Abstract
Colistin has re-emerged as a last-resort antibiotic for treating infections caused by multidrug-resistant (MDR) Gram-negative bacilli (GNB). However, increasing resistance threatens its efficacy. This study aimed to evaluate colistin resistance trends among clinical isolates of Gram-negative bacilli isolated over a five-year period at a large Emergency Hospital in North-Eastern Romania. A total of 23,143 GNB strains were isolated during the study period, including 14,531 Enterobacterales and 8294 non-fermenting Gram-negative bacilli. The percentage of colistin-resistant strains among those analyzed was 3.98%. Species-specific analysis focused on Klebsiella spp., Escherichia coli, Enterobacter spp., Citrobacter spp., Pseudomonas spp., and Acinetobacter spp. Klebsiella spp. exhibited the highest prevalence of colistin resistance, accounting for over 80% of all colistin-resistant strains, with annual resistance rates fluctuating between 12.97% and 21.64%. Colistin resistance among E. coli was low (0.18–1.25%). Citrobacter spp. showed no resistance in the last three years of the study, and Enterobacter spp. maintained relatively stable resistance (3–5%). Resistance in Pseudomonas spp. remained below 1%, while Acinetobacter spp. showed a resistance rate of 5.43%. Several distinct resistance phenotypes were identified among Klebsiella spp., Pseudomonas spp., and Acinetobacter spp. strains, reflecting both endemic and sporadic circulation patterns. The study highlights a persistent presence of colistin resistance, especially in Klebsiella spp., underlining the importance of ongoing surveillance. Despite low resistance in other species, the emergence of resistant strains underscores the need for robust antimicrobial stewardship and infection control policies.
1. Introduction
Antibiotic resistance represents one of the greatest challenges of modern medicine. Among these, resistance to colistin in Gram-negative bacilli has emerged as a particularly alarming issue. The resurgence in the use of colistin has been necessitated by the increasing prevalence of multidrug-resistant Gram-negative pathogens. In recent years, the rate of antibiotic resistance has grown significantly, contributing to therapeutic failure in the treatment of severe infections.
The swift dissemination of multidrug-resistant (MDR) bacteria—including carbapenem-resistant Enterobacterales, Acinetobacter baumannii, and Pseudomonas aeruginosa has emerged as a significant public health concern, particularly in countries where the spread of carbapenem-resistant organisms has become endemic [1,2]. Carbapenemase-producing Enterobacterales (CRE) represent a major public health concern, as these organisms either produce carbapenemases—enzymes belonging to the β-lactamase class that hydrolyze carbapenem antibiotics—or exhibit acquired resistance to carbapenems despite lacking intrinsic mechanisms [3,4]. Given the role of carbapenems as last-line agents for multidrug-resistant infections, the increasing prevalence of CRE has led to a greater reliance on colistin as salvage therapy [5]. This shift has intensified selective pressure, thereby contributing to the emergence and dissemination of colistin-resistant strains, further limiting therapeutic options and complicating infection control efforts [3].
Although carbapenem resistance in Enterobacterales can arise through various mechanisms, the most prevalent is the production of carbapenemase enzymes, which efficiently hydrolyze carbapenem antibiotics [1,6,7]. These carbapenemase-producing strains utilize diverse molecular strategies to inactivate antimicrobial agents [4]. In addition to enzymatic degradation, other notable resistance mechanisms include the overexpression of efflux pumps and alterations in outer membrane porins, which reduce antibiotic permeability and hinder the drugs from reaching their intracellular targets [3].
The resistance mechanisms exhibited by MDR or XDR (extensively drug-resistant) Acinetobacter baumannii complex—recognized as a critical priority pathogen by the World Health Organization [8]—commonly involve reduced outer membrane permeability due to porin loss, constitutive overexpression of efflux pump systems, and the production of various β-lactamases [9].
Pseudomonas aeruginosa exhibits multiple resistance mechanisms, including carbapenemase production (notably VIM and OXA types), overexpression of AmpC β-lactamases, and increased activity of efflux pumps, which collectively contribute to its resistance to imipenem and other β-lactam antibiotics [10].
The extensive application of colistin in clinical practice and agriculture has facilitated the global spread of the mobile colistin resistance gene mcr-1 [11]. Colistin’s bactericidal effect is mediated by electrostatic interactions between its positively charged residues and the negatively charged lipid A of lipopolysaccharide, resulting in disruption of the bacterial outer membrane [11].
Colistin, a pentacationic cyclic lipodecapeptide mainly consisting of colistin A and B, belongs to the polymyxin class of antibiotics. It is effective against most Gram-negative bacteria, except for those from the genera Proteus, Providencia, Morganella, Serratia, Edwardsiella, and Burkholderia, which exhibit intrinsic resistance. Its positive charge facilitates binding to the negatively charged lipopolysaccharide (LPS) in the outer membrane, displacing divalent cations and disrupting membrane stability, ultimately leading to bacterial cell death [12].
For several decades (1960–1990), colistin resistance was rarely observed. However, recent years have seen a notable rise in reports of the emergence, isolation, and dissemination of colistin-resistant Klebsiella pneumoniae (CRKP) in clinical settings across various countries in the Americas, Asia, and Europe [13,14,15,16,17,18]. The increased detection of CRKP has been largely attributed to the renewed clinical use of colistin and polymyxin B [19,20,21,22] as well as the multiclonal transmission of resistant strains among hospitalized patients [16,23]. While the development of CRKP is often linked to prior colistin exposure [21], cases have also been documented in patients with no history of colistin therapy [24,25].
Of particular concern is the rapid emergence of colistin-resistant Escherichia coli, which has gained global attention as a growing public health threat [26].
Several recent studies have reported that Enterobacter strains harbor carbapenemase genes in addition to exhibiting resistance to colistin [27,28,29].
Citrobacter spp. are Gram-negative bacteria commonly present in the environment and in the intestinal tracts of humans and animals. While they are typically susceptible to third-generation cephalosporins, carbapenems, and colistin, an increasing number of studies have reported the presence of antibiotic resistance genes in these organisms. This raises concerns that Citrobacter spp. may serve as a potential reservoir for the dissemination of antimicrobial resistance genes [30].
Colistin remains one of the few effective treatment options for life-threatening infections caused by multidrug- and extensively drug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa [31]. Furthermore, resistance to colistin is relatively uncommon and is primarily mediated by chromosomally encoded mechanisms, which limits its potential for horizontal gene transfer [32].
This study aimed to assess colistin resistance rates over the 2019–2023 period in the most frequently isolated Gram-negative bacilli strains—Klebsiella spp., Escherichia coli, Enterobacter spp., Citrobacter spp., Pseudomonas spp., and Acinetobacter spp.—in order to better understand their resistance profiles in hospitalized patients, and we also investigated the prevalence and distribution of multidrug-resistant (MDR) phenotypes among these isolates, providing a comprehensive overview of resistance patterns.
2. Results
2.1. Isolation and Identification
The results were analyzed by bacterial species, tracking the annual evolution of colistin resistance rates in comparison with other antimicrobial agents.
Between 2019 and 2023, a total of 23,143 Gram-negative bacilli (GNB) strains were isolated at “Sf. Spiridon” County Clinical Emergency Hospital in Iași, Romania. Of these, 14,849 were identified as Enterobacterales, and 8294 were non-fermenting Gram-negative bacilli. Out of the total, 4795 strains were classified as multidrug-resistant Gram-negative bacilli (MDR GNB).
This study illustrates the distribution of Gram-negative bacilli isolates analyzed between 2019 and 2023, with Escherichia coli (8167 isolates) being the most frequently identified organism, followed by Pseudomonas spp. (5964 isolates) and Klebsiella spp. (5390 isolates), all of which are major contributors to hospital-acquired infections. Acinetobacter spp. accounted for 2330 isolates and is particularly notable for its association with multidrug resistance, especially in intensive care settings. Enterobacter spp. and Citrobacter spp. were less frequently isolated, with 1116 and 176 isolates, respectively, though Citrobacter spp. remains clinically relevant in the context of resistant strains. Overall, the data show that E. coli, Pseudomonas, and Klebsiella species represent the majority of isolates, reflecting common patterns observed in clinical practice (Figure 1).
