The Structural Basis of Binding Stability and Selectivity of Sarolaner Enantiomers for Ctenocephalides felis RDL Receptors
Abstract
1. Introduction
2. Results
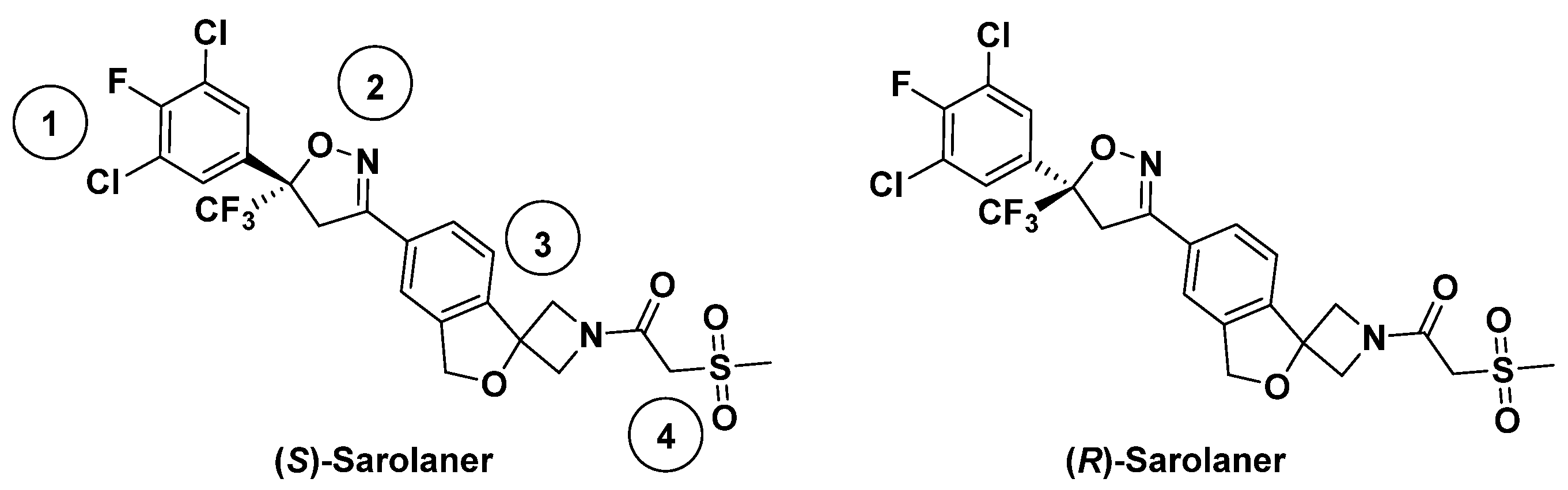
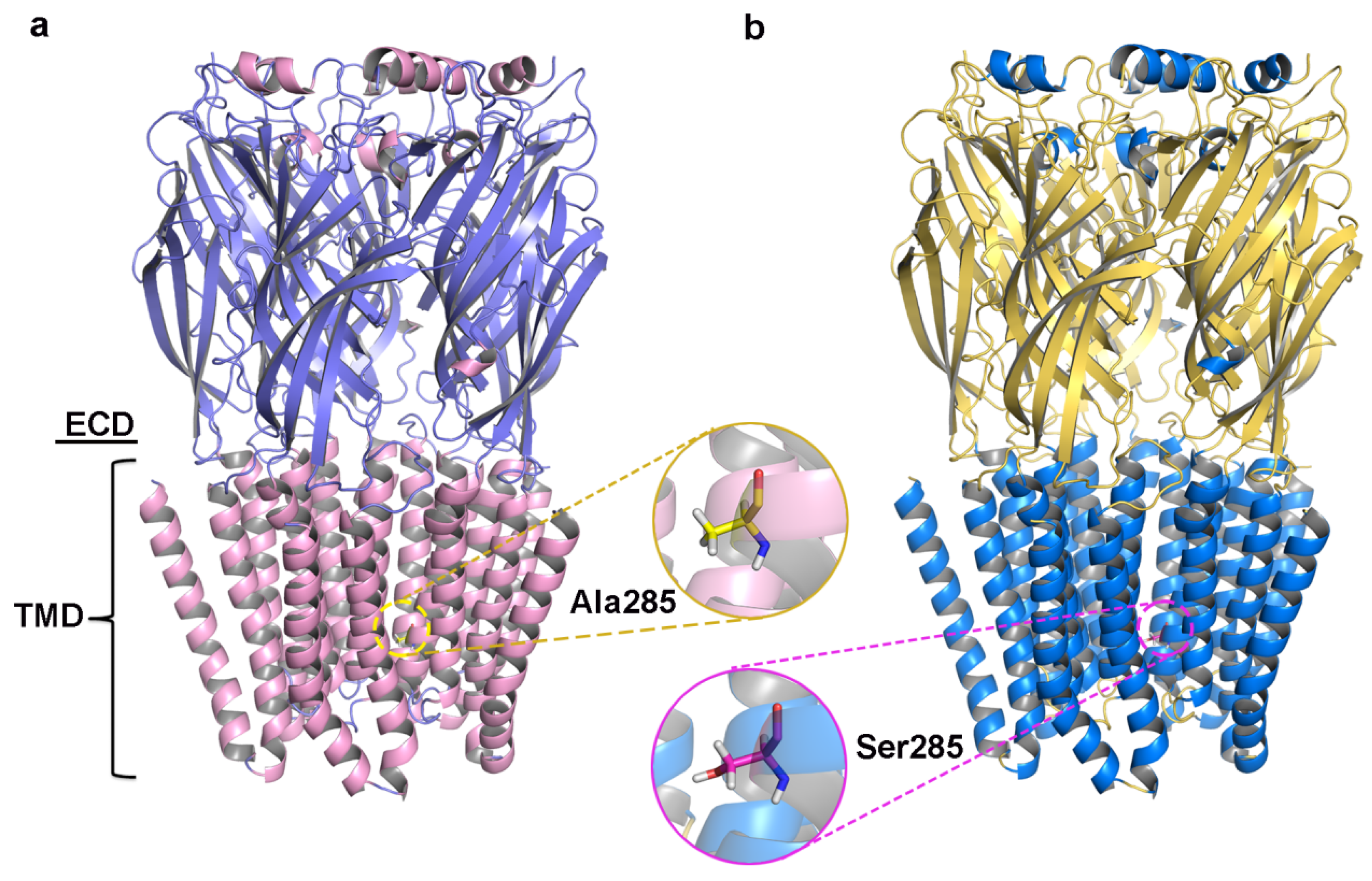
2.1. Prediction and Validation of Receptor Structures
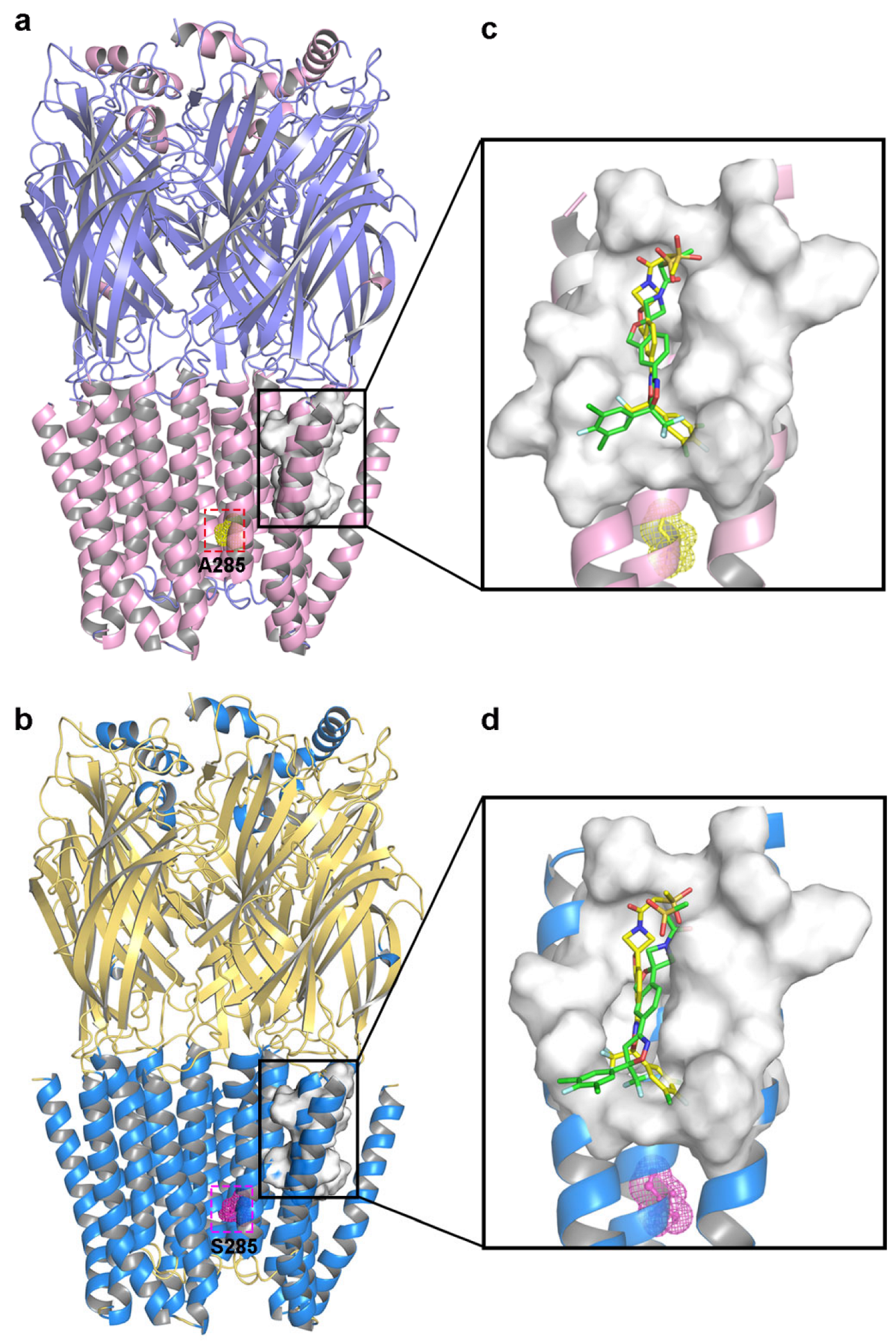
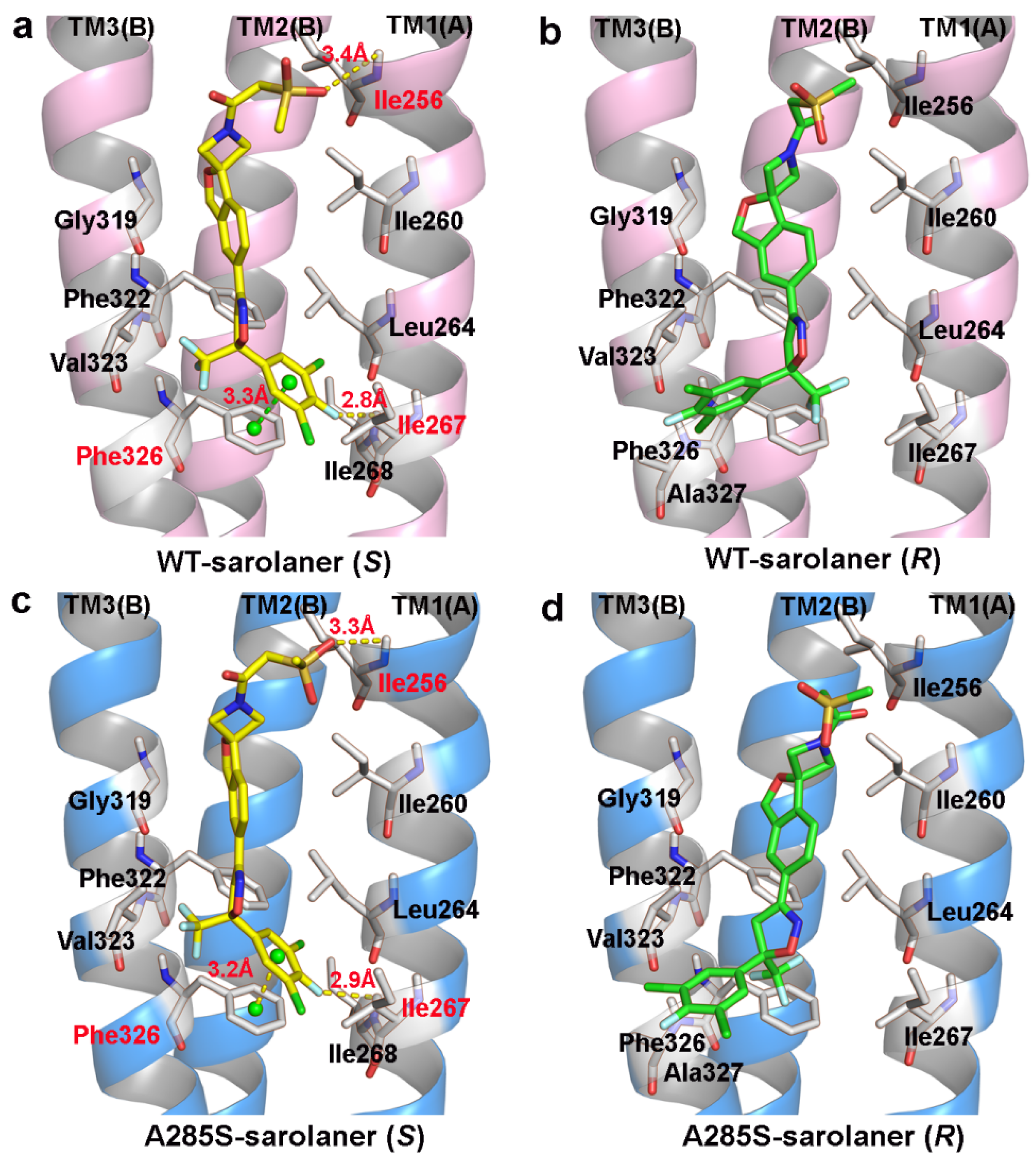
2.2. Molecular Docking
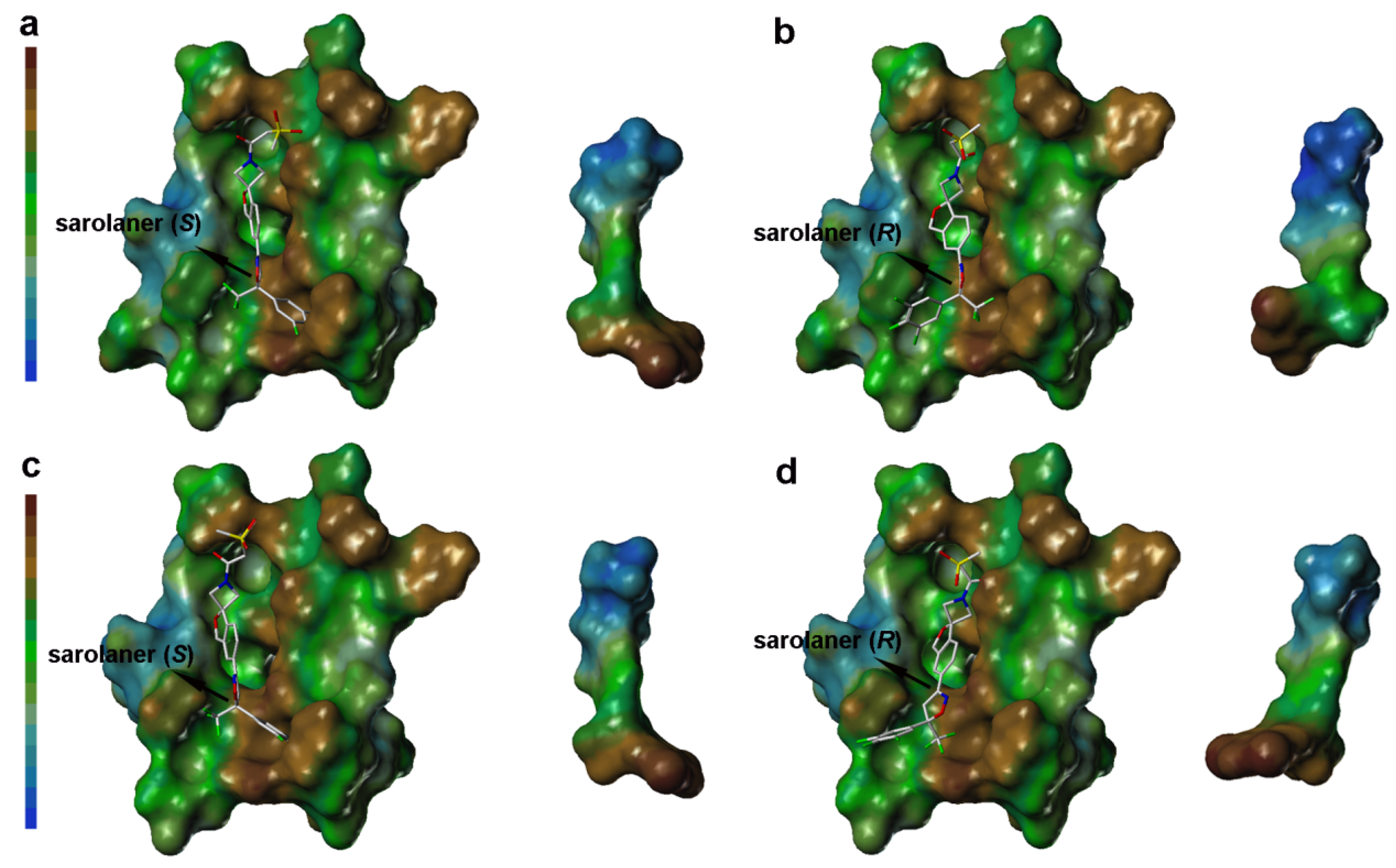
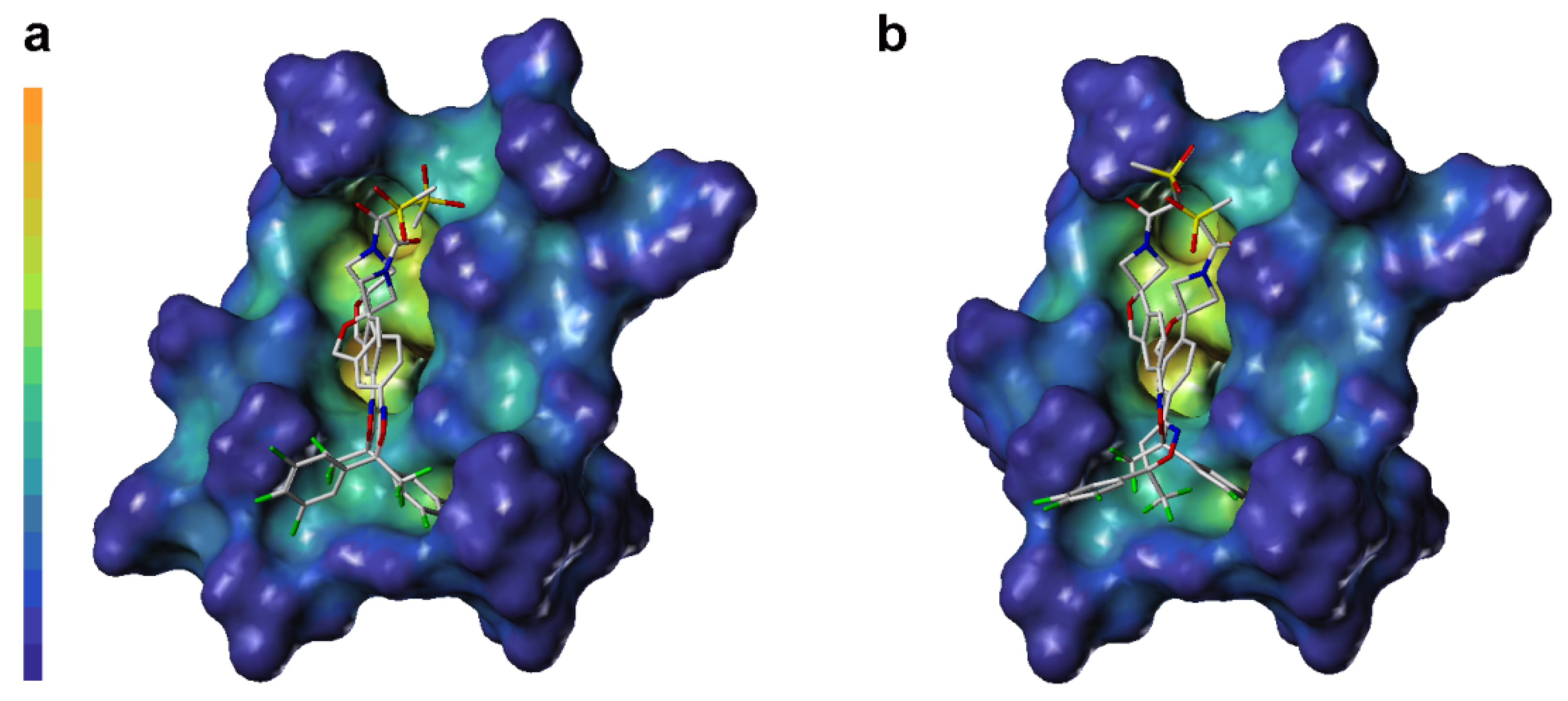
2.3. Computational Analysis of MOLCAD Protein Surface
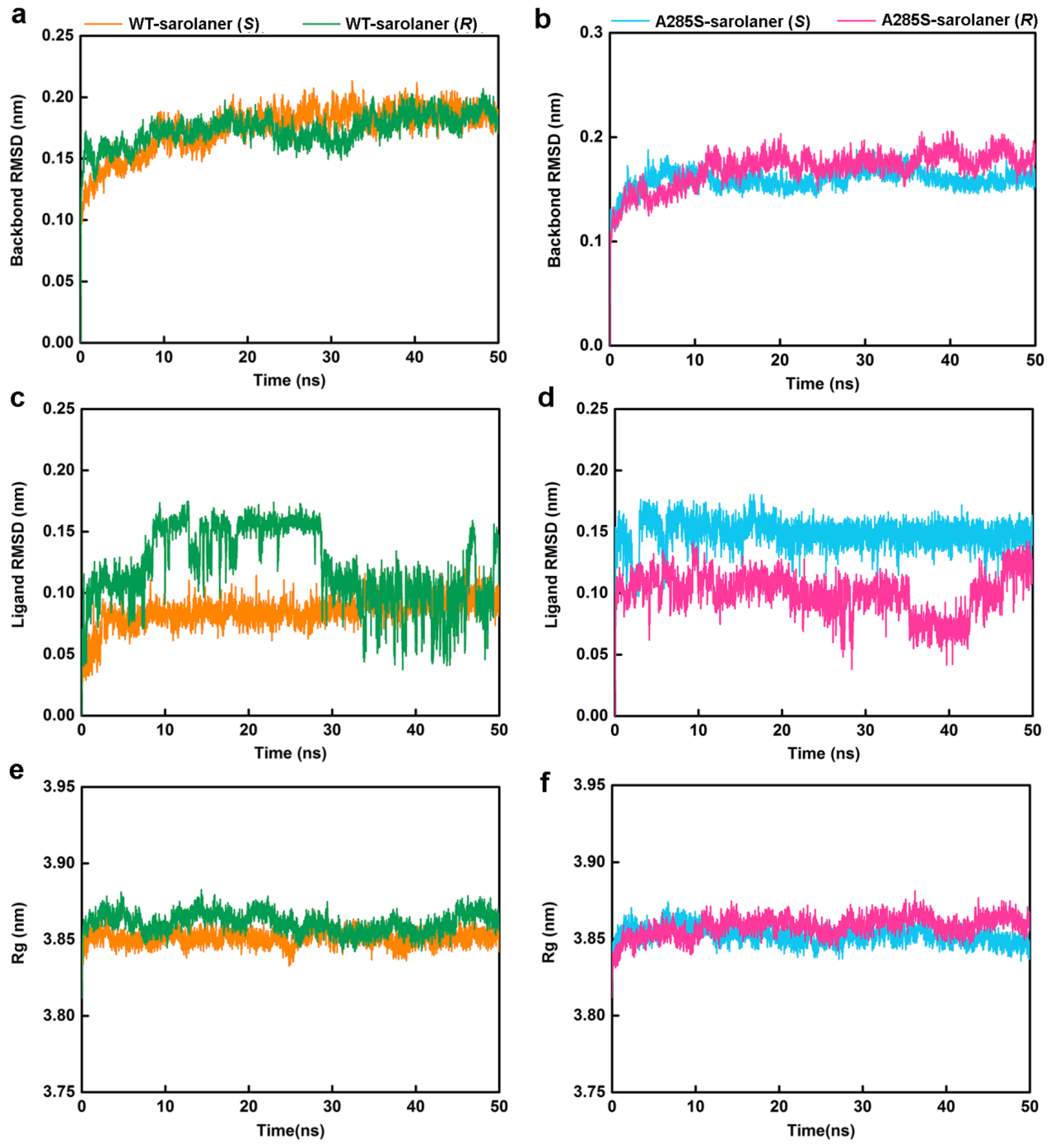
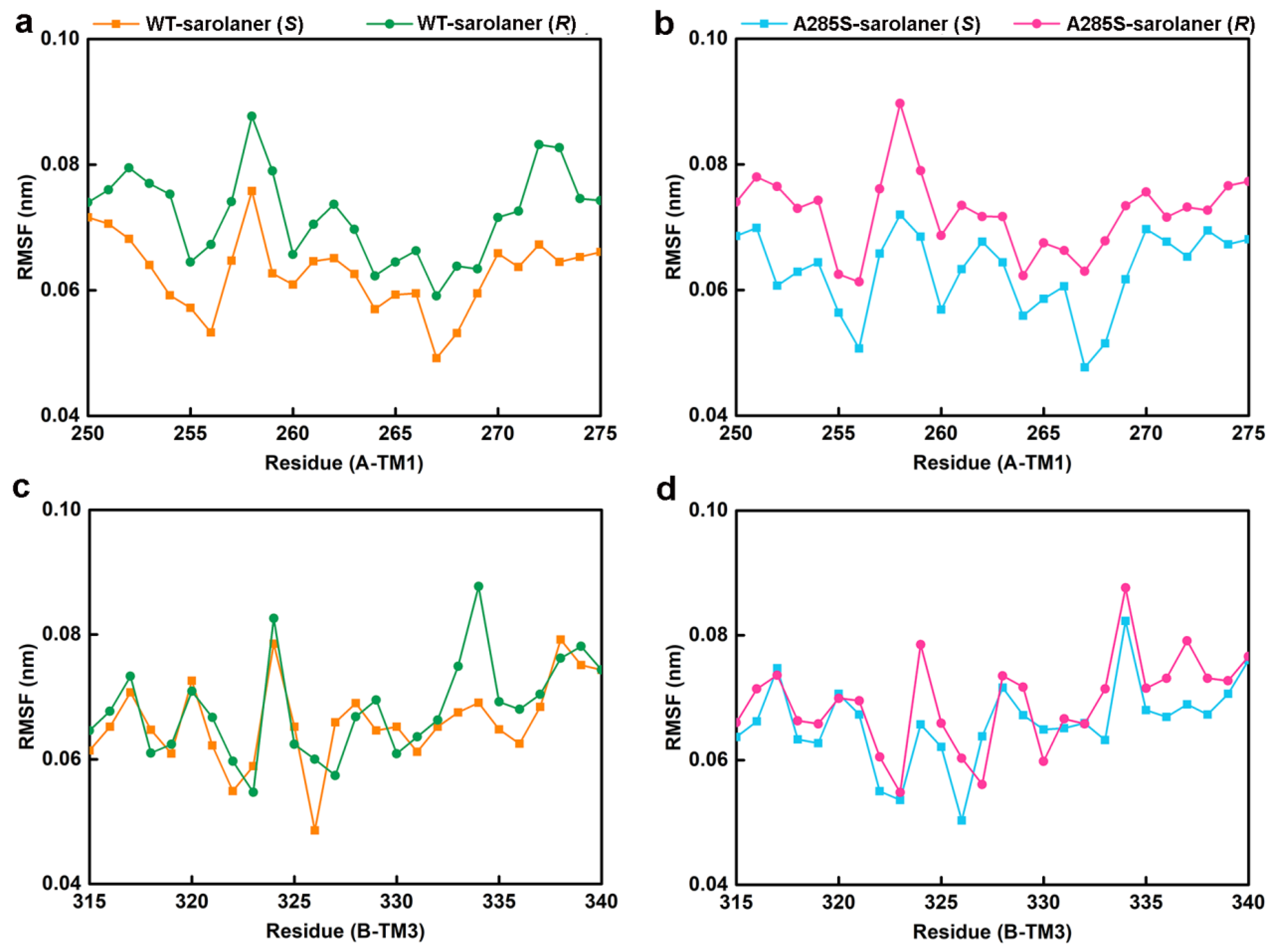
2.4. Molecular Dynamics Simulation
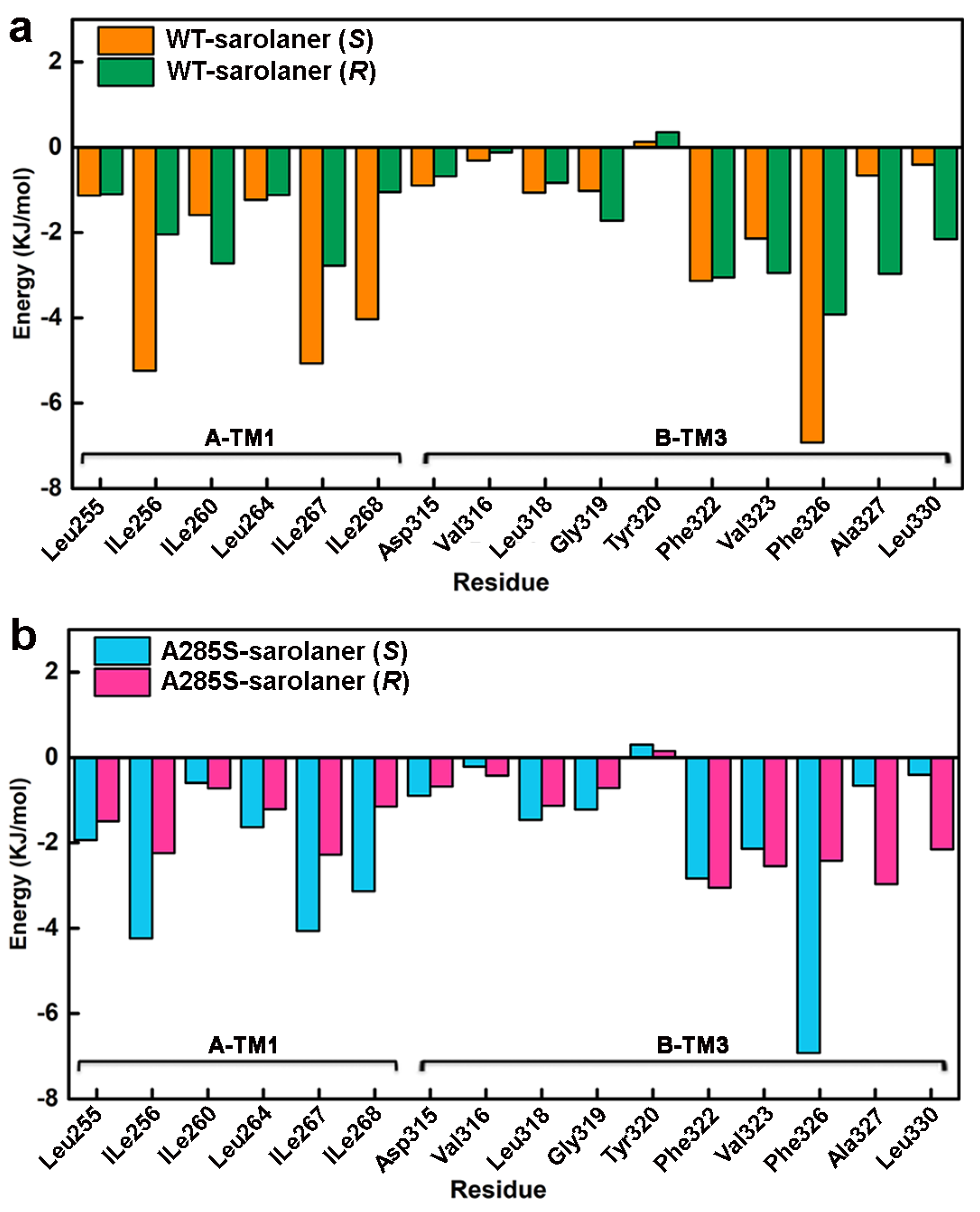
2.5. Binding Free Energy Analysis
3. Materials and Methods
3.1. Protein Structure Prediction
3.2. Ligand Docking
3.3. Receptor Protein Surface Calculation
3.4. Molecular Dynamics Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thompson, A.J.; Lester, H.A.; Lummis, S.C.R. The structural basis of function in Cys-loop receptors. Q. Rev. Biophys. 2010, 43, 449–499. [Google Scholar] [CrossRef]
- Chua, H.C.; Chebib, M. GABA-A receptors and the diversity in their structure and pharmacology. Adv. Pharmacol. 2017, 79, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Knoflach, F.; Bertrand, D. Pharmacological modulation of GABAA receptors. Curr. Opin. Pharmacol. 2021, 59, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Bormann, J. The ‘ABC’ of GABA receptors. Trends Pharmacol Sci. 2000, 21, 16–19. [Google Scholar] [CrossRef]
- Olsen, R.W.; Sieghart, W. International Union of Pharmacology. LXX. Subtypes of γ-aminobutyric acid A receptors: Classification on the basis of subunit composition, pharmacology, and function. update. Pharmacol. Rev. 2008, 60, 243–260. [Google Scholar] [CrossRef]
