. 2025, 16, 2900. [Google Scholar] [CrossRef] [PubMed]
- McClain, C.B.; Vabret, N. SARS-CoV-2: The many pros of targeting PLpro. Signal Transduct. Target. Ther. 2020, 5, 223. [Google Scholar] [CrossRef] [PubMed]
- Barretto, N.; Jukneliene, D.; Ratia, K.; Chen, Z.B.; Mesecar, A.D.; Baker, S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005, 79, 15189–15198. [Google Scholar] [CrossRef] [PubMed]
- Osipiuk, J.; Azizi, S.A.; Dvorkin, S.; Endres, M.; Jedrzejczak, R.; Jones, K.A.; Kang, S.; Kathayat, R.S.; Kim, Y.; Lisnyak, V.G.; et al. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat. Commun. 2021, 12, 743. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.H.; Thai, Q.M.; Pham, M.Q.; Minh, P.T.H.; Phung, H.T.T. Machine learning combines atomistic simulations to predict SARS-CoV-2 Mpro inhibitors from natural compounds. Mol. Divers. 2024, 28, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Van, N.T.H.; Nghi, D.H.; Bach, P.C.; Tuyen, T.T.; Bich, H.T.; Toan, T.Q.; Lam, D.T.; Hoang, L.V.; Quan, P.M.; Minh, P.T.H. Triterpenoids from the leaves of Camellia chrysantha growing in Quang Ninh (Vietnam) and their activities on main protease (Mpro) and ACE2. VJCH 2023, 61, 140–147. [Google Scholar] [CrossRef]
- Tam, N.M.; Nguyen, T.H.; Pham, M.Q.; Hong, N.D.; Tung, N.T.; Vu, V.V.; Quang, D.T.; Ngo, S.T. Upgrading nirmatrelvir to inhibit SARS-CoV-2 Mpro via DeepFrag and free energy calculations. J. Mol. Graph. Model. 2023, 124, 108535. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Xia, H.; Wang, Z.; Xiong, F. Discovery of Severe Acute Respiratory Syndrome Coronavirus 2 Main Protease Inhibitors through Rational Design of Novel Fluorinated 1,3,4-oxadiazole Amide Derivatives: An In-Silico Study. Chem. Biodivers. 2025, 22, e202403179. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.H.; Yang, P.Y.; Zhang, J. Potential Inhibitors Targeting Papain-Like Protease of SARS-CoV-2: Two Birds with One Stone. Front. Chem. 2022, 10, 822785. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Du, X.Y.; Duan, Y.K.; Pan, X.Y.; Sun, Y.F.; You, T.; Han, L.; Jin, Z.M.; Shang, W.J.; Yu, J.; et al. High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors. Protein Cell 2021, 12, 877–888. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.L.; Sacco, M.D.; Xia, Z.L.; Lambrinidis, G.; Townsend, J.A.; Hu, Y.M.; Meng, X.Z.; Szeto, T.; Ba, M.; Zhang, X.J.; et al. Discovery of SARS-CoV-2 Papain-like Protease Inhibitors through a Combination of High-Throughput Screening and a FlipGFP-Based Reporter Assay. ACS Central Sci. 2021, 7, 1245–1260. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Huang, B.; Tang, J.; Liu, S.; Liu, M.; Ye, Y.; Liu, Z.; Xiong, Y.; Zhu, W.; Cao, D.; et al. The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery. Nat. Commun. 2021, 12, 488. [Google Scholar] [CrossRef] [PubMed]