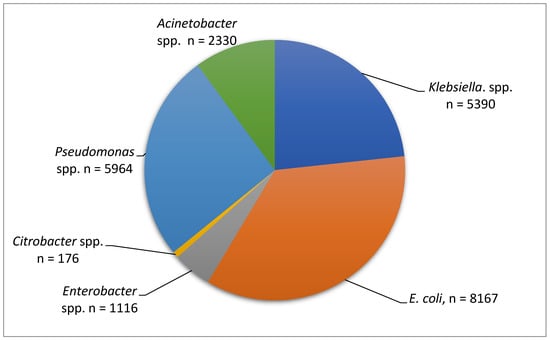
Figure 1.
Number of strains analyzed during the period 2019–2023.
Over the five-year period, the total number of MDR GNB isolates peaked in 2019 and reached its lowest in 2022. Among the individual species, Acinetobacter spp. consistently accounted for a substantial proportion of MDR isolates. In 2023, Acinetobacter spp. represented the highest proportion of MDR isolates relative to the total MDR count, surpassing Klebsiella, E. coli, and Pseudomonas spp. These trends suggest that while overall MDR rates have fluctuated, Acinetobacter spp. remains a dominant and persistent MDR pathogen, warranting continuous surveillance and targeted infection control strategies. At the opposite end of the spectrum, Citrobacter spp. exhibited extremely low MDR rates throughout the study period, with certain years registering no MDR isolates at all (Figure 2).
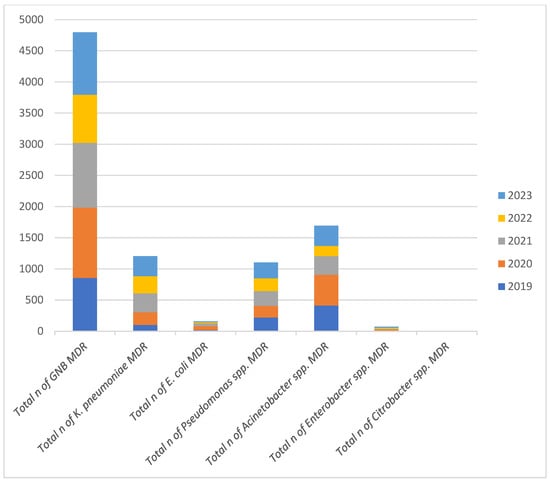
Figure 2.
Annual Distribution of Multidrug-Resistant Gram-Negative Bacilli Isolates (2019–2023). Note: n = number of strains.
The percentage of colistin-resistant strains among those analyzed was 3.98%. Among the Enterobacterales group, the highest number of colistin-resistant isolates were identified as Klebsiella spp. (80.98%), but resistance to colistin was also detected in Escherichia coli (4.80%), Enterobacter spp. (5.43%), and Citrobacter spp. (0.31%) (Figure 3).
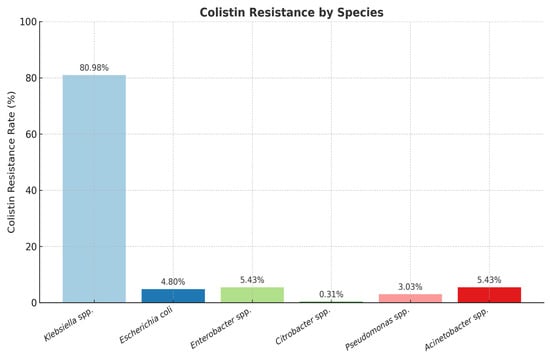
Figure 3.
Distribution of colistin-resistant Gram-negative strains. Note: The number of isolates per species differs and reflects the clinical frequency of isolation during the study period.
While species-specific isolate numbers vary, the data reflect the actual clinical distribution of multidrug-resistant Gram-negative bacilli during the study period in our hospital.
Colistin resistance among the analyzed strains varied but showed a slightly increasing annual trend over the study period across all included species.
The overwhelming majority of resistant isolates were identified as Klebsiella spp. (80.98%), highlighting their significant role in colistin resistance among clinical pathogens.
Other species showed considerably lower proportions, with Enterobacter spp. and Acinetobacter spp. each representing 5.43% of resistant strains.
Escherichia coli represented 4.80% of the colistin-resistant isolates, while Pseudomonas spp. and Citrobacter spp. accounted for 3.03% and 0.31%, respectively. These findings suggest that, although colistin resistance is distributed among various Gram-negative species, Klebsiella spp. remain the most significant source of resistance in our study (Figure 3).
2.2. Klebsiella spp.
Klebsiella spp. isolates exhibited a rising trend in resistance to ciprofloxacin and cephalosporins during the five-year analysis. Although a gradual increase in colistin resistance among Klebsiella spp. was observed over the study period, this trend did not reach statistical significance (p = 0.894), suggesting that the variation may be due to random fluctuation rather than a consistent upward pattern. Colistin-resistant Klebsiella spp. strains were additionally resistant to other classes of antibiotics, including carbapenems, leaving clinicians with very few therapeutic options available (Table 1).

Table 1.
Evolution of antibiotic resistance (%) in Klebsiella spp. strains between 2019 and 2023.
To assess the association between year of isolation and resistance rate for each antibiotic, p-values were calculated using the Pearson Chi-square test. A p-value < 0.05 was considered statistically significant.
Among all tested antibiotics, a statistically significant difference in resistance rates over the five-year period was observed only for meropenem (p < 0.05), indicating temporal variation. For the other antibiotics, the differences were not statistically significant (p > 0.05) (Table 1).
The annual assessment of colistin resistance in Klebsiella spp. isolates over the 2019–2023 period revealed the highest resistance rate in 2020 (21.64%), followed by a significant decline in 2021 to 12.97%. While the rates remained relatively stable in 2019 (12.99%) and 2021, a gradual increase was noted again in 2022 (14.06%) and 2023 (15.82%). Despite year-to-year fluctuations, the overall trend indicates that colistin resistance in Klebsiella spp. remained consistently present at concerning levels over the five-year period. (Table 1).
During the 2019–2023 study period, the number of Klebsiella spp. strains tested for colistin susceptibility increased steadily, ranging from 674 in 2019 to 1282 in 2023. Among these, the number of colistin-resistant strains also showed a gradual rise, from 76 in 2019 to 195 in 2023, reflecting a concerning upward trend in resistance (Figure 4).
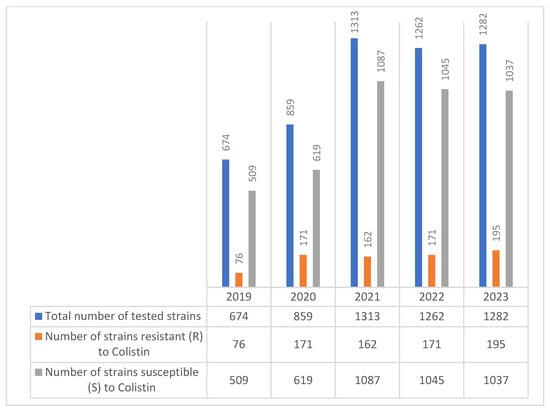
Figure 4.
Evolution of colistin resistance in Klebsiella spp. strains isolated during the 2019–2023 period.
Throughout the study, various MDR Klebsiella spp. phenotypes were repeatedly isolated, suggesting their ongoing endemic presence within the hospital environment (Table 2). Descriptive phenotypic resistance profiles were presented only for Klebsiella spp., Pseudomonas spp., and Acinetobacter spp., as these were the most frequently isolated species and posed the greatest therapeutic challenges, while less frequent isolates such as E. coli, Enterobacter spp., and Citrobacter spp. did not display complex resistance patterns warranting detailed phenotypic analysis. These strains were repeatedly isolated from multiple patients and from a broad spectrum of pathological specimens, including blood, respiratory secretions, urine, and wound exudates. Their recurrent detection across different clinical contexts suggests persistent transmission pathways and adaptation to selective antibiotic pressure within the healthcare setting. The phenotypes displayed limited susceptibility, with some strains remaining susceptible only to amikacin and trimethoprim-sulfamethoxazole while exhibiting intermediate susceptibility to imipenem.

Table 2.
Klebsiella spp. resistance phenotypes.
