Design and Biological Evaluation of hBest1-Containing Bilayer Nanostructures
Abstract
1. Introduction
2. Results
2.1. Design and Self-Association of Lipid Nanostructures Containing hBest1 Protein
2.2. Biological Evaluation of hBest1 Lipid Nanostructures
2.2.1. Metabolic Activity Assessment of MDCK II Cells Treated with hBest1 Lipid Nanostructures
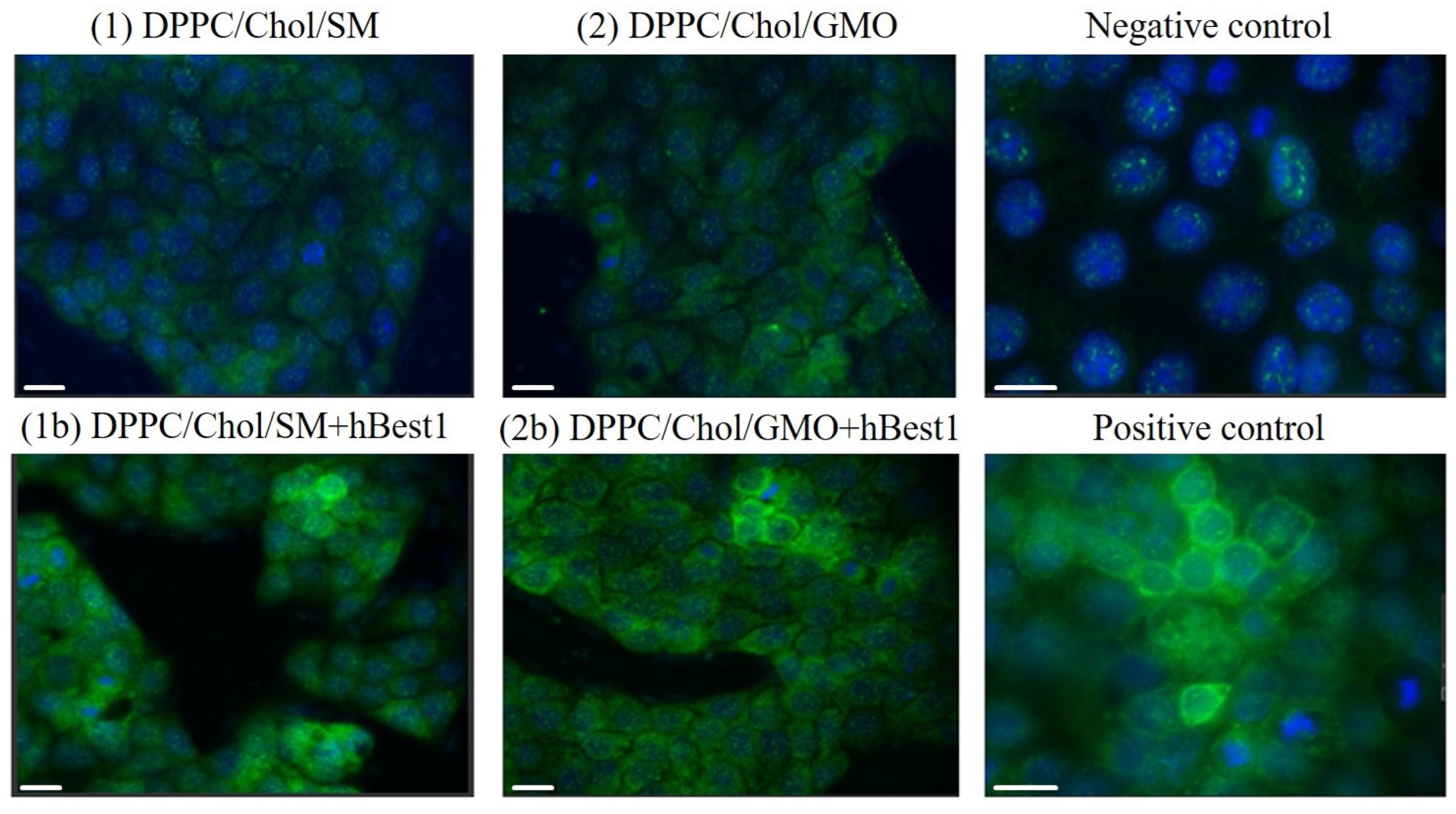
2.2.2. Immunofluorescence Staining of hBest1 Protein in MDCK II Cells Treated with hBest1 Vesicles
3. Discussion
4. Materials and Methods
4.1. Preparation of Nanostructures Containing hBest1 Protein
4.2. Cryogenic Transmission Electron Microscopy (Cryo-TEM) Measurements
4.3. Cell Cultures
4.4. hBest1 Purification from MDCK II-hBest1 Cells
4.5. MTT Assay for Metabolic Activity
4.6. Immunofluorescence Staining of hBest1 Protein
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MDCK | Madin–Darby canine kidney |
| Chol | Cholesterol |
| SM | Sphingomyelin |
| POPC | 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine |
References
- Marquardt, A.; Stohr, H.; Passmore, L.A.; Kramer, F.; Rivera, A.; Weber, B.H. Mutations in a novel gene, VMD2, encoding a protein of unknown properties cause juvenile-onset vitelliform macular dystrophy (Best’s disease). Hum. Mol. Genet. 1998, 7, 1517–1525. [Google Scholar] [CrossRef]
- Petrukhin, K.; Koisti, M.J.; Bakall, B.; Li, W.; Xie, G.; Marknell, T.; Sandgren, O.; Forsman, K.; Holmgren, G.; Andreasson, S.; et al. Identification of the gene responsible for Best macular dystrophy. Nat. Genet. 1998, 19, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Boon, C.J.; Klevering, B.J.; Leroy, B.P.; Hoyng, C.B.; Keunen, J.E.; den Hollander, A.I. The spectrum of ocular phenotypes caused by mutations in the BEST1 gene. Prog. Retin. Eye Res. 2009, 28, 187–205. [Google Scholar] [CrossRef]
- Weingeist, T.A.; Kobrin, J.L.; Watzke, R.C. Histopathology of Best’s macular dystrophy. Arch. Ophthalmol. 1982, 100, 1108–1114. [Google Scholar] [CrossRef] [PubMed]
- Marmorstein, A.D.; Cross, H.E.; Peachey, N.S. Functional roles of bestrophins in ocular epithelia. Prog. Retin. Eye Res. 2009, 28, 206–226. [Google Scholar] [CrossRef]
- Davidson, A.E.; Millar, I.D.; Urquhart, J.E.; Burgess-Mullan, R.; Shweikh, Y.; Parry, N.; O’Sullivan, J.; Maher, G.J.; McKibbin, M.; Downes, S.M.; et al. Missense Mutations in a Retinal Pigment Epithelium Protein, Bestrophin-1, Cause Retinitis Pigmentosa. Am. J. Hum. Human. Genet. 2009, 85, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Burgess, R.; Millar, I.D.; Leroy, B.P.; Urquhart, J.E.; Fearon, I.M.; De Baere, E.; Brown, P.D.; Robson, A.G.; Wright, G.A.; Kestelyn, P.; et al. Biallelic mutation of BEST1 causes a distinct retinopathy in humans. Am. J. Hum. Genet. 2008, 82, 19–31. [Google Scholar] [CrossRef]
- Davidson, A.E.; Millar, I.D.; Burgess-Mullan, R.; Maher, G.J.; Urquhart, J.E.; Brown, P.D.; Black, G.C.; Manson, F.D. Functional characterization of bestrophin-1 missense mutations associated with autosomal recessive bestrophinopathy. Investig. Opthalmology Vis. Sci. 2011, 52, 3730–3736. [Google Scholar] [CrossRef]