- Varghese, A.; Liu, J.; Liu, B.; Guo, W.; Dong, F.; Patterson, T.A.; Hong, H. Analysis of Structures of SARS-CoV-2 Papain-like Protease Bound with Ligands Unveils Structural Features for Inhibiting the Enzyme. Molecules 2025, 30, 491. [Google Scholar] [CrossRef] [PubMed]
- Rut, W.; Lv, Z.; Zmudzinski, M.; Patchett, S.; Nayak, D.; Snipas, S.J.; El Oualid, F.; Huang, T.T.; Bekes, M.; Drag, M.; et al. Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: A framework for anti-COVID-19 drug design. Sci. Adv. 2020, 6, eabd4596. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Rao, Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat. Rev. Microbiol. 2021, 19, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Ewert, W.; Gunther, S.; Miglioli, F.; Falke, S.; Reinke, P.Y.A.; Niebling, S.; Gunther, C.; Han, H.; Srinivasan, V.; Brognaro, H.; et al. Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease. Front. Chem. 2022, 10, 832431. [Google Scholar] [CrossRef] [PubMed]
- Baez-Santos, Y.M.; St John, S.E.; Mesecar, A.D. The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015, 115, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Qin, B.; Chen, P.; Zhu, K.; Hou, P.; Wojdyla, J.A.; Wang, M.; Cui, S. Crystal structure of SARS-CoV-2 papain-like protease. Acta Pharm. Sin. B 2021, 11, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Klemm, T.; Ebert, G.; Calleja, D.J.; Allison, C.C.; Richardson, L.W.; Bernardini, J.P.; Lu, B.G.; Kuchel, N.W.; Grohmann, C.; Shibata, Y.; et al. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020, 39, e106275. [Google Scholar] [CrossRef] [PubMed]
- Garnsey, M.R.; Robinson, M.C.; Nguyen, L.T.; Cardin, R.; Tillotson, J.; Mashalidis, E.; Yu, A.; Aschenbrenner, L.; Balesano, A.; Behzadi, A.; et al. Discovery of SARS-CoV-2 papain-like protease (PLpro) inhibitors with efficacy in a murine infection model. Sci. Adv. 2024, 10, eado4288. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Nance, K.D.; Joshi, D.R.; Kales, S.C.; Ye, L.; Hu, X.; Shamim, K.; Zakharov, A.V. Applications of Machine Learning Approaches for the Discovery of SARS-CoV-2 PLpro Inhibitors. J. Chem. Inf. Model. 2025, 65, 1338–1356. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Ratia, K.; Cooper, L.; Kong, D.; Lee, H.; Kwon, Y.; Li, Y.; Alqarni, S.; Huang, F.; Dubrovskyi, O.; et al. Potent, Novel SARS-CoV-2 PLpro Inhibitors Block Viral Replication in Monkey and Human Cell Cultures. bioRxiv 2021. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhang, J.; Wang, Y.; Liu, Z.; Kelly, R.; Zhou, G.; Fang, H.; Borlak, J.; Tong, W. The liver toxicity knowledge base: A systems approach to a complex end point. Clin. Pharmacol. Ther. 2013, 93, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E.; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Ghoreishi, D.; Gilson, M.K.; et al. AMBER 2018; University of California, San Francisco: San Francisco, CA, USA, 2018. [Google Scholar]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Peters, M.B.; Yang, Y.; Wang, B.; Füsti-Molnár, L.; Weaver, M.N.; Merz, K.M. Structural Survey of Zinc-Containing Proteins and Development of the Zinc AMBER Force Field (ZAFF). J. Chem. Theory Comput. 2010, 6, 2935–2947. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef] [PubMed]
- Davidchack, R.L.; Handel, R.; Tretyakov, M.V. Langevin thermostat for rigid body dynamics. J. Chem. Phys. 2009, 130, 234101. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.C.; Postma, J.P.M.; Vangunsteren, W.F.; Dinola, A.; Haak, J.R. Molecular-Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E., 3rd. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Calleja, D.J.; Kuchel, N.; Lu, B.G.C.; Birkinshaw, R.W.; Klemm, T.; Doerflinger, M.; Cooney, J.P.; Mackiewicz, L.; Au, A.E.; Yap, Y.Q.; et al. Insights into Drug Repurposing, as Well as Specificity and Compound Properties of Piperidine-Based SARS-CoV-2 PLpro Inhibitors. Front. Chem. 2022, 10, 861209. [Google Scholar] [CrossRef] [PubMed]
- Shan, H.; Liu, J.; Shen, J.; Dai, J.; Xu, G.; Lu, K.; Han, C.; Wang, Y.; Xu, X.; Tong, Y.; et al. Development of potent and selective inhibitors targeting the papain-like protease of SARS-CoV-2. Cell Chem. Biol. 2021, 28, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Zhang, X.; Ansari, A.; Jadhav, P.; Tan, H.; Li, K.; Chopra, A.; Ford, A.; Chi, X.; Ruiz, F.X.; et al. Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model. Science 2024, 383, 1434–1440. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, V.; Brognaro, H.; Prabhu, P.R.; de Souza, E.E.; Gunther, S.; Reinke, P.Y.A.; Lane, T.J.; Ginn, H.; Han, H.; Ewert, W.; et al. Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease. Commun. Biol. 2022, 5, 805. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.; Tong, W.; Xie, Q.; Fang, H.; Perkins, R. An in silico ensemble method for lead discovery: Decision forest. SAR QSAR Environ. Res. 2005, 16, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Ratnasinghe, L.D.; Hong, H.; Perkins, R.; Tang, Z.Z.; Hu, N.; Taylor, P.R.; Tong, W. Decision forest analysis of 61 single nucleotide polymorphisms in a case-control study of esophageal cancer; a novel method. BMC Bioinform. 2005, 6 (Suppl. S2), S4. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Liu, J.; Xu, L.; Guo, W.J.; Li, Z.; Khan, M.K.H.; Ge, W.G.; Patterson, T.A.; Hong, H.X. Developing a SARS-CoV-2 main protease binding prediction random forest model for drug repurposing for COVID-19 treatment. Exp. Biol. Med. 2023, 248, 1927–1936. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Hong, H.X.; Thakkar, S.; Chen, M.J.; Tong, W.D. Development of Decision Forest Models for Prediction of Drug-Induced Liver Injury in Humans Using A Large Set of FDA-approved Drugs. Sci. Rep. 2017, 7, 17311. [Google Scholar] [CrossRef]
- Liu, J.; Guo, W.J.; Dong, F.; Aungst, J.; Fitzpatrick, S.; Patterson, T.A.; Hong, H.X. Machine learning models for rat multigeneration reproductive toxicity prediction. Front. Pharmacol. 2022, 13, 1018226. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, J.; Li, Z.; Dong, F.; Guo, W.; Ge, W.; Patterson, T.A.; Hong, H. Developing predictive models for µ opioid receptor binding using machine learning and deep learning techniques. Exp. Biol. Med. 2025, 250, 10359. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Khan, M.K.H.; Guo, W.; Dong, F.; Ge, W.; Zhang, C.; Gong, P.; Patterson, T.A.; Hong, H. Machine learning and deep learning approaches for enhanced prediction of hERG blockade: A comprehensive QSAR modeling study. Expert. Opin. Drug Metab. Toxicol. 2024, 20, 665–684. [Google Scholar] [CrossRef] [PubMed]
- Kar, S.; Roy, K.; Leszczynski, J. Applicability Domain: A Step Toward Confident Predictions and Decidability for QSAR Modeling. Methods Mol. Biol. 2018, 1800, 141–169. [Google Scholar] [PubMed]
- Santos, L.H.; Kronenberger, T.; Almeida, R.G.; Silva, E.B.; Rocha, R.E.O.; Oliveira, J.C.; Barreto, L.V.; Skinner, D.; Fajtova, P.; Giardini, M.A.; et al. Structure-Based Identification of Naphthoquinones and Derivatives as Novel Inhibitors of Main Protease Mpro and Papain-like Protease PLpro of SARS-CoV-2. J. Chem. Inf. Model. 2022, 62, 6553–6573. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, K.; Zhang, Z.; Wei, P.; Zhang, L.; Xu, Y.; Lun, Q.; Ma, Y.; Wu, F.; Zhang, Y.; et al. Investigating Derivatives of Tanshinone IIA Sulfonate Sodium and Chloroxine for Their Inhibition Activities Against the SARS-CoV-2 Papain-like Protease. ACS Omega 2022, 7, 48416–48426. [Google Scholar] [CrossRef] [PubMed]