2.3. Escherichia coli
Over the five-year period, a total of 8167 E. coli strains were tested for colistin susceptibility. While the number of isolates tested increased steadily each year—from 1191 in 2019 to 2281 in 2023—the number of colistin-resistant strains remained low overall, ranging from just 3 cases in 2021 to a peak of 15 in 2019.
Despite the low absolute numbers of resistant strains, the data demonstrate sporadic detection of resistance across all years, with a slight resurgence in 2023 (11 resistant strains) (Figure 5).
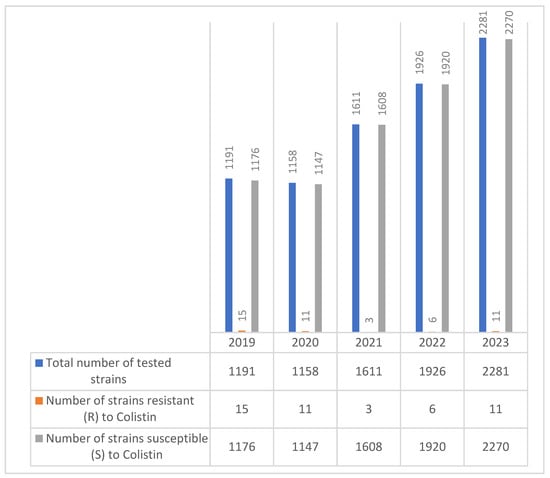
Figure 5.
Evolution of colistin resistance in E. coli strains isolated during the 2019–2023 period.
The colistin-resistant E. coli strains were characterized by susceptibility to other antibiotic classes, indicating that they were not multidrug-resistant (MDR). This pattern suggests a possible plasmid-mediated mechanism of colistin resistance.
In E. coli isolates, a statistically significant decreasing trend in resistance over the five-year period was observed for cefotaxime (p = 0.048), ceftriaxone (p = 0.024), and cefepime (p = 0.046), indicating a potential shift in susceptibility patterns to third- and fourth-generation cephalosporins.
For the remaining antibiotics, resistance rates remained relatively stable, with p- above the 0.05 threshold (Table 3).

Table 3.
Evolution of antibiotic resistance in Escherichia coli strains between 2019 and 2023.
Amoxicillin–clavulanic acid showed variable resistance, peaking in 2020 (39.6%) and decreasing to around 29% by 2023. This fluctuation may reflect shifts in prescription patterns or local strain dynamics (Table 3).
Third-generation cephalosporins (cefotaxime, ceftazidime, and ceftriaxone) showed a clear declining trend in resistance: cefotaxime dropped from 26% to 16.5%; ceftriaxone dropped from 33.3% to 16.75%. This suggests improved susceptibility, potentially due to reduced selection pressure or better control of ESBL-producing strains. Cefepime resistance also declined significantly, reaching 12.5% in 2023 from 19% in 2019 (Table 3).
Imipenem, meropenem, and ertapenem all maintained very low resistance rates, although slight increases were noted in some years (e.g., imipenem reaching 2.89% in 2023). These consistently low values support carbapenems’ continued effectiveness against E. coli, although caution is still warranted due to the potential emergence of carbapenemase-producing strains.
Ciprofloxacin resistance remained moderately high, with a slight decrease from 30.3% in 2020 to 21.7% in 2023. This reflects ongoing selection pressure from widespread fluoroquinolone use.
Colistin resistance remained very low across all years, decreasing from 1.25% in 2019 to 0.48% in 2023, supporting its role as a last-resort treatment.
Trimethoprim–sulfamethoxazole showed consistently high resistance, ranging from approximately 25% to 36%, limiting its utility for empirical treatment.
Nitrofurantoin, commonly used for urinary tract infections, showed very low resistance rates throughout the study period (2–3.75%), confirming its continued effectiveness against E. coli (Table 3).
These findings suggest that colistin resistance in E. coli remains relatively rare but must be closely monitored, especially given the observed fluctuation and potential for increased resistance in clinical environments.
2.4. Enterobacter spp.
Regarding the evolution of colistin resistance in Enterobacter spp. strains over the five-year period (2019–2023), a total of 1116 isolates were tested, with annual strain sizes ranging from 180 to 277. Colistin resistance was reported in each year, with the number of resistant strains varying between 9 and 13 annually (Figure 6).
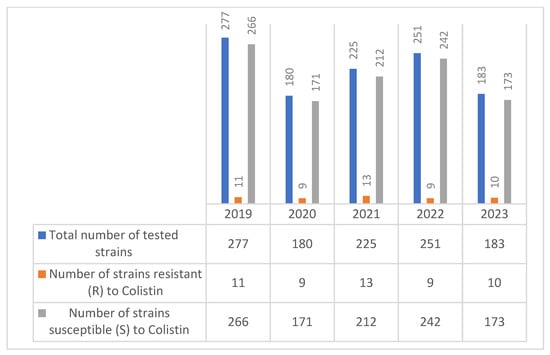
Figure 6.
Evolution of colistin resistance in Enterobacter spp. strains isolated during the 2019–2023 period.
The number of colistin-resistant strains remained relatively stable—peaking at 13 in 2021—and did not show a clear increasing or decreasing trend. Most Enterobacter spp. isolates remained susceptible to colistin.
This data suggests a persistent low-level presence of colistin resistance in Enterobacter spp.
Among the antibiotics tested, only ertapenem showed a statistically significant variation in resistance rates over the study period (p = 0.011), while for the others, the p-values were above the conventional threshold of 0.05, indicating no significant temporal trend (Table 4).

Table 4.
Evolution of antibiotic resistance in Enterobacter spp. strains between 2019 and 2023.
2.5. Citrobacter spp.
According to our study, the number of colistin-resistant Citrobacter spp. strains was low in 2019 and 2020 (2 strains and 1 strain, respectively), and starting from 2021, no colistin-resistant Citrobacter spp. strains were detected (Figure 7).
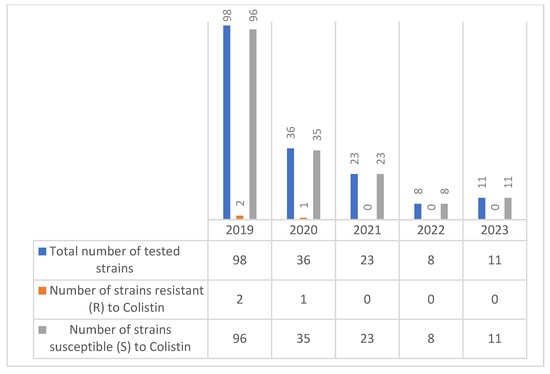
Figure 7.
Evolution of colistin resistance in Citrobacter spp. strains isolated during the period 2019–2023.
In Citrobacter spp., statistically significant changes in resistance rates over the 2019–2023 period were observed for ceftazidime (p = 0.031) and cefotaxime (p = 0.042), suggesting potential shifts in susceptibility patterns for these third-generation cephalosporins. For all other antibiotics, the p-values exceeded 0.05, indicating no significant temporal variation (Table 5).

Table 5.
Evolution of antibiotic resistance in Citrobacter spp. strains between 2019 and 2023.
Resistance to carbapenems was very low throughout the period, and by 2022 and 2023, no resistance was detected for imipenem, meropenem, and ertapenem. This suggests carbapenems remain largely effective against Citrobacter spp. in this clinical context.
The slight elevation in ertapenem resistance in 2021 (8%) could indicate a temporary emergence of a resistant subpopulation.
Colistin resistance was minimal, with low rates (2–2.7%) reported in 2019 and 2020. No resistance was detected in the last three years (2021–2023), indicating stable susceptibility and minimal selective pressure from colistin use among Citrobacter spp. isolates. Resistance levels gradually increased for cefotaxime and ceftazidime, with the most pronounced rise seen in ceftriaxone, reaching 50% in 2023.
Cefepime resistance, while initially low (4% in 2019), appears to have disappeared in recent years (0% in 2022 and 2023), possibly due to limited use.
Resistance to ciprofloxacin fluctuated over the years, with a peak of 28% in 2021, followed by a moderate decline. This variation may reflect shifting use patterns or the sporadic presence of resistant clones.