- Miller, P.S.; Smart, T.G. Binding, activation and modulation of Cys-loop receptors. Trends Pharmacol. Sci. 2010, 31, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.; Steichen, J.C.; Ffrench-Constant, R.H. Conservation of cyclodiene insecticide resistance-associated mutations in insects. Insect Mol. Biol. 2010, 2, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Mcgonigle, I.; Lummis, S.C.R. RDL receptors. Biochem. Soc. Trans. 2009, 37, 1404–1406. [Google Scholar] [CrossRef]
- Liu, G.; Zhou, C.; Zhang, Z.; Wang, C.; Luo, X.; Ju, X.; Zhao, C.; Ozoe, Y. Novel insecticidal 1,6-dihydro-6-iminopyridazine derivatives as competitive antagonists of insect RDL GABA receptors. Pest Manag. Sci. 2022, 78, 2872–2882. [Google Scholar] [CrossRef]
- Amundarain, M.J.; Ribeiro, R.P.; Costabel, M.D.; Giorgetti, A. GABAA receptor family: Overview on structural characterization. Future Med. Chem. 2019, 11, 229–245. [Google Scholar] [CrossRef]
- Miller, P.S.; Aricescu, A.R. Crystal structure of a human GABAA receptor. Nature 2014, 512, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Casida, J.E.; Durkin, K.A. Novel GABA receptor pesticide targets. Pestic. Biochem. Physiol. 2015, 121, 22–30. [Google Scholar] [CrossRef]
- Dantas-Torres, F.; Chomel, B.B.; Otranto, D. Ticks and tick-borne diseases: A One Health perspective. Trends Parasitol. 2012, 28, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.Y.; Hohman, A.E.; Hsu, W.H. Current review of isoxazoline ectoparasiticides used in veterinary medicine. J. Vet. Pharmacol. Ther. 2022, 45, 1–15. [Google Scholar] [CrossRef]