- Marmorstein, A.D.; Marmorstein, L.Y.; Rayborn, M.; Wang, X.; Hollyfield, J.G.; Petrukhin, K. Bestrophin, the product of the Best vitelliform macular dystrophy gene (VMD2), localizes to the basolateral plasma membrane of the retinal pigment epithelium. Proc. Natl. Acad. Sci. USA 2000, 97, 12758–12763. [Google Scholar] [CrossRef]
- Strauss, O. The Retinal Pigment Epithelium in Visual Function. Physiol. Rev. 2005, 85, 845–881. [Google Scholar] [CrossRef]
- Doumanov, J.A.; Zeitz, C.; Dominguez Gimenez, P.; Audo, I.; Krishna, A.; Alfano, G.; Diaz, M.L.; Moskova-Doumanova, V.; Lancelot, M.E.; Sahel, J.A.; et al. Disease-causing mutations in BEST1 gene are associated with altered sorting of bestrophin-1 protein. Int. J. Mol. Sci. 2013, 14, 15121–15140. [Google Scholar] [CrossRef]
- Sun, H.; Tsunenari, T.; Yau, K.W.; Nathans, J. The vitelliform macular dystrophy protein defines a new family of chloride channels. Proc. Natl. Acad. Sci. USA 2002, 99, 4008–4013. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Prussia, A.; Yu, K.; Cui, Y.Y.; Hartzell, H.C. Regulation of bestrophin Cl channels by calcium: Role of the C terminus. J. Gen. Physiol. 2008, 132, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Woo, D.H.; Han, K.S.; Shim, J.W.; Yoon, B.E.; Kim, E.; Bae, J.Y.; Oh, S.J.; Hwang, E.M.; Marmorstein, A.D.; Bae, Y.C.; et al. TREK-1 and Best1 channels mediate fast and slow glutamate release in astrocytes upon GPCR activation. Cell 2012, 151, 25–40. [Google Scholar] [CrossRef]
- Han, K.-S.; Woo, J.; Park, H.; Yoon, B.-J.; Choi, S.; Lee, C.J. Channel-mediated astrocytic glutamate release via Bestrophin-1 targets synaptic NMDARs. Mol. Brain 2013, 6, 4. [Google Scholar] [CrossRef]
- Lee, S.; Yoon, B.E.; Berglund, K.; Oh, S.J.; Park, H.; Shin, H.S.; Augustine, G.J.; Lee, C.J. Channel-mediated tonic GABA release from glia. Science 2010, 330, 790–796. [Google Scholar] [CrossRef] [PubMed]
- Owji, A.P.; Wang, J.; Kittredge, A.; Clark, Z.; Zhang, Y.; Hendrickson, W.A.; Yang, T. Structures and gating mechanisms of human bestrophin anion channels. Nat. Commun. 2022, 13, 3836. [Google Scholar] [CrossRef]
- Dickson, V.K.; Pedi, L.; Long, S.B. Structure and insights into the function of a Ca(2+)-activated Cl(-) channel. Nature 2014, 516, 213–218. [Google Scholar] [CrossRef]
- Yang, T.; Liu, Q.; Kloss, B.; Bruni, R.; Kalathur, R.C.; Guo, Y.; Kloppmann, E.; Rost, B.; Colecraft, H.M.; Hendrickson, W.A. Structure and selectivity in bestrophin ion channels. Science 2014, 346, 355–359. [Google Scholar] [CrossRef]
- Mladenova, K.; Petrova, S.D.; Georgiev, G.A.; Moskova-Doumanova, V.; Lalchev, Z.; Doumanov, J.A. Interaction of Bestrophin-1 with 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) in surface films. Colloids Surf. B Biointerfaces 2014, 122, 432–438. [Google Scholar] [CrossRef]