Amikacin resistance was present in the earlier years (6.5% in 2019, 8% in 2021) but dropped to 0% in 2022 and 2023, suggesting regained effectiveness and potential suitability as part of therapy for resistant Citrobacter infections (Table 5).
During the analyzed period, colistin resistance was also recorded in non-fermenting Gram-negative bacilli strains. In this study, a particular feature of colistin resistance within the hospital was that resistance was initially detected in Enterobacterales strains, followed by its emergence in non-fermenting Gram-negative bacilli.
2.6. Pseudomonas spp.
Pseudomonas aeruginosa colistin-resistant strains were first identified in our hospital in 2019 in small numbers, with a stable trend throughout the study period, peaking at 11 strains (0.67%) in 2023 (Figure 8).
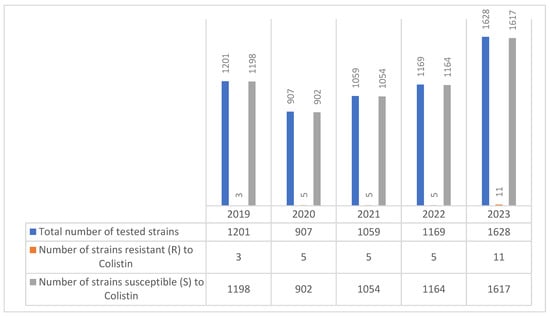
Figure 8.
Evolution of colistin resistance in Pseudomonas spp. strains isolated during the period 2019–2023.
A notable finding was that all strains were susceptible to ceftazidime-avibactam. Among the aminoglycosides, amikacin and tobramycin showed moderate efficacy, with susceptibility rates ranging between 48 and 53%. Resistance rates in Pseudomonas spp. were also recorded for cephalosporins and beta-lactamase inhibitors.
The resistance profile of Pseudomonas spp. over a five-year period highlights several clinically significant trends (Table 6).

Table 6.
Evolution of antibiotic resistance in Pseudomonas spp. strains between 2019 and 2023.
Fluoroquinolones, particularly ciprofloxacin and levofloxacin, exhibited persistently high resistance rates, starting at approximately 60% in 2019 and gradually decreasing to approximately 41.5% and 38%, respectively, by 2023. Although a slight decrease was observed, resistance rates remain considerably high, indicating a reduced effectiveness of this antibiotic class for empirical therapy. Resistance to β-lactams, including piperacillin-tazobactam, ceftazidime, and cefepime, showed a notable decline across the study period. For example:
- Piperacillin-tazobactam resistance decreased from 42% in 2019 to 30% in 2023.
- Ceftazidime dropped from 53.5% to 29%.
- Cefepime fell from 53.8% to 20%. This trend may reflect changes in local prescribing practices or the displacement of resistant clones by more susceptible strains.
Carbapenem resistance (imipenem and meropenem) remained worryingly high, with initial rates exceeding 48% and decreasing only modestly to 38% and 30.8%, respectively, by 2023. These figures suggest persistent carbapenemase activity and ongoing transmission of carbapenem-resistant Pseudomonas strains.
Amikacin showed variable resistance, notably dropping to just 5% in 2020 but rising again in later years, suggesting episodic use or selective pressure shifts.
Ceftazidime-avibactam resistance decreased significantly from 30.9% in 2019 to 12.9% in 2023, supporting its role as a useful option for MDR Pseudomonas, especially given its activity against β-lactamase-producing strains.
Colistin exhibited very low resistance rates, remaining below 1% throughout the study period, underscoring its role as a last-resort therapeutic option. Nevertheless, even low levels of resistance call for caution given the limited treatment alternatives for MDR infections.
Several antibiotics tested against Pseudomonas spp. showed statistically significant changes in resistance rates over the 2019–2023 period, with p-values below 0.05 observed for piperacillin–tazobactam, ceftazidime, cefepime, ciprofloxacin, levofloxacin, and meropenem. These findings suggest a dynamic evolution of antimicrobial resistance in this species.
We identified several MDR Pseudomonas spp. phenotypes that circulated endemically, having been isolated from multiple patients (Table 7).

Table 7.
Pseudomonas spp. resistance phenotypes.
2.7. Acinetobacter spp.
During the period 2019–2023, 2330 MDR Acinetobacter spp. were isolated, with 52 (5.43%) showing resistance to colistin (Figure 9).
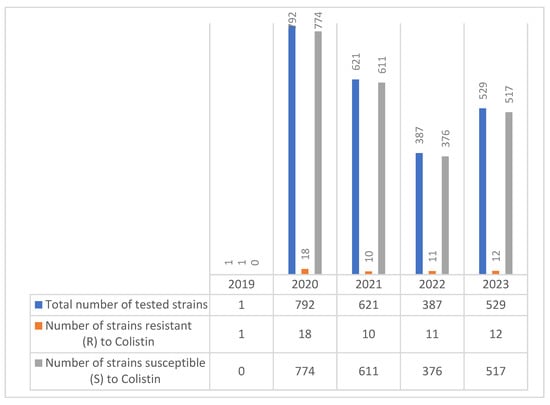
Figure 9.
Colistin resistance in Acinetobacter spp. strains during the 2019–2023 period.
The correlation analysis between year and resistance rates in Acinetobacter spp. isolates did not reveal any statistically significant temporal trends for any of the antibiotics tested (Table 8). Based on the data analyzed, Acinetobacter spp. strains showed marked resistance to meropenem, fluoroquinolones, and cephalosporins, while lower resistance rates were observed for colistin.

Table 8.
Evolution of antibiotic resistance in Acinetobacter spp. strains between 2019 and 2023 period.
Colistin has proven effective in the treatment of infections caused by MDR A. baumannii strains, with over 97% of Acinetobacter spp. strains remaining susceptible to colistin in each of the five years studied (Table 8).
A variety of resistance phenotypes were observed among Acinetobacter spp. isolates, ranging from strains susceptible only to colistin to others retaining susceptibility to multiple agents such as tobramycin, amikacin, and trimethoprim-sulfamethoxazole (Table 9).

Table 9.
Acinetobacter spp. resistance phenotypes.
3. Discussion
The emergence and progression of antimicrobial resistance among Gram-negative bacilli, particularly colistin resistance, present a growing challenge in the clinical management of healthcare-associated infections. This five-year study offers a comprehensive overview of resistance trends in six key bacterial species: Klebsiella spp., Escherichia coli, Enterobacter spp., Citrobacter spp., Pseudomonas spp., and Acinetobacter spp., isolated from hospitalized patients in a tertiary care setting. Klebsiella spp. demonstrated the highest rate of colistin resistance among all tested isolates, representing more than 80% of the total colistin-resistant strains. This observation is consistent with global data, which highlight Klebsiella pneumoniae as a key contributor to colistin resistance, frequently associated with multidrug-resistant or extensively drug-resistant profiles. The high rates of resistance to carbapenems, cephalosporins, and fluoroquinolones further underscore the therapeutic difficulties posed by these isolates.
Our findings indicate that colistin resistance among Klebsiella spp. isolates in our hospital ranged between approximately 13% and 21.6% during the 2019–2023 period, with the highest rate recorded in 2020 (21.64%). These values significantly exceed those observed in earlier epidemiological studies. For instance, in Tunisia, resistance increased from 0.4% to 1.9% between 2005 and 2009 [33], while in Europe, resistance rates rose from 1.1% to 2.2% over the same period [33]. More recent surveillance data have shown elevated rates in specific countries, with Romania reporting up to 25.8%, followed by Greece (19.9%) and Italy (15.4%) [34]. Although the years of analysis differ, the comparison underscores a concerning pattern of colistin resistance emergence and persistence, particularly in regions with high antibiotic selection pressure.
Furthermore, more recent reports have documented a significant escalation in colistin resistance among carbapenem-resistant K. pneumoniae, with rates ranging between 27% and 71% in various regions across Asia, South Africa, and South America [35].
In contrast, Escherichia coli strains showed notably lower rates of colistin resistance, with most resistant strains exhibiting susceptibility to other antibiotic classes. This susceptibility profile suggests that plasmid-mediated resistance mechanisms, such as those associated with mcr genes, may be involved, rather than chromosomal mutations typically linked with broader resistance. Although rare, the presence of such strains warrants ongoing monitoring due to their potential for horizontal gene transfer and wider dissemination [36,37,38].