- Cho, C.; Li, S.G.; Lalonde, T.J.; Yang, K.S.; Yu, G.; Qiao, Y.; Xu, S.; Ray Liu, W. Drug Repurposing for the SARS-CoV-2 Papain-Like Protease. ChemMedChem 2022, 17, e202100455. [Google Scholar] [CrossRef] [PubMed]
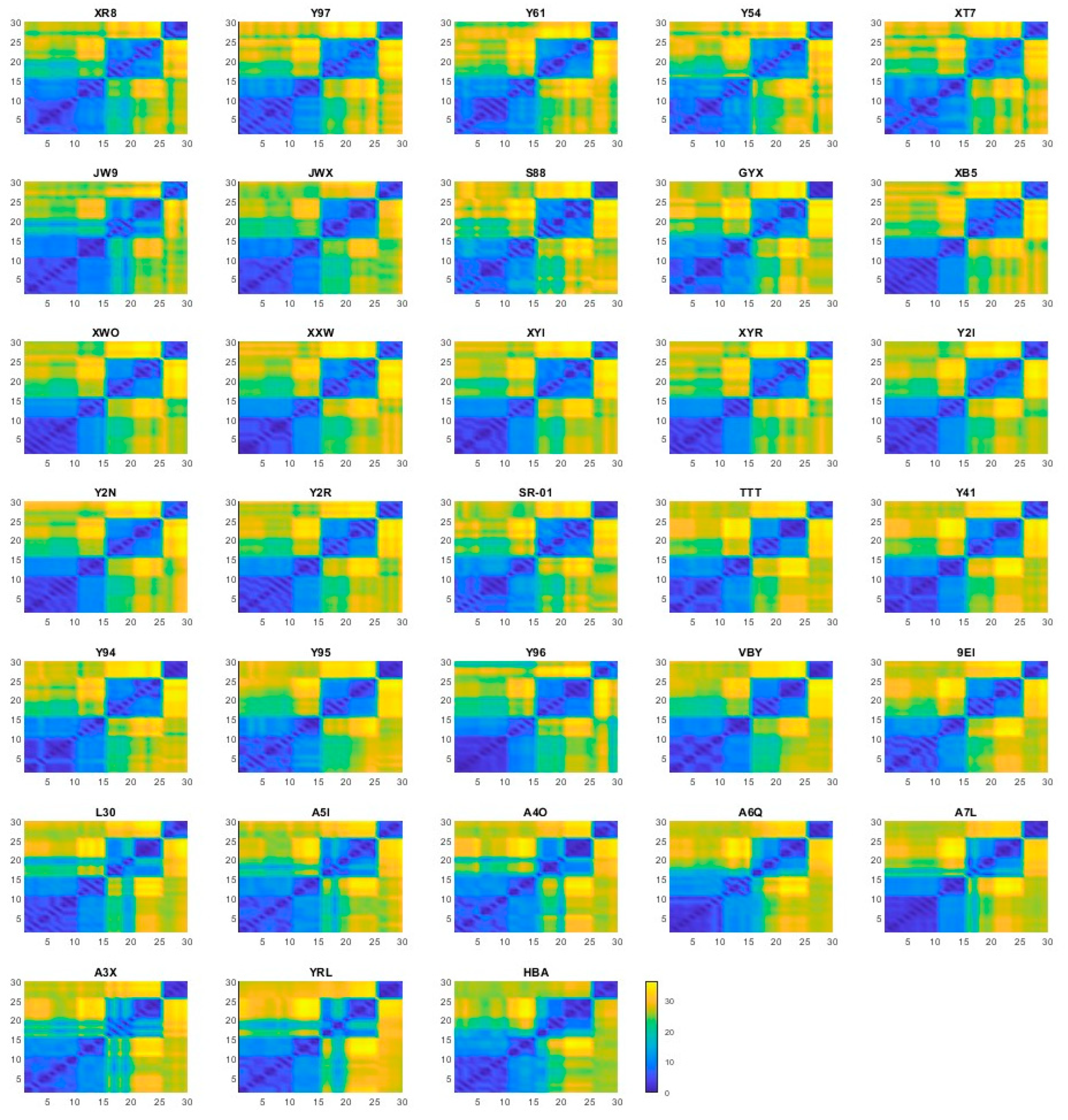

| Ligand | Binding Site | Prediction | Average for S4 | Average for SUb2 |
|---|---|---|---|---|
| XR8 | S4 | S4 | −8.740 | −7.553 |
| Y97 | S4 | S4 | −8.560 | −7.291 |
| Y61 | S4 | S4 | −8.80 | −7.394 |
| Y54 | S4 | S4 | −8.647 | −7.459 |
| XT7 | S4 | S4 | −9.080 | −7.486 |
| JW9 | S4 | S4 | −7.433 | −6.583 |
| JWX | S4 | S4 | −8.173 | −7.673 |
| S88 | S4 | S4 | −8.36 | −8.104 |
| GYX | S4 | S4 | −9.013 | −8.176 |
| XB5 | S4 | S4 | −8.480 | −7.018 |
| XWO | S4 | S4 | −8.793 | −6.893 |
| XXW | S4 | S4 | −8.407 | −6.950 |
| XYI | S4 | S4 | −8.780 | −7.161 |
| XYR | S4 | S4 | −8.793 | −7.037 |
| Y2I | S4 | S4 | −8.687 | −7.187 |
| Y2N | S4 | S4 | −8.553 | −6.728 |
| Y2R | S4 | S4 | −8.393 | −6.837 |
| SR-01 | S4 | S4 | −8.727 | −8.114 |
| TTT | S4 | S4 | −8.547 | −7.415 |
| Y41 | S4 | S4 | −8.693 | −7.631 |
| Y94 | S4 | S4 | −8.500 | −7.550 |
| Y95 | S4 | S4 | −9.120 | −7.877 |
| Y96 | S4 | S4 | −8.647 | −7.303 |
| VBY | S4 | S4 | −8.560 | −7.4225 |
| 9EI | S4 | SUb2 | −7.707 | −7.857 |
| L30 | S4 | SUb2 | −7.440 | −7.659 |
| A5I | SUb2 | SUb2 | −5.853 | −5.997 |
| A4O | SUb2 | SUb2 | −5.913 | −6.088 |
| T2 | SUb2 | SUb2 | −5.987 | −6.068 |
| A7L | SUb2 | SUb2 | −5.987 | −6.262 |
| A3X | SUb2 | S4 | −6.140 | −5.890 |
| YRL | SUb2 | SUb2 | −4.967 | −5.548 |
| HBA | SUb2 | SUb2 | −4.720 | −5.233 |
| Docking Score From | Rank | Importance Contribution (%) | ||
|---|---|---|---|---|
| Docking Site | Cluster | Top Pose | ||
| S4 | 2 | 4 | 1 | 7.45 |
| S4 | 2 | 3 | 2 | 7.15 |
| S4 | 2 | 2 | 3 | 6.92 |
| S4 | 2 | 5 | 4 | 6.65 |
| S4 | 2 | 1 | 5 | 5.01 |
| S4 | 1 | 2 | 6 | 4.92 |
| SUb2 | 2 | 2 | 7 | 4.56 |
| S4 | 1 | 1 | 8 | 4.49 |
| SUb2 | 2 | 5 | 9 | 3.64 |
| S4 | 3 | 2 | 10 | 3.63 |
| SUb2 | 2 | 1 | 11 | 3.42 |
| Drug | ATC Code | DrugBank ID | Use |
|---|---|---|---|
| Darifenacin | G04BD10 | DB00496 | Treat overactive bladder |
| Penbutolol | C07AA23 | DB01359 | Treat hypertension |
| Zafirlukast | R03DC01 | DB00549 | Treat asthma |
| Cromoglicic acid | R03BC01 | DB01003 | Treat asthma and allergies |
| Ponatinib | L01XE24 | DB08901 | Treat leukemia |
| S. No. | Ligand | Binding Pocket | PDB ID | Reference |