- Asahi, M.; Yamato, K.; Ozoe, F.; Ozoe, Y. External amino acid residues of insect GABA receptor channels dictate the action of the isoxazoline ectoparasiticide fluralaner. Pest Manag. Sci. 2023, 79, 4078–4082. [Google Scholar] [CrossRef]
- Gonçalves, I.L.; das Neves, G.M.; Kagami, L.P.; Eifler-Lima, V.L.; Merlo, A.A. Discovery, development, chemical diversity and design of isoxazoline-based insecticides. Bioorg. Med. Chem. 2021, 30, 115934. [Google Scholar] [CrossRef]
- Asahi, M.; Kobayashi, M.; Kagami, T.; Nakahira, K.; Furukawa, Y.; Ozoe, Y. Fluxametamide: A novel isoxazoline insecticide that acts via distinctive antagonism of insect ligand-gated chloride channels. Pestic. Biochem. Physiol. 2018, 151, 67–72. [Google Scholar] [CrossRef]
- Selzer, P.M.; Epe, C. Antiparasitics in animal health: Quo vadis? Trends Parasitol. 2021, 37, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Selzer, P.M. Isoxazolines: A novel chemotype highly effective on ectoparasites. ChemMedChem 2016, 11, 270–276. [Google Scholar] [CrossRef]
- Rufener, L.; Danelli, V.; Bertrand, D.; Sager, H. The novel isoxazoline ectoparasiticide lotilaner (Credelio™): A non-competitive antagonist specific to invertebrates γ-aminobutyric acid-gated chloride channels (GABACls). Parasites Vectors 2017, 10, 530. [Google Scholar] [CrossRef]
- Shoop, W.L.; Hartline, E.J.; Gould, B.R.; Waddell, M.E.; McDowell, R.G.; Kinney, J.B.; Lahm, G.P.; Long, J.K.; Xu, M.; Wagerle, T.; et al. Discovery and mode of action of afoxolaner, a new isoxazoline parasiticide for dogs. Vet. Parasitol. 2014, 201, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Taenzler, J.; Wengenmayer, C.; Williams, H.; Fourie, J.; Zschiesche, E.; Roepke, R.K.A.; Heckeroth, A.R. Onset of activity of fluralaner (BRAVECTO™) against Ctenocephalides felis on dogs. Parasites Vectors 2014, 7, 567. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Becskei, C.; De Bock, F.; Illambas, J.; Cherni, J.A.; Fourie, J.J.; Lane, M.; Mahabir, S.P.; Six, R.H. Efficacy and safety of a novel oral isoxazoline, sarolaner (Simparica™), for the treatment of sarcoptic mange in dogs. Vet. Parasitol. 2016, 222, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Curtis, M.P.; Vaillancourt, V.; Goodwin, R.M.; Chubb, N.A.L.; Howson, W.; McTier, T.L.; Pullins, A.; Zinser, E.W.; Meeus, P.F.M.; Woods, D.J.; et al. Design and synthesis of sarolaner, a novel, once-a-month, oral isoxazoline for the control of fleas and ticks on dogs. Bioorg. Med. Chem. Lett. 2016, 26, 1831–1835. [Google Scholar] [CrossRef]
- McTier, T.L.; Chubb, N.; Curtis, M.P.; Hedges, L.; Inskeep, G.A.; Knauer, C.S.; Menon, S.; Mills, B.; Pullins, A.; Zinser, E.; et al. Discovery of sarolaner: A novel, orally administered, broad-spectrum, isoxazoline ectoparasiticide for dogs. Vet. Parasitol. 2016, 222, 3–11. [Google Scholar] [CrossRef]
- Toutain, C.E.; Seewald, W.; Jung, M. The intravenous and oral pharmacokinetics of lotilaner in dogs. Parasite. Vector. 2017, 10, 522. [Google Scholar] [CrossRef]
- Geurden, T.; Becskei, C.; Grace, S.; Strube, C.; Doherty, P.; Liebenberg, J.; Mahabir, S.P.; Slootmans, N.; Lloyd, A.; Six, R.H. Efficacy of a novel oral formulation of sarolaner (Simparica™) against four common tick species infesting dogs in Europe. Vet. Parasitol. 2016, 222, 33–36. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Zheng, W.; Zhang, C.; Li, Y.; Pearce, R.; Bell, E.W.; Zhang, Y. Folding non-homologous proteins by coupling deep-learning contact maps with I-TASSER assembly simulations. Cell Rep. Methods 2021, 1, 100014. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021, 596, 583–586. [Google Scholar] [CrossRef]