The predominance of colistin resistance among K. pneumoniae isolates in our hospital, compared to E. coli, aligns with findings from studies conducted in other countries, including China, Iran, and India [39,40,41].
Enterobacter spp. and Citrobacter spp. displayed low and relatively stable rates of colistin resistance throughout our study period. The absence of colistin resistance in Citrobacter spp. from 2021 onward is encouraging and may reflect limited selective pressure or effective antimicrobial stewardship.
The results of our study indicate that, over the five-year period (2019–2023), colistin resistance among Enterobacter spp. remained relatively stable, with annual resistance counts ranging from 9 to 13 out of a total of 1116 tested strains—corresponding to yearly resistance rates of approximately 3% to 5%. This persistent yet moderate level of resistance did not show a clear upward or downward trend over time. These findings are in line with those reported by Mirelis et al., who found that while the overall colistin resistance rate among Enterobacteriaceae was 0.67%, Enterobacter cloacae exhibited a considerably higher resistance rate of 4.2%, exceeding those of Escherichia coli and Klebsiella pneumoniae [42].
Throughout the five-year observation period, our findings revealed that colistin resistance among Citrobacter spp. remained exceptionally low, with only isolated cases identified in 2019 and 2020 (2–2.7%), and no resistant strains detected in the subsequent years (2021–2023). This trend suggests stable susceptibility and limited selective pressure associated with colistin use in our clinical setting. In contrast, a recent study by Puljko et al., conducted in 2024, reported a higher resistance rate of 5% among Citrobacter isolates, indicating possible regional or institutional differences in antimicrobial usage patterns or resistance dynamics [43].
Pseudomonas spp. and Acinetobacter spp. remain prominent nosocomial pathogens due to their intrinsic resistance mechanisms and ability to acquire additional resistance determinants [44].
Our data show consistently low colistin resistance in Pseudomonas spp. isolates, remaining below 1% throughout 2019–2023. This finding reinforces the continued efficacy of colistin as a last-line therapeutic agent in our institution. However, even minimal resistance remains a cause for concern due to the limited treatment alternatives available for multidrug-resistant infections. In contrast, international data reveal a more variable resistance landscape. For instance, a recent study in Ardabil, Iran, reported a colistin resistance rate of 9% in P. aeruginosa, marking the first such report from that region [45]. Other studies across Iran and globally have documented resistance rates ranging from 0% to over 21%—including 21.3% in Egypt, 6.3% in Brazil, 7.4% in South Korea, and 7.2% in Peru [46,47,48,49]. Lower resistance rates were reported in regions such as Shiraz, Iran (0%), and Tehran, Iran (0%), whereas other Iranian cities like Isfahan and Hamadan reported rates of 3.9% to 7% [50,51,52,53,54,55]. These discrepancies underscore the influence of local antimicrobial stewardship practices, infection control measures, and selective pressure on colistin resistance dynamics.
Although colistin resistance remained low, below 1% for Pseudomonas spp. and around 2–3% for Acinetobacter spp., the high resistance rates to carbapenems and fluoroquinolones observed in our study are concerning and underscore the ongoing challenges in managing infections caused by multidrug-resistant Gram-negative bacilli. Notably, Pseudomonas spp. showed a gradual improvement in susceptibility to β-lactams, possibly reflecting shifts in antimicrobial usage or infection control successes. Meanwhile, Acinetobacter spp. exhibited persistently high resistance to nearly all tested antibiotics, reinforcing the clinical reliance on colistin as a last-resort agent.
Among the Acinetobacter spp. isolates analyzed in our research, only 52 out of 2330 strains were resistant to colistin, reflecting a relatively low overall resistance rate of 5.43%. Annual resistance rates fluctuated between 0% and 2.84%, suggesting limited but persistent occurrence of colistin resistance within this group of multidrug-resistant pathogens. Notably, over 97% of Acinetobacter spp. strains remained susceptible to colistin in each of the five years analyzed, underscoring its continued clinical utility in our setting.
These findings are in line with earlier data from Mohammadi et al. [56], who reported a 2% resistance rate in a 2017 meta-analysis of 448 A. baumannii strains. However, more recent studies have demonstrated increasing resistance rates globally. A meta-analysis by Ciftci et al. in Turkey revealed a rise in colistin resistance from 2.94% (2011–2015) to 13.42% (2016–2020), with an overall resistance rate of 7.9% [57]. Additionally, individual reports from India, Pakistan, Turkey, and Sweden have shown even higher colistin resistance rates of 10.1%, 9.6%, 28%, and 36.36%, respectively [58,59,60,61].
In addition to colistin resistance, our data revealed complex and diverse MDR phenotypes among Klebsiella spp., Pseudomonas spp., and Acinetobacter spp. isolates. The presence of strains resistant to nearly all tested antibiotic classes, including carbapenems, aminoglycosides, and fluoroquinolones, underscores the growing challenge of treating infections caused by these pathogens. Particularly concerning were Klebsiella spp. phenotypes that remained susceptible only to last-resort agents such as colistin. These patterns are consistent with known resistance mechanisms such as carbapenemase production (e.g., KPC, OXA-48), efflux pump overexpression, and porin loss [62].
The clinical implications are significant, as treatment options become increasingly limited, often requiring combination therapies or off-label use of less validated agents. These findings align with international reports and highlight the importance of local surveillance data to guide empirical therapy and stewardship efforts.
Overall, our study reveals that while colistin resistance remains relatively low in most Gram-negative species (except Klebsiella spp.), the threat of emerging resistance persists.
4. Materials and Methods
4.1. Isolation and Identification
A retrospective-prospective study was conducted over a five-year period (2019–2023) at “Sf. Spiridon” County Clinical Emergency Hospital in Iași, Romania. During this time, a total of 23,143 Gram-negative bacilli (GNB) strains were isolated, of which 14,849 were classified as Enterobacterales and 8294 as non-fermenting GNB. Out of the total GNB isolates, 4795 strains were identified as MDR, and 910 were colistin-resistant. All isolates were identified using matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI-TOF MS) (Bruker Daltonik GmbH, Bremen, Germany) or conventional biochemical assays.
All clinical specimens analyzed in this study were obtained from hospitalized patients as part of standard diagnostic and therapeutic protocols at “Sf. Spiridon” County Clinical Emergency Hospital, Iași, Romania. No additional sampling was performed for research purposes. The analysis was conducted exclusively on samples routinely submitted to the microbiology laboratory, which included wound cultures, endotracheal aspirates, urine, blood, serous fluids, abscess contents, and other pathological specimens. These were collected from a broad range of hospital departments, including internal medicine wards, surgical units, and intensive care departments.
4.2. Antimicrobial Susceptibility
Antimicrobial susceptibility testing was performed using the broth microdilution method, in accordance with the EUCAST guidelines applicable at the time of testing (versions 9.0–13.0, 2019–2023), using an automated system (MICRONAUT-S, Merlin, Bornheim, Germany). For colistin, susceptibility testing was specifically conducted using the standard broth microdilution method, the only method recommended by EUCAST and CLSI for reliable results. Minimum inhibitory concentrations (MICs) were interpreted according to EUCAST clinical breakpoints. For quality control, standard reference strains such as Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were included in each testing series [63,64,65,66,67]. Antibiogram results were analyzed using the Statistical Package for the Social Sciences (SPSS® Statistics, version 25; IBM Corp., Armonk, NY, USA). Associations between the year of isolation and resistance rates for each antibiotic were evaluated using the Pearson Chi-square test. A p-value of <0.05 was considered indicative of statistical significance.
For colistin, both the Clinical and Laboratory Standards Institute (CLSI) and EUCAST recognize broth microdilution, performed in accordance with ISO 20776 [68], as the gold standard method for susceptibility testing [69].
In our study, the definition of multidrug resistance (MDR) follows the criteria proposed by Magiorakos et al. in collaboration with the European Centre for Disease Prevention and Control (ECDC) and the Centers for Disease Control and Prevention (CDC). An isolate was considered MDR if it showed acquired non-susceptibility to at least one agent in three or more antimicrobial categories relevant to that species [70].