|---|---|---|---|---|
| 1 | GRL0617 | S3 and S4 | 7JIR | [43] |
| 2 | XR8-24 | 7LBS | [61] | |
| 3 | XR8-65 | 7LOS | [61] | |
| 4 | XR8-69 | 7LLZ | [61] | |
| 5 | XR8-83 | 7LLF | [61] | |
| 6 | XT7 | 7LBR | [61] | |
| 7 | PLP_Snyder441 | 7JN2 | - | |
| 8 | PLP_Snyder494 | 7KOJ | - | |
| 9 | PLP_Snyder495 | 7JIT | [43] | |
| 10 | PLP_Snyder496 | 7KOK | - | |
| 11 | PLP_Snyder530 | 7JIW | [43] | |
| 12 | PLP_Snyder608 | 7SGU | - | |
| 13 | PLP_Snyder630 | 7SGW | - | |
| 14 | Jun9-72-2 | 7SDR | - | |
| 15 | Jun9-84-3 | 7SQE | - | |
| 16 | 3k | 7TZJ | [75] | |
| 17 | S43 | 7E35 | [76] | |
| 18 | Jun12682 | 8UOB | [77] | |
| 19 | Jun11941 | 8UUF | [77] | |
| 20 | Jun12303 | 8UUG | [77] | |
| 21 | Jun12199 | 8UUH | [77] | |
| 22 | Jun12162 | 8UUU | [77] | |
| 23 | Jun12197 | 8UUV | [77] | |
| 24 | Jun12145 | 8UUW | [77] | |
| 25 | Jun12129 | 8UUY | [77] | |
| 26 | SR-01 | 8JUX | - | |
| 27 | A5I | SUb2 | 7QCH | [55] |
| 28 | T2 | 7QCI | [55] | |
| 29 | A7L | 7QCK | [55] | |
| 30 | A4O | 7QCJ | [55] | |
| 31 | A3X | 7QCM | [55] | |
| 32 | YRL | 7OFS | [78] | |
| 33 | HBA | 7OFT | [78] |
| Structure in MD Simulation | Representative Structure | Box Dimension (Å) |
|---|---|---|
| PDB ID 7LBR | R1 | 19.875 × 23.625 × 24.375 |
| R2 | 22.125 × 23.625 × 22.875 | |
| R3 | 25.875 × 26.625 × 21.375 | |
| PDB ID 7QCI | R1 | 18.375 × 22.125 × 22.125 |
| R2 | 18.375 × 19.875 × 23.625 | |
| R3 | 20.625 × 22.875 × 22.125 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varghese, A.; Liu, J.; Patterson, T.A.; Hong, H. Integrating Molecular Dynamics, Molecular Docking, and Machine Learning for Predicting SARS-CoV-2 Papain-like Protease Binders. Molecules 2025, 30, 2985. https://doi.org/10.3390/molecules30142985
Varghese A, Liu J, Patterson TA, Hong H. Integrating Molecular Dynamics, Molecular Docking, and Machine Learning for Predicting SARS-CoV-2 Papain-like Protease Binders. Molecules. 2025; 30(14):2985. https://doi.org/10.3390/molecules30142985
Chicago/Turabian StyleVarghese, Ann, Jie Liu, Tucker A. Patterson, and Huixiao Hong. 2025. "Integrating Molecular Dynamics, Molecular Docking, and Machine Learning for Predicting SARS-CoV-2 Papain-like Protease Binders" Molecules 30, no. 14: 2985. https://doi.org/10.3390/molecules30142985
APA StyleVarghese, A., Liu, J., Patterson, T. A., & Hong, H. (2025). Integrating Molecular Dynamics, Molecular Docking, and Machine Learning for Predicting SARS-CoV-2 Papain-like Protease Binders. Molecules, 30(14), 2985. https://doi.org/10.3390/molecules30142985