- Zheng, X.; Liu, G.; Ozoe, Y.; Ju, X. Mechanistic insights into the selectivity of bicyclophosphorothionate antagonists for housefly versus rat GABA receptors. Pest Manag. Sci. 2024, 80, 1382–1399. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Rullmannn, J.A.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [PubMed]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, R.; Haas, J.; Schwede, T. QMEANDisCo-distance constraints applied on model quality estimation. Bioinformatics 2019, 36, 1765–1771. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Gao, Y.; Chen, Y.; Liu, G.; Ju, X. Identification of the fipronil resistance associated mutations in Nilaparvata lugens GABA receptors by molecular modeling. Molecules 2019, 24, 4116. [Google Scholar] [CrossRef]
- Massey, I.; Yadav, S.; Kumar, D.; Maharia, R.S.; Kumari, K.; Singh, P. An insight for the inhibition of anxiolytic and anti-convulsant effects in zebrafish using the curcumins via exploring molecular docking and molecular dynamics simulations. Mol. Divers. 2025, 29, 439–455. [Google Scholar] [CrossRef]
- Shafiee, Z.; Karami, L.; Akbari, M.; Rezaee, E.; Maaza, M.; and Tabatabai, S.A. Insights into the molecular mechanism of triazolopyrimidinone derivatives effects on the modulation of α1β2γ2 subtype of GABAA receptor: An in silico approach. Arch. Biochem. Biophys. 2022, 729, 109380. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R.; Lynn, A. g_mmpbsa--a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef]