- Andreeva, T.D.; Petrova, S.D.; Mladenova, K.; Moskova-Doumanova, V.; Topouzova-Hristova, T.; Petseva, Y.; Mladenov, N.; Balashev, K.; Lalchev, Z.; Doumanov, J.A. Effects of Ca(2+), Glu and GABA on hBest1 and composite hBest1/POPC surface films. Colloids Surf. B Biointerfaces 2018, 161, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Mladenov, N.; Petrova, S.D.; Mladenova, K.; Bozhinova, D.; Moskova-Doumanova, V.; Topouzova-Hristova, T.; Videv, P.; Veleva, R.; Kostadinova, A.; Staneva, G.; et al. Miscibility of hBest1 and sphingomyelin in surface films—A prerequisite for interaction with membrane domains. Colloids Surf. B Biointerfaces 2020, 189, 110893. [Google Scholar] [CrossRef]
- Videv, P.; Mladenov, N.; Andreeva, T.; Mladenova, K.; Moskova-Doumanova, V.; Nikolaev, G. Condensing Effect of Cholesterol on hBest1/POPC and hBest1/SM Langmuir Monolayers. Membranes 2021, 11, 52. [Google Scholar] [CrossRef]
- Videv, P.; Mladenova, K.; Andreeva, T.D.; Park, J.H.; Moskova-Doumanova, V.; Petrova, S.D.; Doumanov, J.A. Cholesterol Alters the Phase Separation in Model Membranes Containing hBest1. Molecules 2022, 27, 4267. [Google Scholar] [CrossRef]
- Alavi, M.; Karimi, N.; Safaei, M. Application of Various Types of Liposomes in Drug Delivery Systems. Adv. Pharm. Bull. 2017, 7, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, S.M. Liposome Techniques for Synthesis of Biomimetic Lipid Membranes. In Nanobiotechnology of Biomimetic Membranes; Martin, D.K., Ed.; Springer: Boston, MA, USA, 2007; pp. 75–87. [Google Scholar]
- Meikle, T.G.; Drummond, C.J.; Separovic, F.; Conn, C.E. Chapter Three—Membrane-Mimetic Inverse Bicontinuous Cubic Phase Systems for Encapsulation of Peptides and Proteins. In Advances in Biomembranes and Lipid Self-Assembly; Iglič, A., Garcia-Sáez, A., Rappolt, M., Eds.; Academic Press: Cambridge, MA, USA, 2017; Volume 25, pp. 63–94. [Google Scholar]
- Bakardzhiev, P.; Forys, A.; Trzebicka, B.; Andreeva, T.; Rangelov, S. Unprecedented formation of sterically stabilized phospholipid liposomes of cuboidal morphology. Nanoscale 2021, 13, 15210–15214. [Google Scholar] [CrossRef]
- Neuhaus, F.; Mueller, D.; Tanasescu, R.; Balog, S.; Ishikawa, T.; Brezesinski, G.; Zumbuehl, A. Vesicle Origami: Cuboid Phospholipid Vesicles Formed by Template-Free Self-Assembly. Angew. Chem. Int. Ed. Engl. 2017, 56, 6515–6518. [Google Scholar] [CrossRef]
- Rieder, A.A.; Koller, D.; Lohner, K.; Pabst, G. Optimizing rapid solvent exchange preparation of multilamellar vesicles. Chem. Phys. Lipids 2015, 186, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, T.; Husen, P.; Brewer, J.; Bagatolli, L.A.; Hansen, P.L.; Ipsen, J.H.; Mouritsen, O.G. Preparing giant unilamellar vesicles (GUVs) of complex lipid mixtures on demand: Mixing small unilamellar vesicles of compositionally heterogeneous mixtures. Biochim. Biophys. Acta 2015, 1848, 3175–3180. [Google Scholar] [CrossRef]