5. Conclusions
This study highlights the emergence and progressive increase in colistin resistance, as well as resistance to commonly used antibiotics among Gram-negative bacilli strains. Within the framework of this study, we provide a comprehensive overview of antibiotic resistance in the North-East region of Romania.
The analysis of the collected data allowed us to evaluate the current profile of colistin resistance among clinical isolates of Gram-negative bacilli.
Our research emphasizes the importance of assessing the local epidemiological context and the need for continuous monitoring of antibiotic resistance, with the aim of ensuring rapid diagnosis and appropriate antimicrobial prescription.
Moreover, the study reveals a diverse range of multidrug-resistant (MDR) phenotypes circulating in our region, particularly among Klebsiella spp., Pseudomonas spp., and Acinetobacter spp., underscoring the therapeutic challenges posed by these pathogens.
Author Contributions
Conceptualization, C.G.T. and M.A.V.; Methodology, M.A.V., C.G.T. and M.D.; Data extraction, syntesis and interpretation M.A.V., C.G.T., M.D., S.D. and A.N.C.; Writing-original draft preparation, M.A.V. and C.G.T.; Writing-review and editing M.A.V. and C.G.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by a Doctoral Grant from the University of Medicine and Pharmacy ‘Grigore T. Popa’, Iași, Romania.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee the University of Medicine and Pharmacy “Grigore T. Popa” Iasi, Romania (IRB number: 99) and by the Hospital Ethics Committee (IRB number: 30).
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Munoz-Price, L.S.; Poirel, L.; Bonomo, R.A.; Schwaber, M.J.; Daikos, G.L.; Cormican, M.; Cornaglia, G.; Garau, J.; Gniadkowski, M.; Hayden, M.K.; et al. Clinical Epidemiology of the Global Expansion of Klebsiella Pneumoniae Carbapenemases. Lancet Infect. Dis. 2013, 13, 785–796. [Google Scholar] [CrossRef]
- Maragakis, L.L.; Perl, T.M. Antimicrobial Resistance: Acinetobacter baumannii: Epidemiology, Antimicrobial Resistance, and Treatment Options. Clin. Infect. Dis. 2008, 46, 1254–1263. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Tamma, P.D.; Aitken, S.L.; Bonomo, R.A.; Mathers, A.J.; van Duin, D.; Clancy, C.J. Infectious Diseases Society of America 2023 Guidance on the Treatment of Antimicrobial Resistant Gram-Negative Infections. Clin. Infect. Dis. 2023. ahead of print. [Google Scholar] [CrossRef]
- Vardakas, K.Z.; Tansarli, G.S.; Rafailidis, P.I.; Falagas, M.E. Carbapenems versus Alternative Antibiotics for the Treatment of Bacteraemia Due to Enterobacteriaceae Producing Extended-Spectrum-Lactamases: A Systematic Review and Meta-Analysis. J. Antimicrob. Chemother. 2012, 67, 2793–2803. [Google Scholar] [CrossRef]
- Codjoe, F.; Donkor, E. Carbapenem Resistance: A Review. Med. Sci. 2017, 6, 1. [Google Scholar] [CrossRef]
- Tzouvelekis, L.S.; Markogiannakis, A.; Psichogiou, M.; Tassios, P.T.; Daikos, G.L. Carbapenemases in Klebsiella Pneumoniae and Other Enterobacteriaceae: An Evolving Crisis of Global Dimensions. Clin. Microbiol. Rev. 2012, 25, 682–707. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Piperaki, E.-T.; Tzouvelekis, L.S.; Miriagou, V.; Daikos, G.L. Carbapenem-Resistant Acinetobacter Baumannii: In Pursuit of an Effective Treatment. Clin. Microbiol. Infect. 2019, 25, 951–957. [Google Scholar] [CrossRef] [PubMed]
- Kao, C.-Y.; Chen, S.-S.; Hung, K.-H.; Wu, H.-M.; Hsueh, P.-R.; Yan, J.-J.; Wu, J.-J. Overproduction of Active Efflux Pump and Variations of OprD Dominate in Imipenem-Resistant Pseudomonas Aeruginosa Isolated from Patients with Bloodstream Infections in Taiwan. BMC Microbiol. 2016, 16, 107. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Brunel, J.-M.; Dubus, J.-C.; Reynaud-Gaubert, M.; Rolain, J.-M. Colistin: An Update on the Antibiotic of the 21st Century. Expert Rev. Anti Infect. Ther. 2012, 10, 917–934. [Google Scholar] [CrossRef] [PubMed]
- Mammina, C.; Bonura, C.; Di Bernardo, F.; Aleo, A.; Fasciana, T.; Sodano, C.; Saporito, M.A.; Verde, M.S.; Tetamo, R.; Palma, D.M. Ongoing Spread of Colistin-Resistant Klebsiella Pneumoniae in Different Wards of an Acute General Hospital, Italy, June to December 2011. Eurosurveillance 2012, 17, 202248. [Google Scholar] [CrossRef]
- Bogdanovich, T.; Adams-Haduch, J.M.; Tian, G.-B.; Nguyen, M.H.; Kwak, E.J.; Muto, C.A.; Doi, Y. Colistin-Resistant, Klebsiella Pneumoniae Carbapenemase (KPC)-Producing Klebsiella Pneumoniae Belonging to the International Epidemic Clone ST258. Clin. Infect. Dis. 2011, 53, 373–376. [Google Scholar] [CrossRef]
- Suh, J.-Y.; Son, J.S.; Chung, D.R.; Peck, K.R.; Ko, K.S.; Song, J.-H. Nonclonal Emergence of Colistin-Resistant Klebsiella Pneumoniae Isolates from Blood Samples in South Korea. Antimicrob. Agents Chemother. 2010, 54, 560–562. [Google Scholar] [CrossRef]
- Marchaim, D.; Chopra, T.; Pogue, J.M.; Perez, F.; Hujer, A.M.; Rudin, S.; Endimiani, A.; Navon-Venezia, S.; Hothi, J.; Slim, J.; et al. Outbreak of Colistin-Resistant, Carbapenem-Resistant Klebsiella pneumoniae in Metropolitan Detroit, Michigan. Antimicrob. Agents Chemother. 2011, 55, 593–599. [Google Scholar] [CrossRef]
- Kontopoulou, K.; Protonotariou, E.; Vasilakos, K.; Kriti, M.; Koteli, A.; Antoniadou, E.; Sofianou, D. Hospital Outbreak Caused by Klebsiella Pneumoniae Producing KPC-2 β-Lactamase Resistant to Colistin. J. Hosp. Infect. 2010, 76, 70–73. [Google Scholar] [CrossRef]
- Arduino, S.M.; Quiroga, M.P.; Ramírez, M.S.; Merkier, A.K.; Errecalde, L.; Di Martino, A.; Smayevsky, J.; Kaufman, S.; Centrón, D. Transposons and Integrons in Colistin-Resistant Clones of Klebsiella Pneumoniae and Acinetobacter Baumannii with Epidemic or Sporadic Behaviour. J. Med. Microbiol. 2012, 61, 1417–1420. [Google Scholar] [CrossRef]
- Cannatelli, A.; D’Andrea, M.M.; Giani, T.; Di Pilato, V.; Arena, F.; Ambretti, S.; Gaibani, P.; Rossolini, G.M. In Vivo Emergence of Colistin Resistance in Klebsiella Pneumoniae Producing KPC-Type Carbapenemases Mediated by Insertional Inactivation of the PhoQ/PhoP MgrB Regulator. Antimicrob. Agents Chemother. 2013, 57, 5521–5526. [Google Scholar] [CrossRef]
- Lopez-Camacho, E.; Gomez-Gil, R.; Tobes, R.; Manrique, M.; Lorenzo, M.; Galvan, B.; Salvarelli, E.; Moatassim, Y.; Salanueva, I.J.; Pareja, E.; et al. Genomic Analysis of the Emergence and Evolution of Multidrug Resistance during a Klebsiella Pneumoniae Outbreak Including Carbapenem and Colistin Resistance. J. Antimicrob. Chemother. 2014, 69, 632–636. [Google Scholar] [CrossRef]