- Zheng, X.J.; Wang, C.C.; Zhai, N.; Luo, X.G.; Liu, G.Y.; Ju, X.L. In Silico screening of novel α1-GABAA receptor PAMs towards schizophrenia based on combined modeling studies of imidazo 1,2-a -pyridines. Int. J. Mol. Sci. 2021, 22, 9645. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, Y.; Wu, F.; Pei, J.; Luo, X.; Ju, X.; Zhao, C.; Liu, G. Exploring the interaction mechanism of desmethyl-broflanilide in insect GABA receptors and screening potential antagonists by In Silico smulations. J. Agric. Food Chem. 2020, 68, 14768–14780. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Caballero, J. The latest automated docking technologies for novel drug discovery. Expert Opin. Drug Dis. 2021, 16, 625–645. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.L.; Yang, H.H.; Wu, F.S.; Luo, X.G.; Sun, Q.; Feng, W.L.; Ju, X.L.; Liu, G.Y. Exploration of N-arylsulfonyl-indole-2-carboxamide derivatives as novel fructose-1,6-bisphosphatase inhibitors by molecular simulation. Int. J. Mol. Sci. 2022, 23, 10259. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.N. Surflex: Fully automatic flexible molecular docking using a molecular similarity-based search engine. J. Med. Chem. 2003, 46, 499–511. [Google Scholar] [CrossRef]
- Zhou, T.; Wu, W.; Ma, S.; Chen, J.; Huang, J.; Qiao, X. Effects of RDL GABA receptor point mutants on susceptibility to Meta-diamide and isoxazoline insecticides in Drosophila melanogaster. Insects 2024, 15, 334. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, Q.; Sheng, C.; Liu, G.; Zhang, K.; Jia, Z.; Liu, Y.; Ma, L.; Zhou, Y.; Zhao, C. G3′MTMD3 in the insect GABA receptor subunit, RDL, confers resistance to broflanilide and fluralaner. PLoS Genet. 2023, 19, e1010814. [Google Scholar] [CrossRef]
- Lill, M.A.; Danielson, M.L. Computer-aided drug design platform using PyMOL. J. Comput. Aided Mol. Des. 2011, 25, 13–19. [Google Scholar] [CrossRef]