- Akbarzadeh, A.; Rezaei-Sadabady, R.; Davaran, S.; Joo, S.W.; Zarghami, N.; Hanifehpour, Y.; Samiei, M.; Kouhi, M.; Nejati-Koshki, K. Liposome: Classification, preparation, and applications. Nanoscale Res. Lett. 2013, 8, 102. [Google Scholar] [CrossRef]
- Sharma, A.; Sharma, U.S. Liposomes in drug delivery: Progress and limitations. Int. J. Pharm. 1997, 154, 123–140. [Google Scholar] [CrossRef]
- Sahoo, S.K.; Labhasetwar, V. Nanotech approaches to drug delivery and imaging. Drug Discov. Today 2003, 8, 1112–1120. [Google Scholar] [CrossRef] [PubMed]
- Gabizon, A.; Goren, D.; Cohen, R.; Barenholz, Y. Development of liposomal anthracyclines: From basics to clinical applications. J. Control Release 1998, 53, 275–279. [Google Scholar] [CrossRef]
- Large, D.E.; Abdelmessih, R.G.; Fink, E.A.; Auguste, D.T. Liposome composition in drug delivery design, synthesis, characterization, and clinical application. Adv. Drug Deliv. Rev. 2021, 176, 113851. [Google Scholar] [CrossRef]
- Bozzuto, G.; Molinari, A. Liposomes as nanomedical devices. Int. J. Nanomed. 2015, 10, 975–999. [Google Scholar] [CrossRef]
- Chang, H.I.; Yeh, M.K. Clinical development of liposome-based drugs: Formulation, characterization, and therapeutic efficacy. Int. J. Nanomed. 2012, 7, 49–60. [Google Scholar]
- Bulbake, U.; Doppalapudi, S.; Kommineni, N.; Khan, W. Liposomal Formulations in Clinical Use: An Updated Review. Pharmaceutics 2017, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Conn, C.E.; Darmanin, C.; Sagnella, S.M.; Mulet, X.; Greaves, T.L.; Varghese, J.N.; Drummond, C.J. Incorporation of the dopamine D2L receptor and bacteriorhodopsin within bicontinuous cubic lipid phases. 1. Relevance to in meso crystallization of integral membrane proteins in monoolein systems. Soft Matter 2010, 6, 4828–4837. [Google Scholar] [CrossRef]
- Conn, C.E.; Mulet, X.; Moghaddam, M.J.; Darmanin, C.; Waddington, L.J.; Sagnella, S.M.; Kirby, N.; Varghese, J.N.; Drummond, C.J. Enhanced uptake of an integral membrane protein, the dopamine D2L receptor, by cubic nanostructured lipid nanoparticles doped with Ni(ii) chelated EDTA amphiphiles. Soft Matter 2011, 7, 567–578. [Google Scholar] [CrossRef]
- Sakla, M.; Breitinger, U.; Breitinger, H.G.; Mansour, S.; Tammam, S.N. Delivery of trans-membrane proteins by liposomes; the effect of liposome size and formulation technique on the efficiency of protein delivery. Int. J. Pharm. 2021, 606, 120879. [Google Scholar] [CrossRef]
- Ramadan, S.; Tammam, S.N.; Shetab Boushehri, M.A.; Breitinger, H.G.; Breitinger, U.; Mansour, S.; Lamprecht, A. Liposomal delivery of functional transmembrane ion channels into the cell membranes of target cells; a potential approach for the treatment of channelopathies. Int. J. Biol. Macromol. 2020, 153, 1080–1089. [Google Scholar] [CrossRef] [PubMed]
- Yeagle, P.L. Chapter 7—Structures of Lipid Assemblies. In The Membranes of Cells, 3rd ed.; Yeagle, P.L., Ed.; Academic Press: Boston, MA, USA, 2016; pp. 115–154. [Google Scholar]