- Strenger, V.; Gschliesser, T.; Grisold, A.; Zarfel, G.; Feierl, G.; Masoud, L.; Hoenigl, M.; Resch, B.; Müller, W.; Urlesberger, B. Orally Administered Colistin Leads to Colistin-Resistant Intestinal Flora and Fails to Prevent Faecal Colonisation with Extended-Spectrum β-Lactamase-Producing Enterobacteria in Hospitalised Newborns. Int. J. Antimicrob. Agents 2011, 37, 67–69. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kontopidou, F.; Plachouras, D.; Papadomichelakis, E.; Koukos, G.; Galani, I.; Poulakou, G.; Dimopoulos, G.; Antoniadou, A.; Armaganidis, A.; Giamarellou, H. Colonization and Infection by Colistin-Resistant Gram-Negative Bacteria in a Cohort of Critically Ill Patients. Clin. Microbiol. Infect. 2011, 17, E9–E11. [Google Scholar] [CrossRef] [PubMed]
- Antoniadou, A.; Kontopidou, F.; Poulakou, G.; Koratzanis, E.; Galani, I.; Papadomichelakis, E.; Kopterides, P.; Souli, M.; Armaganidis, A.; Giamarellou, H. Colistin-Resistant Isolates of Klebsiella Pneumoniae Emerging in Intensive Care Unit Patients: First Report of a Multiclonal Cluster. J. Antimicrob. Chemother. 2007, 59, 786–790. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Hu, F.; Zhang, X.; Xu, X.; Liu, Y.; Zhu, D.; Wang, H. Independent Emergence of Colistin-Resistant Enterobacteriaceae Clinical Isolates without Colistin Treatment. J. Clin. Microbiol. 2011, 49, 4022–4023. [Google Scholar] [CrossRef]
- Tóth, Á.; Damjanova, I.; Puskás, E.; Jánvári, L.; Farkas, M.; Dobák, A.; Böröcz, K.; Pászti, J. Emergence of a Colistin-Resistant KPC-2-Producing Klebsiella Pneumoniae ST258 Clone in Hungary. Eur. J. Clin. Microbiol. Infect. Dis. 2010, 29, 765–769. [Google Scholar] [CrossRef]
- Li, X.; Hu, H.; Zhu, Y.; Wang, T.; Lu, Y.; Wang, X.; Peng, Z.; Sun, M.; Chen, H.; Zheng, J.; et al. Population Structure and Antibiotic Resistance of Swine Extraintestinal Pathogenic Escherichia Coli from China. Nat. Commun. 2024, 15, 5811. [Google Scholar] [CrossRef]
- Tarumoto, N.; Kodana, M.; Watanabe, N.; Sakai, J.; Imai, K.; Yamaguchi, T.; Taji, Y.; Ebihara, Y.; Murakami, T.; Maeda, T.; et al. First Report of the Isolation of BlaIMI-1-Producing Colistin-Heteroresistant Enterobacter Cloacae in Japan, September 2016. J. Infect. Chemother. 2018, 24, 941–943. [Google Scholar] [CrossRef]
- Uechi, K.; Tada, T.; Shimada, K.; Nakasone, I.; Kirikae, T.; Fujita, J. Emergence of a Carbapenem-Resistant and Colistin-Heteroresistant Enterobacter Cloacae Clinical Isolate in Japan. J. Infect. Chemother. 2019, 25, 285–288. [Google Scholar] [CrossRef]
- Chavda, B.; Lv, J.; Hou, M.; Chavda, K.D.; Kreiswirth, B.N.; Feng, Y.; Chen, L.; Yu, F. Coidentification of Mcr-4.3 and Bla NDM-1 in a Clinical Enterobacter Cloacae Isolate from China. Antimicrob. Agents Chemother. 2018, 62, e00649. [Google Scholar] [CrossRef]
- Phuadraksa, T.; Wichit, S.; Songtawee, N.; Tantimavanich, S.; Isarankura-Na-Ayudhya, C.; Yainoy, S. Emergence of Plasmid-Mediated Colistin Resistance Mcr-3.5 Gene in Citrobacter Amalonaticus and Citrobacter Sedlakii Isolated from Healthy Individual in Thailand. Front. Cell. Infect. Microbiol. 2023, 12, 1067572. [Google Scholar] [CrossRef]
- Bialvaei, A.Z.; Samadi Kafil, H. Colistin, Mechanisms and Prevalence of Resistance. Curr. Med. Res. Opin. 2015, 31, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Olaitan, A.O.; Morand, S.; Rolain, J.-M. Mechanisms of Polymyxin Resistance: Acquired and Intrinsic Resistance in Bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef] [PubMed]
- Mezghani Maalej, S.; Rekik Meziou, M.; Mahjoubi, F.; Hammami, A. Epidemiological Study of Enterobacteriaceae Resistance to Colistin in Sfax (Tunisia). Med. Mal. Infect. 2012, 42, 256–263. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. ECDC Surveillance Report: Antimicrobial Resistance Surveillance in Europe 2014; ECDC: Solna, Sweden, 2014; Available online: https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2017 (accessed on 7 July 2025).
- Binsker, U.; Käsbohrer, A.; Hammerl, J.A. Global Colistin Use: A Review of the Emergence of Resistant Enterobacterales and the Impact on Their Genetic Basis. FEMS Microbiol. Rev. 2022, 46, fuab049. [Google Scholar] [CrossRef]
- Trongjit, S.; Assavacheep, P.; Samngamnim, S.; My, T.H.; An, V.T.T.; Simjee, S.; Chuanchuen, R. Plasmid-Mediated Colistin Resistance and ESBL Production in Escherichia Coli from Clinically Healthy and Sick Pigs. Sci. Rep. 2022, 12, 2466. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Fu, Y.; Xiong, Z.; Ma, Y.; Wei, Y.; Qu, X.; Zhang, H.; Zhang, J.; Liao, M. Highly Prevalent Multidrug-Resistant Salmonella From Chicken and Pork Meat at Retail Markets in Guangdong, China. Front. Microbiol. 2018, 9, 2104. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Abbas, M.; Rehman, M.U.; Wang, M.; Jia, R.; Chen, S.; Liu, M.; Zhu, D.; Zhao, X.; Gao, Q.; et al. Updates on the Global Dissemination of Colistin-Resistant Escherichia Coli: An Emerging Threat to Public Health. Sci. Total Environ. 2021, 799, 149280. [Google Scholar] [CrossRef]
- Gysin, M.; Hon, P.Y.; Tan, P.; Sengduangphachanh, A.; Simmalavong, M.; Hinfonthong, P.; Kaewphanderm, N.; Pham, T.D.; Nguyen, T.H.; Haldimann, K.; et al. Apramycin Susceptibility of Multidrug-Resistant Gram-Negative Blood Culture Isolates in Five Countries in Southeast Asia. Int. J. Antimicrob. Agents 2022, 60, 106659. [Google Scholar] [CrossRef]
- Kaza, P.; Mahindroo, J.; Veeraraghavan, B.; Mavuduru, R.S.; Mohan, B.; Taneja, N. Evaluation of Risk Factors for Colistin Resistance among Uropathogenic Isolates of Escherichia Coli and Klebsiella Pneumoniae: A Case–Control Study. J. Med. Microbiol. 2019, 68, 837–847. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Y.; Qi, X.; Wang, R.; Jin, L.; Zhao, M.; Zhang, Y.; Wang, Q.; Chen, H.; Wang, H. Molecular Epidemiology of Colistin-Resistant Enterobacteriaceae in Inpatient and Avian Isolates from China: High Prevalence of Mcr -Negative Klebsiella Pneumoniae. Int. J. Antimicrob. Agents 2017, 50, 536–541. [Google Scholar] [CrossRef]
- Prim, N.; Turbau, M.; Rivera, A.; Rodríguez-Navarro, J.; Coll, P.; Mirelis, B. Prevalence of Colistin Resistance in Clinical Isolates of Enterobacteriaceae: A Four-Year Cross-Sectional Study. J. Infect. 2017, 75, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Puljko, A.; Barišić, I.; Dekić Rozman, S.; Križanović, S.; Babić, I.; Jelić, M.; Maravić, A.; Udiković-Kolić, N. Molecular Epidemiology and Mechanisms of Carbapenem and Colistin Resistance in Klebsiella and Other Enterobacterales from Treated Wastewater in Croatia. Environ. Int. 2024, 185, 108554. [Google Scholar] [CrossRef]