- Patel, B.; Patel, A.; Patel, A.; Bhatt, H. CoMFA, CoMSIA, molecular docking and MOLCAD studies of pyrimidinone derivatives to design novel and selective tankyrase inhibitors. J. Mol. Struct. 2020, 1221, 128783. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Silva, A.W.S.D.; Vranken, W.F. ACPYPE-Antechamber python parser interface. BMC Res. Notes 2012, 5, 367. [Google Scholar] [CrossRef] [PubMed]
- And, K.T.; Kitao, A. Effects of water model and simulation box size on protein diffusional motions. J. Phys. Chem. B. 2007, 111, 11870–11872. [Google Scholar] [CrossRef]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.; Wang, J.; Li, Y.; Wang, W. Assessing the performance of MM-PBSA and MM-GBSA methods. 7. Entropy effects on the performance of end-point binding free energy calculation approaches. J. Chem. Inf. Model. 2011, 51, 69–82. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 2715–2725. [Google Scholar] [CrossRef]
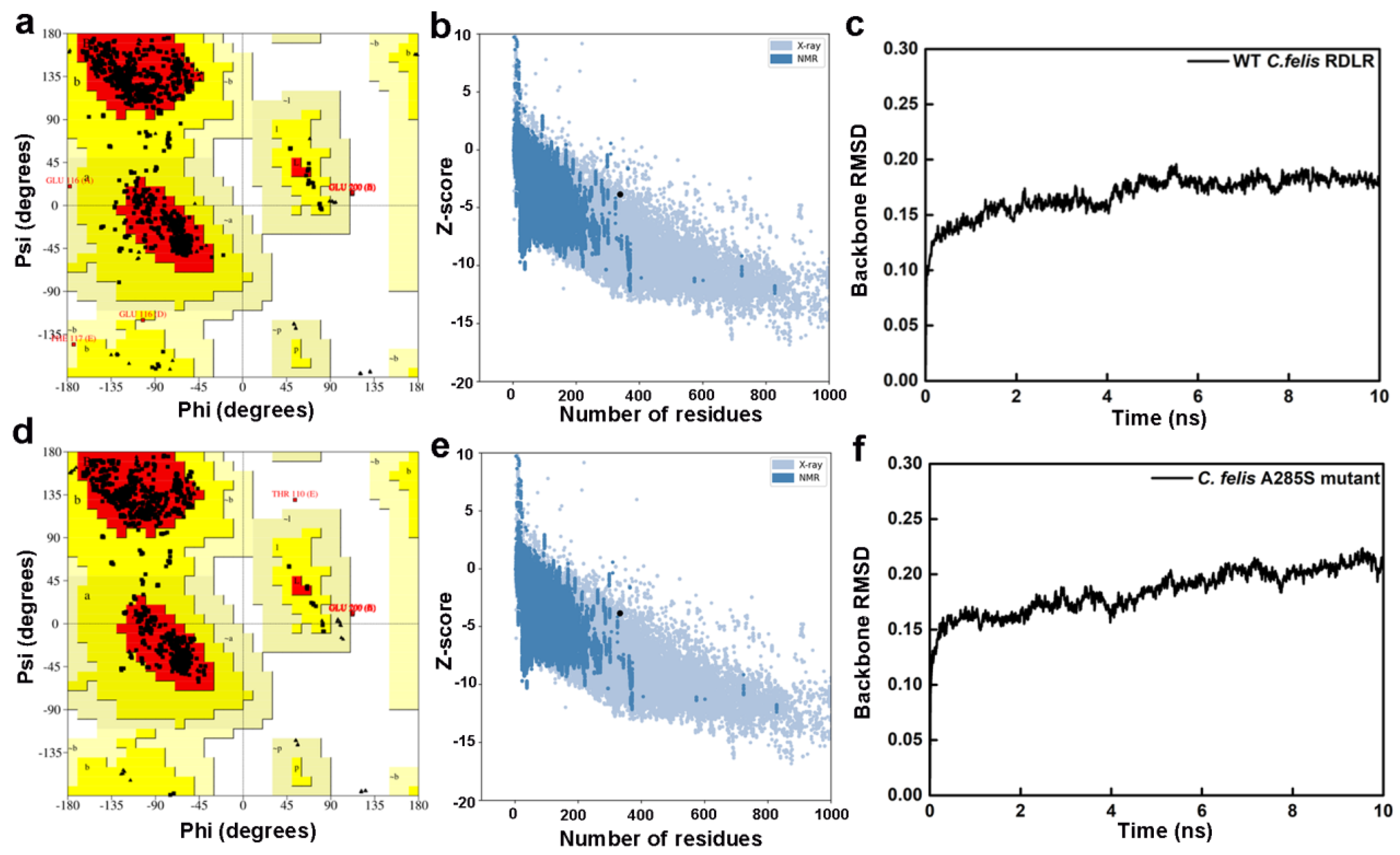
| Method | Model | Proportion of the Residues in Different Regions (%) | Z-Score | |||
|---|---|---|---|---|---|---|
| Most Favored Regions | Additional Allowed Regions | Generously Allowed Regions | Disallowed Allowed Regions | |||
| Swiss-Model | WT-RDLR | 93.5 | 6.0 | 0.2 | 0.3 | −3.86 |
| A285S-RDLR | 94.2 | 5.4 | 0.0 | 0.4 | −3.85 | |
| I-TASSER | WT-RDLR | 73.2 | 19.1 | 5.7 | 2.1 | −3.47 |
| A285S-RDLR | 77.6 | 16.6 | 4.4 | 1.5 | −3.78 | |
| AlphaFold2 | WT-RDLR | 83.7 | 11.0 | 3.6 | 1.6 | −4.22 |
| A285S-RDLR | 86.0 | 9.0 | 2.6 | 2.4 | −4.05 | |
| Protein-Ligand | Total_Score | Binding Energy (kJ/mol) | Intermolecular Interactions |
|---|---|---|---|
| WT-Sarolaner (S) | 5.35 | −30.54 | 2H-bond: Ile256, Ile267 1π-π stacking: Phe326 |