- Conn, C.E.; Drummond, C.J. Nanostructured bicontinuous cubic lipid self-assembly materials as matrices for protein encapsulation. Soft Matter 2013, 9, 3449–3464. [Google Scholar] [CrossRef]
- Demel, R.A.; Bruckdorfer, K.R.; van Deenen, L.L. Structural requirements of sterols for the interaction with lecithin at the air water interface. Biochim. Biophys. Acta 1972, 255, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Demiel, R.A.; Guerts van Kessel, W.S.; van Deenen, L.L. The properties of polyunsaturated lecithins in monolayers and liposomes and the interactions of these lecithins with cholesterol. Biochim. Biophys. Acta 1972, 266, 26–40. [Google Scholar] [CrossRef]
- Yeagle, P.L. Chapter 9—Cholesterol and Related Sterols: Roles in Membrane Structure and Function. In The Membranes of Cells, 3rd ed.; Yeagle, P.L., Ed.; Academic Press: Boston, MA, USA, 2016; pp. 189–218. [Google Scholar]
- Bakardzhiev, P.; Toncheva-Moncheva, N.; Mladenova, K.; Petrova, S.; Videv, P.; Moskova-Doumanova, V.; Topouzova-Hristova, T.; Doumanov, J.; Rangelov, S. Assembly of amphiphilic nucleic acid-polymer conjugates into complex superaggregates: Preparation, properties, and in vitro performance. Eur. Polym. J. 2020, 131, 109692. [Google Scholar] [CrossRef]
- Mladenova, K.; Moskova-Doumanova, V.; Tabashka, I.; Petrova, S.; Lalchev, Z.; Doumanov, J. Establishment and characterization of stably transfected mdck cell line, expressing hbest1 protein. Bulg. J. Agric. Sci. 2013, 19, 159–162. [Google Scholar]
- Mayer, L.D.; Bally, M.B.; Hope, M.J.; Cullis, P.R. Techniques for encapsulating bioactive agents into liposomes. Chem. Phys. Lipids 1986, 40, 333–345. [Google Scholar] [CrossRef]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakardzhiev, P.; Koleva, T.; Mladenova, K.; Videv, P.; Moskova-Doumanova, V.; Forys, A.; Pusz, S.; Andreeva, T.; Petrova, S.; Rangelov, S.; et al. Design and Biological Evaluation of hBest1-Containing Bilayer Nanostructures. Molecules 2025, 30, 2948. https://doi.org/10.3390/molecules30142948
Bakardzhiev P, Koleva T, Mladenova K, Videv P, Moskova-Doumanova V, Forys A, Pusz S, Andreeva T, Petrova S, Rangelov S, et al. Design and Biological Evaluation of hBest1-Containing Bilayer Nanostructures. Molecules. 2025; 30(14):2948. https://doi.org/10.3390/molecules30142948
Chicago/Turabian StyleBakardzhiev, Pavel, Teodora Koleva, Kirilka Mladenova, Pavel Videv, Veselina Moskova-Doumanova, Aleksander Forys, Sławomira Pusz, Tonya Andreeva, Svetla Petrova, Stanislav Rangelov, and et al. 2025. "Design and Biological Evaluation of hBest1-Containing Bilayer Nanostructures" Molecules 30, no. 14: 2948. https://doi.org/10.3390/molecules30142948
APA StyleBakardzhiev, P., Koleva, T., Mladenova, K., Videv, P., Moskova-Doumanova, V., Forys, A., Pusz, S., Andreeva, T., Petrova, S., Rangelov, S., & Doumanov, J. (2025). Design and Biological Evaluation of hBest1-Containing Bilayer Nanostructures. Molecules, 30(14), 2948. https://doi.org/10.3390/molecules30142948