- Lupo, A.; Haenni, M.; Madec, J.-Y. Antimicrobial Resistance in Acinetobacter Spp. and Pseudomonas Spp. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Jafari-Ramedani, S.; Nazari, M.; Arzanlou, M.; Peeri-Dogaheh, H.; Sahebkar, A.; Khademi, F. Prevalence and Molecular Characterization of Colistin Resistance in Pseudomonas Aeruginosa Isolates: Insights from a Study in Ardabil Hospitals. BMC Microbiol. 2024, 24, 152. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Baky, R.M.; Masoud, S.M.; Mohamed, D.S.; Waly, N.G.; Shafik, E.A.; Mohareb, D.A.; Elkady, A.; Elbadr, M.M.; Hetta, H.F. Prevalence and Some Possible Mechanisms of Colistin Resistance Among Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa. Infect. Drug Resist. 2020, 13, 323–332. [Google Scholar] [CrossRef]
- Rossi, F.; Girardello, R.; Cury, A.P.; Di Gioia, T.S.R.; de Almeida, J.N.; da Silva Duarte, A.J. Emergence of Colistin Resistance in the Largest University Hospital Complex of São Paulo, Brazil, over Five Years. Braz. J. Infect. Dis. 2017, 21, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Wi, Y.M.; Choi, J.-Y.; Lee, J.-Y.; Kang, C.-I.; Chung, D.R.; Peck, K.R.; Song, J.-H.; Ko, K.S. Emergence of Colistin Resistance in Pseudomonas Aeruginosa ST235 Clone in South Korea. Int. J. Antimicrob. Agents 2017, 49, 767–769. [Google Scholar] [CrossRef]
- Zarate, M.; Barrantes Salinas, D.; Cuicapuza, D.; Velasquez, J.; Fernández, N.; Salvatierra, G.; Tamariz, J. Frecuencia de Resistencia a La Colistina En Pseudomonas Aeruginosa: Primer Reporte En El Perú. Rev. Peru. Med. Exp. Salud. Publica 2021, 38, 308–312. [Google Scholar] [CrossRef]
- Farajzadeh Sheikh, A.; Shahin, M.; Shokoohizadeh, L.; Halaji, M.; Shahcheraghi, F.; Ghanbari, F. Molecular Epidemiology of Colistin-Resistant Pseudomonas Aeruginosa Producing NDM-1 from Hospitalized Patients in Iran. Iran. J. Basic Med. Sci. 2019, 22, 38–42. [Google Scholar] [CrossRef]
- Aghazadeh, M.; Goli, H.R.; Nahaei, M.R.; Rezaee, M.A.; Hasani, A.; Kafil, H.S. Emergence of Colistin Resistant Pseudomonas aeruginosa at Tabriz Hospitals, Iran. Iran. J. Microbiol. 2016, 8, 62. [Google Scholar]
- Heidari, L.; Darvishi, M.; Pharm Sci, P.J.; Sepahvand, S.; Jafari, R. Molecular Analysis of PmrA and PmrB Genes in Colistin-Resistant Pseudomonas Aeruginosa Strains via PCR Method. Pak. J. Pharm. Sci. 2019, 32, 1175–1177. [Google Scholar] [PubMed]
- Malekzadegan, Y.; Abdi, A.; Heidari, H.; Moradi, M.; Rastegar, E.; Sedigh Ebrahim-Saraie, H. In Vitro Activities of Colistin, Imipenem and Ceftazidime against Drug-Resistant Pseudomonas Aeruginosa and Acinetobacter Baumannii Isolates in the South of Iran. BMC Res. Notes 2019, 12, 301. [Google Scholar] [CrossRef]
- Tahmasebi, H.; Dehbashi, S.; Arabestani, M. Prevalence and Molecular Typing of Colistin-Resistant Pseudomonas aeruginosa (CRPA) Among β-Lactamase-Producing Isolates: A Study Based on High-Resolution Melting Curve Analysis Method. Infect. Drug Resist. 2020, 13, 2943–2955. [Google Scholar] [CrossRef]
- Talebi, G.; Hakemi-Vala, M. Survey on Some Carbapenems and Colistin Resistance Genes Among Pseudomonas Aeruginosa Isolates from Burn and Cystic Fibrosis Patients, Tehran, Iran. Arch. Clin. Infect. Dis. 2019, 14, e93651. [Google Scholar] [CrossRef]
- Mohammadi, M.; Khayat, H.; Sayehmiri, K.; Soroush, S.; Sayehmiri, F.; Delfani, S.; Bogdanovic, L.; Taherikalani, M. Synergistic Effect of Colistin and Rifampin Against Multidrug Resistant Acinetobacter Baumannii: A Systematic Review and Meta-Analysis. Open Microbiol. J. 2017, 11, 63–71. [Google Scholar] [CrossRef]
- Ciftci, İ.H.; Kilbaş, İ.; Kilbaş, E.P.K. The War against the Resistance of Acinetobacter Baumannii: A Meta-Analysis in Turkey. Erciyes Med. J. 2022, 44, 447–454. [Google Scholar] [CrossRef]
- Hameed, F.; Khan, M.A.; Muhammad, H.; Sarwar, T.; Bilal, H.; Rehman, T.U. Plasmid-Mediated Mcr-1 Gene in Acinetobacter Baumannii and Pseudomonas Aeruginosa: First Report from Pakistan. Rev. Soc. Bras. Med. Trop. 2019, 52, e20190237. [Google Scholar] [CrossRef]
- Singhal, L.; Sharma, M.; Verma, S.; Kaur, R.; Britto, X.B.; Kumar, S.M.; Ray, P.; Gautam, V. Comparative Evaluation of Broth Microdilution with Polystyrene and Glass-Coated Plates, Agar Dilution, E-Test, Vitek, and Disk Diffusion for Susceptibility Testing of Colistin and Polymyxin B on Carbapenem-Resistant Clinical Isolates of Acinetobacter baumannii. Microb. Drug Resist. 2018, 24, 1082–1088. [Google Scholar] [CrossRef]
- Çağlan, E.; Nigiz, Ş.; Sancak, B.; Gür, D. Resistance and Heteroresistance to Colistin among Clinical Isolates of Acinetobacter baumannii. Acta Microbiol. Immunol. Hung. 2019, 67, 107–111. [Google Scholar] [CrossRef]
- Matuschek, E.; Åhman, J.; Webster, C.; Kahlmeter, G. Antimicrobial Susceptibility Testing of Colistin—Evaluation of Seven Commercial MIC Products against Standard Broth Microdilution for Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter Spp. Clin. Microbiol. Infect. 2018, 24, 865–870. [Google Scholar] [CrossRef]
- Logan, L.K.; Weinstein, R.A. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J. Infect. Dis. 2017, 215, S28–S36. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST) Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 13.1. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_13.1_Breakpoint_Tables.pdf (accessed on 3 June 2025).
- The European Committee on Antimicrobial Susceptibility Testing (EUCAST) Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 12.0. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_12.0_Breakpoint_Tables.pdf (accessed on 3 June 2025).
- The European Committee on Antimicrobial Susceptibility Testing (EUCAST) Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 11.0. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_11.0_Breakpoint_Tables.pdf (accessed on 3 June 2025).
- The European Committee on Antimicrobial Susceptibility Testing (EUCAST) Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 10.0. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_10.0_Breakpoint_Tables.pdf (accessed on 3 June 2025).
- The European Committee on Antimicrobial Susceptibility Testing (EUCAST) Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 9.0. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.pdf (accessed on 3 June 2025).
- ISO 20776-1:2006; Clinical Laboratory Testing and In Vitro Diagnostic Test Systems—Susceptibility Testing of Infectious Agents and Evaluation of Performance of Antimicrobial Susceptibility Test Devices. Part 1: Reference Method for Testing the In Vitro Activity of Antimicrobial Agents against Rapidly Growing Aerobic Bacteria Involved in Infectious Diseases. International Organization for Standardization: Geneva, Switzerland, 2006.
- Clinical and Laboratory Standards Institute Performance Standard for Antimicrobial Susceptibility Testing, 26th ed.; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2022.
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).