| WT-Sarolaner (R) | 3.42 | −19.52 | - |
| A285S-Sarolaner (S) | 5.18 | −29.54 | 2H-bond: Ile256, Ile267 1π-π stacking: Phe326 |
| A285S-Sarolaner (R) | 3.08 | −17.57 | - |
| Complex | ΔEvdW (kJ/mol) | ΔEele (kJ/mol) | ΔGPB (kJ/mol) | ΔGSA (kJ/mol) | ΔGbinding (kJ/mol) |
|---|---|---|---|---|---|
| WT-sarolaner (S) | −216.5 ± 9.4 | −30.6 ± 10.3 | 123.9 ± 14.1 | −17.8 ± 0.8 | −140.9 ± 13.1 |
| WT-sarolaner (R) | −202.9 ± 13.0 | −35.1 ± 9.2 | 151.0 ± 21.2 | −21.4 ± 0.9 | −108.5 ± 12.9 |
| A285S-sarolaner (S) | −219.2 ± 134 | −33.1 ± 9.9 | 138.1 ± 10.6 | −20.7 ± 1.0 | −134.9 ± 12.1 |
| A285S-sarolaner (R) | −191.6 ± 16.0 | −47.9 ± 11.7 | 159.5 ± 20.4 | −19.5 ± 0.9 | −99.5 ± 16.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, X.; Wang, X.; Ju, X.; Ma, Z.; Liu, G. The Structural Basis of Binding Stability and Selectivity of Sarolaner Enantiomers for Ctenocephalides felis RDL Receptors. Molecules 2025, 30, 2756. https://doi.org/10.3390/molecules30132756
Zheng X, Wang X, Ju X, Ma Z, Liu G. The Structural Basis of Binding Stability and Selectivity of Sarolaner Enantiomers for Ctenocephalides felis RDL Receptors. Molecules. 2025; 30(13):2756. https://doi.org/10.3390/molecules30132756
Chicago/Turabian StyleZheng, Xiaojiao, Xin Wang, Xiulian Ju, Zhichao Ma, and Genyan Liu. 2025. "The Structural Basis of Binding Stability and Selectivity of Sarolaner Enantiomers for Ctenocephalides felis RDL Receptors" Molecules 30, no. 13: 2756. https://doi.org/10.3390/molecules30132756
APA StyleZheng, X., Wang, X., Ju, X., Ma, Z., & Liu, G. (2025). The Structural Basis of Binding Stability and Selectivity of Sarolaner Enantiomers for Ctenocephalides felis RDL Receptors. Molecules, 30(13), 2756. https://doi.org/10.3390/molecules30132756






